Abstract
This study presents the complete sequence of Vanilla pompona chloroplast genome. This 148,009 bp long genome consist of 107 genes out of which 30 of them are tRNA, 4 rRNA. Fairly long inverted repeat regions (IR) and large single-copy (LSC) of length 29,807 and 86,358 bp, respectively, were detected. This means an exceptionally short single-copy (SSC) region with only 2037 bp. This truncation of the SSC is due to multiple translocation of ndh genes to the mitochondrion as in majority of the Orchidaceae and especially in the genus Vanilla. The phylogeny presented here meaningfully places Vanillon within the orchid family.
Keywords: Chloroplast genome, de novo assembly, genome skimming, phylogenomics, plastid evolution
Vanilla is the world’s most popular flavour and, by unit mass, is among the most valuable of the spice crops (Bory et al. 2008). The main source of vanilla aroma is Vanilla planifolia Andrews, and it is the second most expensive spice next only to saffron (Ranadive 1994). The world production of vanilla was estimated to be about 8032 tons in 2014 (FAOSTAT 2017). Two other species V. tahitensis J.W.Moore and V. pompona Schiede are also cultivated. Vanilla pompona is extremely rare, grown only in Guadeloupe, Martinique and Dominica and used only by the cosmetic and perfume industries due to its floral/perfumery/heliotropin-like flavour (Ehlers and Pfister 1997). Besides being economically important, Orchidaceae are an appealing family for evolutionary studies due to multiple translocation of the ndh genes from the chloroplast to the mitochondrion (Lin et al. 2015). The loss or dysfunctioning of these genes in the plastome seems especially bold in Vanilloideae since their plastid genome exhibit only one functional ndh gene.
We extracted DNA according to Shi et al. (2012) from 20 g fresh Vanillon leaves collected in Kaisaniemi Botanical Garden, University of Helsinki, Finland (60.1753; 24.9460; voucher M. Christenhusz 00YY-845). Paired-end libraries of 2 × 150 bp were prepared with Illumina TruSeq DNA Sample prep kit and sequencing was carried out on an Illumina MiSeq platform. Raw reads were filtered with Trimmomatic (Bolger et al. 2014), and de novo assembly of the plastid genome was carried out with the Geneious assembler platform (Kearse et al. 2012). We annotated the genome using DOGMA (Wyman et al. 2004) following manual correction in Geneious. Here, we report the complete chloroplast sequence of Vanilla pompona to provide resources for taxonomic studies and plant breeding.
The complete chloroplast genome of Vanilla pompona (GenBank accession MF197310) has a total length of 148,009 bp which is divided by two IR regions of 29,807 bp. This genome comprises of 107 genes and has 34% overall GC content. The genes are classified into 30 tRNA, 4 rRNA and 75 coding-protein genes. From the 11 ndh plastome encoded NAD(P)H dehydrogenase complex genes (Ueda et al. 2012), only ndhB was detected in the plastid genome of V. pompona. This indicates the loss or pseudogenization of the rest. This independent and complex pattern of loss of functional ndh genes in Orchidaceae is not tied with a significant evolutionary events (Kim et al. 2015), but justifies the exceptionally short length of SSC in the Vanilloideae.
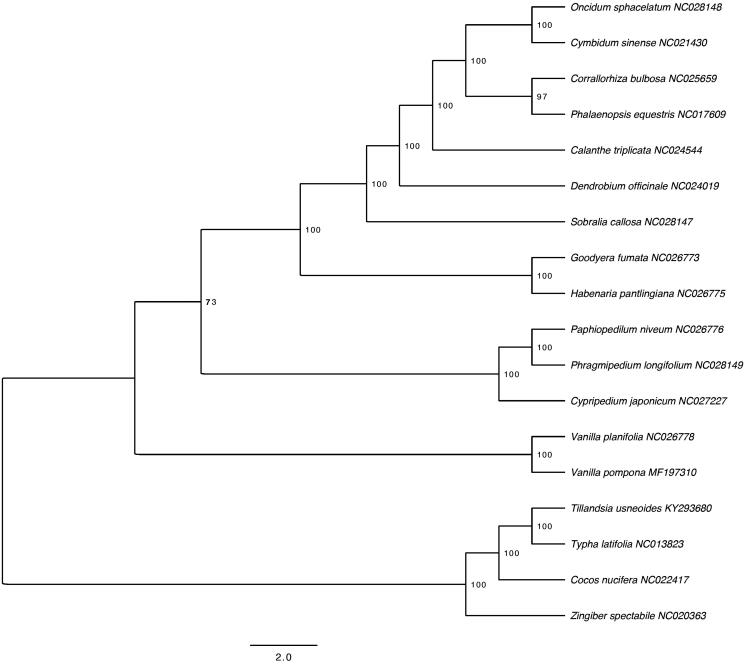
Using the RAxMLv8.0 (Stamatakis 2014) the best scoring ML tree with 10,000 bootstrap replicates was calculated under GTR-GAMMA after running jModelTest2 (Darriba et al. 2012) including 18 representative species in Orchidaceae (Figure 1). As expected Vanilla pompona was resolved as sister to V. planifolia and together they formed a basal clade to Cypripedioideae, Orchidoideae, and Epidendroideae. We also made phylogenetic analysis using parsimony as an optimality criterion. The same matrix as with ML was analyzed using Nona (Goloboff 1994) within the WinClada shell (Nixon 2002).
Figure 1.
The ML tree of 17 selected Orchidaceae chloroplast sequences plus Vanilla pompona. The values on the node show the bootstraps of 10,000 replicates and scale is substitution per site. New paragraph: use this style when you need to begin a new paragraph.
We expect this sequence to clarify the taxonomic status of Vanilla pompona, and provide additional genomic resources for vanilla breeding.
Acknowledgements
We thank the staff and colleagues of the Viikki Biocenter who kindly contributed reagents, materials and analyses tools for our study.
Disclosure statement
The authors claim no conflict of interest.
References
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bory S, Lubinsky P, Risterucci A-M, Noyer J-L, Grisoni M, Duval M-F, Besse P.. 2008. Patterns of introduction and diversification of Vanilla planifolia (Orchidaceae) in Reunion Island (Indian Ocean). Am J Bot. 95:805–815. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers D, Pfister M.. 1997. Compounds of Vanillons (Vanilla pompona Schiede). J Ess Oil Res. 9:427–431. [Google Scholar]
- FAOSTAT 2017. FAO statistical databases. Rome, Italy: Food and Agriculture Organization of the United Nations. [accessed 2017 Sep 1]. http://www.fao.org [Google Scholar]
- Goloboff P. 1994. NONA: a tree searching program. [accessed 2017 Sep 1]. www.cladistics.com [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, et al. . 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinfo. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim JS, Moore MJ, Neubig KM, Williams NH, Whitten WM, Kim JH.. 2015. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across Orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS One. 10:e0142215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CS, Chen JW, Huang YT, Chan MT, Daniell H, Chang WJ, Hsu CT, Liao DC, Wu FH, et al. . 2015. The location and translocation of ndh genes of chloroplast origin in Orchidaceae family. Nat Com. 5:9040. doi: 10.1038/srep09040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon KC. 2002. WinClada 1.00.08. [accessed 2017 Sep 1]. www.cladistics.com [Google Scholar]
- Ranadive AS. 1994. Vanilla-cultivation, curing, chemistry, technology and commercial products In: George C, editor. Spices, herbs and edible fungi. New York, Tokyo: Elsevier; p. 517–604. [Google Scholar]
- Shi C, Hu N, Huang H, Gao J, Zhao Y-J, Gao L-Z.. 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS One. 7:e31468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Kuniyoshi T, Yamamoto H, Sugimoto K, Ishizaki K, Kohchi T, Nishimura Y, Shikanai T.. 2012. Composition and physiological function of the chloroplast NADH dehydrogenase-like complex in Marchantia polymorpha. Plant J. 42:683–693. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]