Abstract
In this study, we analyzed the complete mitochondrial genome sequence of the corn planthopper, Peregrinus maidis. The complete mitogenome sequence of P. maidis was observed to be a circular molecule 16,279 bp long and consisting of 13 protein-coding genes (PCG), 2 ribosomal RNA (rRNA) genes and 22 transfer RNA (tRNA) genes (GenBank accession no. MG049917). The nucleotide composition is biased toward adenine and thymine (77.8% A + T). The A + T-rich region was found between rrnS and trnI, and this entire region was 1596 bp long.
Keywords: Delphacidae, mtDNA, phylogenetic relationship
The corn planthopper, Peregrinus maidis (Ashmead 1890) is a widely distributed destructive insect that causes significant yield losses not only by feeding on vascular tissues via piercing-sucking mouthparts, but also by transmitting viruses between crops (Lastra and Esparza 1976; Nault and Ammar 1989; Yao et al. 2013). Despite its economic importance, the mitogenome sequence of P. maidis so far remained unknown. Therefore, we sequenced the complete mitochondrial DNA genome of P. maidis to provide more comprehensive data for this species and also for its relationship within the family Delphacidae.
Adult P. maidis males were collected from a maize field in Xiashi Town (N 22°07′17.39″ and E 106°54′0.89″), Guangxi, China, in August 2014. Voucher specimens were deposited in the Key Laboratory of Plant Protection Resources and Pest Management of Ministry of Education, Entomological Museum, Northwest A&F University (NWAFU). The complete mitochondrial genome of P. maidis was determined by using next-generation sequencing (NGS).
The P. maidis mitochondrial genome is 16,279 bp (Genbank accession no. MG049917) in length with a total A + T content of 77.8% that is heavily biased toward the A and T nucleotides. It encodes the complete set of 37 genes which are usually found in animal mitogenomes. The gene arrangement in the mitochondrial genome of P. maidis is conserved, similar to other mitogenomes in Delphacidae, with the exception of Nilaparvata lugens (Zhang et al. 2013). In the mitogenome of P. maidis, a total of 19 bp overlaps have been found at nine gene junctions (trnQ and trnM share a nucleotide; nad2 and trnC share two nucleotides; atp8 and atp6 share four nucleotides; trnR and trnN, trnN and trnS1, trnS1 and trnE, trnE and trnF share one nucleotide; nad4 and nad4L share seven nucleotides; and cytb and trnS2 share one nucleotide). The mitogenome is loose and has a total of 488 bp intergenic sequences without the putative A + T-rich region. The intergenic sequences are at 15 locations ranging from 1 to 338 bp, with the longest one located between trnP and nad6. The A + T-rich region of the P. maidis is 1596 bp long and located between the rrnS and trnI genes. The A + T content of this region is 86.2%.
All 22 tRNA genes usually found in the mitogenomes of insects are present in P. maidis. The nucleotide length of tRNA genes ranges from 55 bp (trnH) to 71 bp (trnK), and A + T content ranges from 70.4% (trnK) to 89.3% (trnS2). These two rRNA genes have been identified on the N-strand in the P. maidis mitogenome.
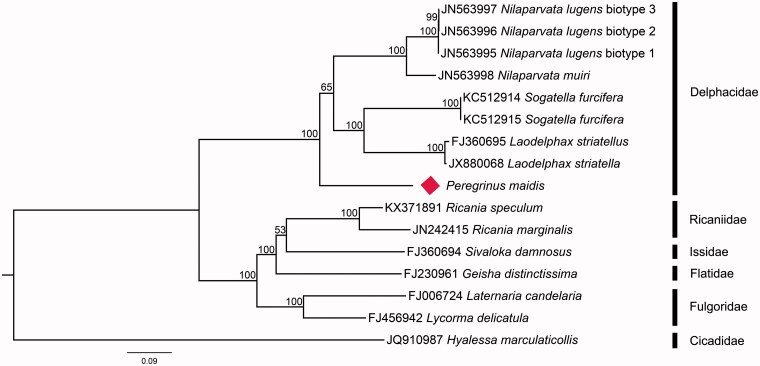
We analyzed the nucleotide sequences of 13 PCGs using the maximum likelihood (ML) method to understand the phylogenetic relationship of P. maidis with other fulgoroids. The mitogenome sequence of Hyalessa maculaticollis (GenBank accession no. JQ910987) was used as outgroup. The result shows that P. maidis belongs to the superfamily Fulgoroidea and is clustered into a branch of Delphacidae (Figure 1). The family Delphacidae is monophyletic and P. maidis is supported as the sister group to the remaining delphacids.
Figure 1.
The maximum-likelihood (ML) phylogenetic tree of P. maidis and other fulgorids. The numbers beside the nodes are percentages of 1000 bootstrap values. Alphanumeric terms indicate the GenBank accession numbers.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ashmead WH. 1890. The corn delphacid, Delphax maidis. Psyche. 5:321–324. [Google Scholar]
- Lastra JR, Esparza J.. 1976. Multiplication of Vesicular stomatitis virus in the leafhopper Peregrinus maidis (Ashm.), a vector of a plant rhabdovirus . J Gen Virol. 32:139–142. [DOI] [PubMed] [Google Scholar]
- Nault LR, Ammar ED.. 1989. Leafhopper and planthopper transmission of plant viruses. Annu Rev Entomol. 34:503–529. [Google Scholar]
- Yao J, Rotenberg D, Afsharifar A, Barandoc-Alviar K, Whitfield AE.. 2013. Development of RNAi methods for Peregrinus maidis, the corn planthopper. PLoS One. 8:e70243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KJ, Zhu WC, Rong X, Zhang YK, Ding XL, Liu J, Chen DS, Du Y, Hong XY.. 2013. The complete mitochondrial genomes of two rice planthoppers, Nilaparvata lugens and Laodelphax striatellus: conserved genome rearrangement in Delphacidae and discovery of new characteristics of atp8 and tRNA genes. BMC Genomics. 14:417. [DOI] [PMC free article] [PubMed] [Google Scholar]