Abstract
In this study, we sequenced and analyzed the complete mitochondrial genome of Culex gelidus. The mitogenome is 15,600 bp long, and contains 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and a control region. The gene order and composition are identical with those mitogenomes reported in other mosquito species. The whole base composition is A (39.8%), T (39.2%), G (8.8%), and C (12.2%). The PCGs have the initiation codon ATN except for COI with a TCG, and possess the complete termination codon TAA or incomplete T. The phylogenetic analysis of known Culex mitogenome sequences was carried out based on the nucleotide sequences of 13 PCGs, and the result showed that Cx. P. pipiens, Cx. P. pallens, and Cx. p. quinquefasciatus were claded into the Pipiens Complex in the Pipiens Group, and Cx. gelidus might be inappropriate to be classified into the Pipiens Group. Whether the mosquito species group with Cx. gelidus as type should be regarded as a Gelidus Group or Subgroup and its taxonomic position need to be elucidated with more molecular data.
Keywords: Mitochondrial genome, Culex gelidus, sequence analysis, Culicidae, phylogeny
Culex gelidus is an important vector of Japanese encephalitis virus and can potentially transmit West Nile, Kunjin, Murray Valley encephalitis, and Ross River viruses (Sudeep et al. 2015). It is mainly distributed in India and Southeast Asia (Sudeep et al. 2015), and its taxonomic position has long been disputed (Lu 1997; Harbach 2011; Harbach et al. 2012). The genus Culex is cosmopolitan distribution and its many species are important vectors of mosquito-borne diseases, including epidemic encephalitis, and lymphatic filariasis (Lu 1997). The phylogeny of the Culex is still unsettled (Harbach 2011), and the knowledge of mitochondrial genome (mitogenome) in the genus is quite limited. Up to date, there have been four species or subspecies with mitogenome sequences available in GenBank in the genus, Cx. P. pipiens, Cx. Quinquefasciatus, Cx. tritaeniorhynchus, and Cx. p. pallens. There are four mitogenome sequences for Cx. p. pipiens from different populations (three of these completely identical), one for Cx. p. pallens, and two for each of Cx. quinquefasciatus and Cx. tritaeniorhynchu with mutual nucleotide sequence differences. In addition, the mitogenome sequences of four other species (Cx. camposi, Cx. coronator, Cx. usquatus, and Cx. usquatissimus) in the genus have also been reported but their sequences are still unavailable in GenBank (Demari-Silva et al. 2015).
For mitogenome, the nucleotide sequences of protein-coding genes (PCGs) are thought to be most suitable as markers to elucidate the phylogenetic relationships in the genus (Luo et al. 2016). In the paper, we report the complete mitochondrial genome of Cx. gelidus, and reconstructed the phylogeny of species/subspecies using the PCG nucleotide sequences of seven known mitogenome sequences.
Using the method from Zou et al. (2015), the total DNA was extracted from the species of mosquitoes, which were collected from Shangsi County, Guangxi Province, China (22° 9.28′N, 108° 08.58′E). The total DNA was stored at −80 °C refrigerator in Institute of Entomology and Molecular Biology, Chongqing Normal University. The All polymerase chain reactions were carried out as described in Luo et al. (2016). Eighteen primer pairs (Zhang et al. 2013) were used for the amplification of the complete mitogenome.
The complete mitogenome of Cx. gelidus is a double-stranded circular molecule of 15 600 bp in size, containing 13 PCGs, 22 tRNA genes, two rRNA genes, and a control region (CR). The whole nucleotide composition is 39.8% A, 39.2% T, 8.8% G, and 12.2% C, presenting an obvious A + T bias (79.0%). All genes of the mitogenome are encoded on heavy strand except for four PCGs genes (ND1, ND4, ND4L, and ND5), nine tRNA genes (trnQ, trnC, trnY, trnS, trnF, trnH, trnP, trnL, and trnV) and two rRNA genes. Except for COI starting with TCG, other PCGs use ATG, ATA or ATC as the initiation codon. Eight PCGs (ND2, APT8, ATP6, COIII, ND3, ND5, ND6, and Cytb) stop with the complete terminate codon TAA, and the rest have incomplete stop codon T. All tRNAs vary from 66 to 72 bp in length. The overall AT content of 22 tRNAs is 79.6%. Among the 22 tRNAs, trnE has the highest AT content 90.9% and trnR has the lowest AT content 67.2%. There are two rRNAs in the entire mitogenome of Cx. gelidus, 16S rRNA with a length of 1338 bp and 12S rRNA with a length of 805 bp. The 16S rRNA is assumed to fill up the blanks between trnL and trnV, and the 12S rRNA is located between trnV and the CR. The CR is 722 bp long with a high AT content 90.6%. We identified two tandem repeat sequences in the CR, the 18 bp of conservative T-stretch region, and the 87 bp of sequence duplicated from the AT unit, (AT)n.
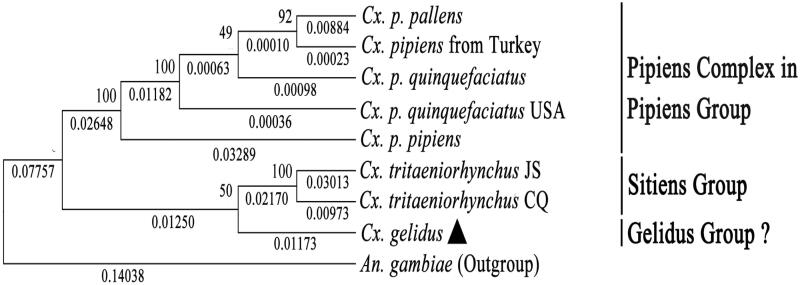
We constructed the Maximum Likelihood (ML) tree of Cx. gelidus and seven other Culex mitogenome sequences available in NCBI based on the nucleotide sequences of the 13 PCGs, with the An. Gambiae as outgroup. As shown in the phylogenetic tree (Figure 1), the eight Culex mitogenome sequences were divided into three clades: the Pipiens Complex clade with five species/subspecies and 100% of bootstrap support, the Sitiens Group clade with only two mitogenomes of Cx. tritaeniorhynchus and supported by a 100% bootstrap, and the Cx. gelidus clade with an only 50% bootstrap support to group with Sitiens Group. In the Pipiens Complex clade, there is a quite small genetic distance (0.00010) between Cx. p. pallens and Cx. pipiens from Turkey with 92% of bootstrap support. The separation of two mitogenomes of Cx. p. quinquefasciatus were located between Cx. p. pallens and Cx. p. pipiens with a non-significant supported bootstrap of 49% and a genetic distance of 0.00063. The Cx. p. pipiens was linked to other mitogenome sequences in the Pipiens Complex clade with 100% of bootstrap support and a genetic distance of 0.01182. The relationships of the five mitogenome sequences in the Pipiens Complex clade are consistent with the previous study (Luo et al. 2016), and it further demonstrates that Cx. p. pallens, Cx. p. pipiens, and Cx. p. quinquefasciatus were in a monophyly and they should have the same taxonomic status.
Figure 1.
The ML tree was constructed using the nucleotide sequences of the 13 PCGs of the Cx. gelidus and seven other Culex mitogenome sequences available in GenBank. The best DNA evolution model, GTR + G + I, detected by Model Test was used for the construction of the phylogenetic tree. The bootstrap values for 1000 replicates were indicated above each node of the tree, and the phylogenetic distances were marked beneath each node. Species/subspecies with GenBank mitogenome accession numbers in bracket: Cx. p. pallens (KT851543.1), Cx. pipiens from Turkey (HQ724616.1), Cx. p. quinquefasciatus (GU188856.1), Cx. p. quinquefasciatus from USA (HQ724617.1), Cx. p. pipiens (NC-015079.1), Cx. tritaeniorhynchus JS (NC-028616.1), Cx. tritaeniorhynchus CQ (KT852976.1), Cx. gelidus (KX753344), An. gambiae (NC-002084).
The Cx. gelidus as a type species was a classified to an independent group, Gelidus Group, based on the morphological characteristics (Lu 1997), and subsequently Gelidus Group was degraded as the Gelidus Subgroup, designated to Pipiens Group (Harbach 2011), and Sitiens Group (Harbach et al. 2012), respectively, both based on morphological characteristics. In the phylogenetic analysis based COI sequences, Cx. gelidus and Cx. sitiens (Sitiens Group) were claded with a low bootstrap value, 24% (Wang et al. 2012). In the present study, Cx. gelidus seemed to have closer relationship with Cx. tritaeniorhynchus (genetic distance 0.02170) than Pipiens Complex (genetic distance 0.02648 + 0.01250); therefore, Cx. gelidus might be inappropriate to be classified into the Pipiens Group. Cx. gelidus was grouped into the Cx. tritaeniorhynchus clade, but with only 50% bootstrap support and 0.01250 of genetic distance. Due to only two species (Cx. tritaeniorhynchus and Cx. gelidus) included in the analysis in the Sitiens Group + Cx. gelidus clade, whether the mosquito species with Cx. gelidus as type should be regarded as a Gelidus Group or Subgroup and its taxonomic position need to be elucidated with more molecular data.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Demari-Silva B, Foster PG, Oliveira TMPD, Bergo ES, Sanabani SS, Pessoa R, Sallum MAM.. 2015. Mitochondrial genomes and comparative analyses of Culex camposi, Culex coronator, Culex usquatus and Culex usquatissimus (Diptera: Culicidae), members of the Coronator group. BMC Genomics. 16:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbach RE. 2011. Classification within the cosmopolitan genus Culex (Diptera: Culicidae): the foundation for molecular systematics and phylogenetic research. Acta Tropica. 120:1–14. [DOI] [PubMed] [Google Scholar]
- Harbach RE, Kitching IJ, Lorna Culverwell D, Dubois J, Linton Y-M.. 2012. Phylogeny of mosquitoes of tribe Culicini (Diptera: Culicidae) based on morphological diversity. Zoologica Scripta. 41:499–514. [Google Scholar]
- Lu BL. 1997. Fauna Sinica. Insecta. Diptera: Culicidae 1. Vol. 8 China: Science Press. [Google Scholar]
- Luo QC, Hao YJ, Meng FX, Li TJ, Ding YR, Hua YQ, Chen B.. 2016. The mitochondrial genomes of Culex pipiens pallens and Culex tritaeniorhynchus (Diptera: Culicidae) and comparison analysis with two other Culex species. Parasites Vectors. 9:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudeep AB, Ghodke YS, George RP, Ingale VS, Dhaigude SD, Gokhale MD.. 2015. Vectorial capacity of Culex gelidus (Theobald) mosquitoes to certain viruses of public health importance in India. J Vector Borne Diseases. 52:153–158. [PubMed] [Google Scholar]
- Wang G, Li CX, Guo XX, Xing D, Dong YD, Wang ZM, Zhang YM, Liu M, Zheng Z, Zhang H, et al. . 2012. Identifying the main mosquito species in China based on DNA barcoding. PLoS One. 7:e47051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang NX, Zhang YJ, Yu G, Chen B.. 2013. Structure characteristics of the mitochondrial genomes of Diptera and design and application of universal primers for their sequencing. Acta Entomologica Sinica. 56:398–407. [Google Scholar]
- Zou YL, Ding YR, Luo QC, Chen B.. 2015. The extraction method of mosquito mitochondrial genome DNA. Chinese J Vector Biol Control. 26:333–336. [Google Scholar]