Abstract
We report the mitochondrial genomes of two freshwater sponges, Spongilla lacustris and Ephydatia cf. muelleri. The genomes contain 14 protein-coding genes and are similar in structure to other published mitochondrial genomes from freshwater sponges. The E. cf. muelleri described here is remarkably similar in coding regions to the published genome, but differs in number and length of hairpin-forming repeats between genes.
Keywords: Freshwater, porifera, sponge, sympatric
Sponges are a diverse animal phylum containing an estimated 15,000 species that perform important physiological and ecological roles in aquatic ecosystems. While the majority of species inhabit marine environments, members of the order Spongillida are found in freshwater habitats around the world. Previous analysis of the mitochondrial protein CO1 suggested that freshwater sponges originated from a single, relatively recent radiation (Itskovich et al. 2006). Here we report the complete mitochondrial (mt) genomes of two freshwater demosponges from Lake Constance in Germany, Spongilla lacustris – the type genus and species of the largest family of freshwater sponges, and the co-sequenced Ephydatia cf. muelleri var. bodenseensis.
We sequenced the genome of Spongilla lacustris gemmules using the Illumina Moleculo (TruSeq) artificial long-read technology. The long reads were mapped using Kearse et al. (2012) to the published mt genome of Ephydatia muelleri (Wang & Lavrov 2008) allowing for indels and up to 20% mismatch. Mapped reads were then assembled using the built-in assembler in Geneious R8, producing a single contig of 28,044 bases with a coverage of 317×. The published cox1 gene from S. lacustris (EU000572.1) matched to the assembled version with 99% identity (two substitutions), confirming the species. From the mapping of the long reads, we found reads with unusually high identity (99%) to the published mt sequence of E. muelleri. These reads were separated and assembled into a distinct mt genome of 23,946 bp at 41× coverage with 99% identity to the published E. muelleri. This finding suggests that S. lacustris and E. muelleri occur sympatrically and are almost indistinct to divers.
Gene annotation was done using the MFannot web server (Valach et al. 2014). Both genomes contain the same 14 protein-coding genes, 25 tRNAs and two ribosomal RNAs, all in the same order as all other published freshwater sponges: Lubomirskia baicalensis (Lavrov 2010), Rezinkovia echinata, Baikalospongia intermedia, Swartschewskia papyracea, Corvomeyenia sp. (Lavrov et al. 2012), Eunapius subterraneus (Plese et al. 2012) and Ephydatia fluviatilis (Imesek et al. 2013).
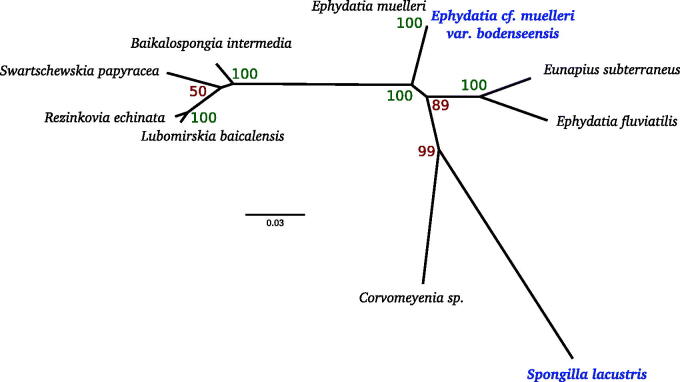
Among the species compared, the lowest sequence identities were 68.2% between S. lacustris and both L. baicalensis and B. intermedia. When comparing only the coding sequences, the minimum sequence identity was 97.7% between S. lacustris and Corvomeyenia sp. Within the entire clade, the lowest coding sequence identity was 96.4% between Corvomeyenia sp. and E. fluviatilis, showing that sequence divergence is low in this order (Figure 1). The E. cf. muelleri var bodenseensis differs from the published E. muelleri by only three nucleotides substitutions within the coding regions and one nucleotide substitution in trnT, while the overall identity is 99.4% between the two variants. The intergenic regions vary considerably with most of the differences occurring as insertions of repetitive hairpin-forming elements (RHEs), as seen in other sponges (Erpenbeck et al. 2009; Lavrov et al. 2012).
Figure 1.
Phylogenetic tree of mitochondrial sequences from freshwater sponges. Whole mitochondrial genomic nucleotide sequences were used for all species. Alignment was made with MAFFT using LINSI parameters, and tree was generated using RAXML 8.2 (Exelixis Lab, Tucson, AZ) using the GTRGAMMA model with 100 bootstrap replicates. Species in bold were sequenced in this study. Species accessions are as follows: L. baicalensis, NC_013760.1; R. echinata, NC_018360.1; B. intermedia, NC_018343.1; S. papyracea, JQ302308; Corvomeyenia sp., JQ302311; E. subterraneus, NC_016431.1; E. muelleri, NC_010202.1; E. fluviatilis, JN209966.
This Whole Genome Shotgun project has been deposited in ENA under the accession numbers LT158503 and LT158504. Specimens and live cultures are kept at the Department of Earth and Environmental Sciences, Paleontology & Geobiology, Ludwig-Maximilians-Universität München.
Acknowledgments
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Erpenbeck D, Voigt O, Wörheide G, Lavrov DV.. 2009. The mitochondrial genomes of sponges provide evidence for multiple invasions by Repetitive Hairpin-forming Elements (RHE). BMC Genomics. 10:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imesek M, Plese B, Lukic-Bilela L, Lelo S, Cetkovic H.. 2013. Mitochondrial genomes of the genus Ephydatia Lamouroux, 1816: can palindromic elements be used in species-level studies? Org Divers Evol. 13:127–134. [Google Scholar]
- Itskovich VB, Belikov SI, Efremova SM, Masuda Y, Krasko A, Schroeder HC, Mueller WEG.. 2006. Monophyletic origin of freshwater sponges in ancient lakes based on partial structures of COXI gene. Hydrobiologia. 568:155–159. [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Mentjies P., & Drummond A (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Bioinformatics, 28(12), 1647-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov DV. 2010. Rapid proliferation of repetitive palindromic elements in mtDNA of the endemic Baikalian sponge Lubomirskia baicalensis. Mol Biol Evol. 27:757–760. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Maikova OO, Pett W, Belikov SI.. 2012. Small inverted repeats drive mitochondrial genome evolution in Lake Baikal sponges. Gene. 505:91–99. [DOI] [PubMed] [Google Scholar]
- Plese B, Lukic-Bilela L, Bruvo-Maaric B, Harcet M, Imesek M, Bilandzija H, Cetkovic H.. 2012. The mitochondrial genome of stygobitic sponge Eunapius subterraneus: mtDNA is highly conserved in freshwater sponges. Hydrobiologia. 687:49–59. [Google Scholar]
- Valach M, Burger G, Gray MW, Lang BF.. 2014. Widespread occur-rence of organelle genome-encoded 5S rRNAs including permuted molecules. Nucleic Acids Res. 42:13764–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lavrov DV.. 2008. Seventeen new complete mtDNA sequences reveal extensive mitochondrial genome evolution within the demospongiae. PLoS ONE. 3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]