Abstract
Cucumber (Cucumis sativus var. sativus) is one of the economically important vegetable crops. In this study, we characterized the complete chloroplast genome sequence of inbred line ID YHB bred from Korean solid green-type cucumber variety, through de novo assembly using next-generation sequencing. The chloroplast genome is 155,501 bp long and has typical quadripartite structures and gene contents as found in reported cucumber chloroplast genomes. Interestingly, sequence comparison revealed a novel 24-bp deletion present only in the chloroplast genome of the inbred line. Phylogenetic analysis confirmed that the inbred line was closely grouped with cucumber cultivars.
Keywords: Cucumis sativus, cucumber, chloroplast genome, novel deletion
The Cucumis genus belongs to the Cucurbitaceae family and consists of about 30 species distributed in tropical and temperate regions (Lu and Jeffrey 2011). Among those species, cucumber is one of the most economically important vegetable crops in the world (Pitrat et al. 1999; Acquaah 2012). Several complete chloroplast genome sequences have been reported in cucumber cultivars (Kim et al. 2006; Plader et al. 2007; Chung et al. 2007). Although various genomic resources have been available for cucumber, the plant species is known to have a narrow genetic base, which makes breeding of this species difficult (Acquaah 2012). On this account, we characterized the complete chloroplast genome sequence of inbred line ID YHB bred from Korean solid green-type cucumber variety.
Genomic DNA was extracted from fresh leaves collected from the inbred line cultivated in a greenhouse of Seoul National University (Seoul, Korea, deposited specimen no. CT604). Illumina paired-end (PE) library with 750-bp insert size was constructed and sequenced using an Illumina MiSeq platform by LabGenomics (www.labgenomics.co.kr, Korea). Complete chloroplast genome sequence was generated by de novo assembly using high quality PE reads of about 500 Mb, as described previously (Kim et al. 2015). The chloroplast genome was initially annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq-app.html) and manually confirmed using Artemis annotation tool (Rutherford et al. 2000) with BLASTN searches.
Complete chloroplast genome sequence (GenBank accession no. MF095790) of inbred line ID YHB is 155,501 bp long and has typical quadripartite structure consisting of large single copy (LSC) region of 86,877 bp, small single-copy (SSC) region of 18,250 bp, and a pair of inverted repeats (IRa and IRb) of 25,187 bp. The genome contains a total of 115 genes, including 80 protein-coding genes, 31 tRNA genes, and four rRNA genes.
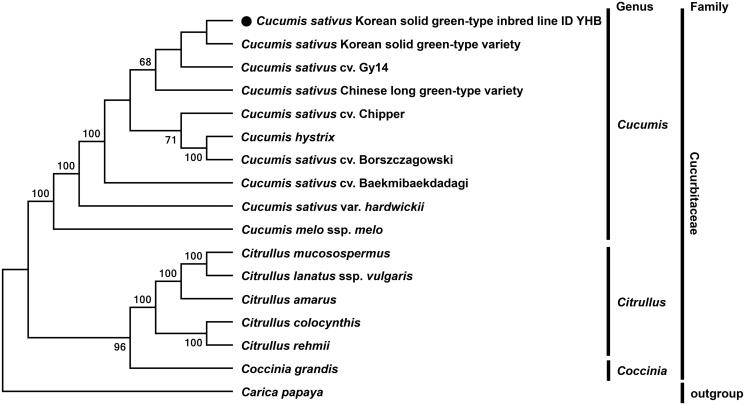
Phylogenetic analysis was carried out based on multiple alignment of complete chloroplast genome sequences and confirmed that inbred line ID YHB was grouped with Cucumis species such as cucumber cultivars, wild cucumber and melon, where the line was much closed to another variety of Korean solid green-type cucumber (Figure 1). Interestingly, sequence comparison revealed a novel 24-bp deletion within ycf1 genes in the YHB chloroplast genome but not in reported chloroplast genomes of other cucumbers. The 24-bp deletion resulted from copy number variation of 24-bp tandem repeats, which was confirmed by PCR amplification and nucleotide sequencing.
Figure 1.
Phylogenetic tree of inbred line ID YHB and other species in the Cucurbitaceae family. Tree was generated by multiple alignment of complete chloroplast genome sequences using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html) and a neighbour-joining (NJ) analysis using MEGA 6.0 (Tamura et al. 2013). Numbers in the nodes are bootstrap support values (>50%) from 1000 replicates. Chloroplast genome sequences used for this tree are: Cucumis sativus Korean solid green-type inbred line ID YHB, MF095790; Cucumis sativus Korean solid green-type variety, KX231327; Cucumis sativus cv. Gy14, DQ865975; Cucumis sativus Chinese long green-type variety, KX231328; Cucumis sativus cv. Chipper, DQ865976; Cucumis hystrix, KF957866; Cucumis sativus cv. Borszczagowski, AJ970307; Cucumis sativus cv. Baekmibaekdadagi, DQ119058; Cucumis sativus var. hardwickii, MF536709; Cucumis melo ssp. melo, JF412791; Citrullus mucosospermus, KY430686; Citrullus lanatus ssp. vulgaris, KY014105; Citrullus amarus, MF536694; Citrullus colocynthis, MF357889; Citrullus rehmii, MF536695; Coccinia grandis, KX147312; Carica papaya, EU431223.
In conclusion, the chloroplast sequence and polymorphic site identified in this study will contribute to enriching genetic resources of cucumber.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Acquaah G. 2012. Principles of plant genetics and breeding: breeding cucumber. New York: John Wiley & Sons, Ltd; p. 679–681. [Google Scholar]
- Chung SM, Gordon VS, Staub JE.. 2007. Sequencing cucumber (Cucumis sativus L.) chloroplast genomes identifies differences between chilling-tolerant and -susceptible cucumber lines. Genome. 50:215–225. [DOI] [PubMed] [Google Scholar]
- Kim JS, Jung JD, Lee JA, Park HW, Oh KH, Jeong WJ, Choi DW, Liu JR, Cho KY.. 2006. Complete sequence and organization of the cucumber (Cucumis sativus L. cv. Baekmibaekdadagi) chloroplast genome. Plant Cell Rep. 25:334–340. [DOI] [PubMed] [Google Scholar]
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Jeffrey C.. 2011. Cucurbitaceae In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 19 Beijing (China) and St. Louis (MO): Science Press and Missouri Botanical Garden Press; Available from: http://efloras.org/florataxon.aspx?flora_id =2&taxon_id =10233. [Google Scholar]
- Pitrat M, Chauvet M, Foury C.. 1999. Diversity, history, and production of cultivated cucurbits. Acta Hort. 492:21–28. [Google Scholar]
- Plader W, Yukawa Y, Sugiura M, Malepszy S.. 2007. The complete structure of the cucumber (Cucumis sativus L.) chloroplast genome: its composition and comparative analysis. Cell Mol Biol Lett. 12:584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B.. 2000. Artemis: sequence visualization and annotation. Bioinformatics. 16:944–945. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]