Abstract
In this study, we determined the complete chloroplast sequence of Habenaria radiata (Thunb.) Spreng. (Orchidaceae) (NCBI acc. no. KX871237), an endangered plant species protected by the national law of Korea. The gene order and gene content of the H. radiata plastome are similar to those of typical angiosperm plastomes. The 11 ndh genes, which are usually lost in orchid plastomes, are intact in the H. radiata plastome. The complete plastome is 155,353 bp in length and consists of a large single copy of 84,833 bp and a small single copy of 17,718 bp, separated by two inverted repeats of 26,401 bp. The plastome contains 113 genes, of which 79 are protein-coding genes, 30 are tRNA genes, and four are rRNA genes. Sixteen genes contain one intron and two genes (clpP, ycf3) have two introns. A total of 76 simple sequence repeat (SSR) loci, which consist of 58 mono-SSR, 17 di-SSR, and 1 tri-SSR, are scattered along the H. radiata plastome. Some of these plastome SSR and high sequence divergent regions may be useful for development of genetic markers for the populations of H. radiata and other congeneric species. Phylogenetic analysis identified the sister relationship between H. radiata and H. pantlingiana within the tribe Orchideae.
Keywords: Habenaria radiata, Orchidaceae, plastome, endangered species
Habenaria radiata (Thunb.) Spreng., one of the terrestrial orchids in the genus Habenaria, is native to Korea and Japan (Lee 2011). It is a single-stemmed, erect plant, up to 40 cm tall, with small root tubers. H. radiata was relatively common in moisture-rich areas on mountain slopes or in grasslands until a few decades ago. The species has been often targeted by orchid collectors and hobbyist because of the beautiful bird-like floral structure. As a result, the number of natural populations of H. radiata has decreased rapidly in recent years. Only a few populations have been sporadically reported from the remote mountain areas of Korea. Therefore, the species was designated as endangered species, and it was protected by the national law of Korea. The genus Habenaria, comprising about 835 species (Chase et al. 2015), belongs to the subfamily Orchidoideae of the family Orchidaceae in the order Asparagales (APG IV 2016). In order to develop genetic markers of H. radiata for conservation studies, we sequenced and analysed the plastome of this species. The plastome data will be useful to evaluate the genetic diversity and the phylogenetic position of H. radiata and related taxa.
The seeds of H. radiata were originally collected from a natural population in the National Arboretum of Korea. The leaf material of H. radiata was collected from a single individual that was cultivated in a laboratory flask from the seeds. A voucher specimen and DNA sample were deposited in the Korea University Herbarium (KUS 2015-1271) and the Plant DNA Bank in Korea (PDBK 2015-1271), respectively. Fresh leaves were ground into powder in liquid nitrogen, and total DNA was extracted by using G-spin™II for Plant Genomic DNA extraction kit (iNtRON, Seongnam, Korea). The complete plastome was generated using Illumina MiSeq (San Diego, CA), and assembled with Geneious 6.1.8 (Kearse et al. 2012). The average coverage of the sequence was 962 times that of the plastome size. Annotations were performed using the National Center for Biotechnology Information (NCBI) BLAST and tRNAscan-SE programs (Lowe and Eddy 1997). The complete plastome sequence was submitted to the NCBI database under accession number KX871237.
The gene order and gene content of the H. radiata were similar to those of a typical angiosperm such as Panax, Nicotina, and Sesamum (Shinozaki et al. 1986; Kim and Lee 2004; Yi and Kim 2012). The orchid plastomes usually lost the ndh genes (Chang et al. 2006; Wu et al. 2010; Lin et al. 2015). However, the H. radiata plastome retained all of the ndh genes. The plastome of H. radiata contains only 113 unique genes, including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. Sixteen genes had a single intron, whereas the clpP and ycf3 genes had two introns. The length of the complete plastome of H. radiata was 155,353 bp, and it was composed of a large single copy (LSC) of 84,833 bp, a small single copy (SSC) of 17,718 bp, and two inverted repeats (IRs) each 26,401 bp long. The length of the H. radiata plastome was approximately 10 kb longer than that of the typical orchid plastome. The average AT content of the plastome was 63.5%. We identified a total of 76 simple sequence repeat (SSR) loci, which consisted of 58 mono-SSR, 17 di-SSR, and 1 tri-SSR, scattered along the plastome. Some of these plastome SSRs may be useful for development of genetic markers for the populations of H. radiata.
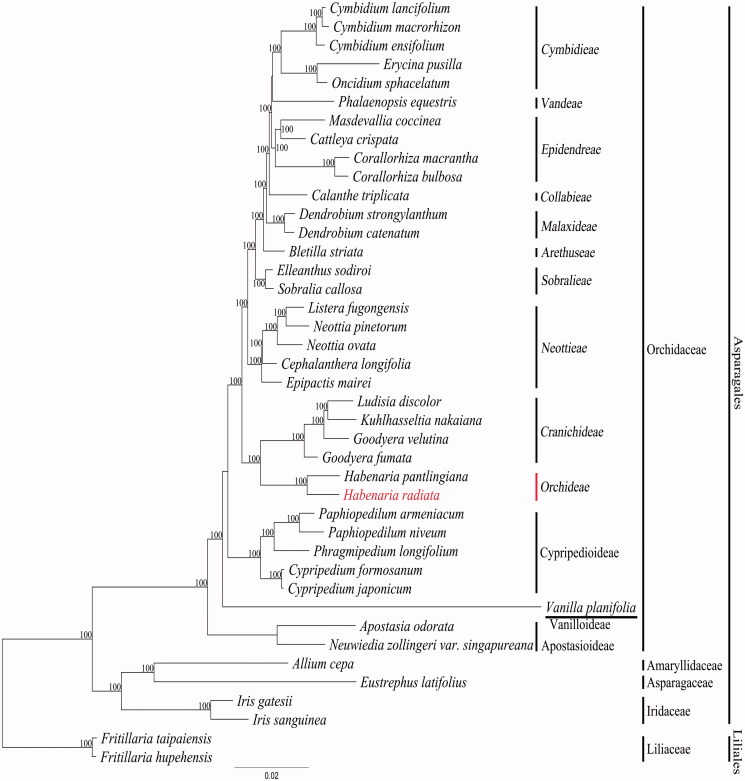
For phylogenetic tree construction, 82 gene sequences from 44 taxa were aligned using MUSCLE implemented in Geneious 6.1.8. The aligned data matrix was 70,020 bp in length. The data matrix was subjected to phylogenetic analysis using RAxML v. 7.7.1 (Stamatakis et al. 2008). An ML tree was obtained with an ML optimization likelihood value of –314504.317667. A sister group relationship between the two Habenaria species was supported by 100% bootstrap support (Figure 1). In order to clarify more precisely the phylogenetic relationships of Habenaria and other Orchidaceae members, we need additional whole plastome data from diverse genera and species of the tribe Orchideae.
Figure 1.
A maximum likelihood (ML) tree of Orchidaceae indicating the phylogenetic position of Habenaria radiata. The ML tree is reconstructed from the sequences of 78 protein coding genes and four rRNA genes for 41 plastome sequences. The number above or below of each node indicates bootstrap supporting value from 100 replications. Genbank accession numbers of taxa are shown below: Allium cepa (NC024813), Apostasia odorata (NC030722), Bletilla striata (NC028422), Calanthe triplicata (NC024544), Cattleya crispata (NC026568), Cephalanthera longifolia (NC030704), Corallorhiza bulbosa (NC025659), Corallorhiza macrantha (NC025660), Cymbidium ensifolium (NC028525), Cymbidium lancifolium (NC029712), Cymbidium macrorhizon (KY354040), Cypripedium formosanum (NC026772), Cypripedium japonicum (NC027227), Dendrobium catenatum (NC024019), Dendrobium strongylanthum (NC027691), Elleanthus sodiroi (NC027266), Epipactis mairei (NC030705), Erycina pusilla (NC018114), Eustrephus latifolius (NC025305), Fritillaria hupehensis (NC024736), Fritillaria taipaiensis (NC023247), Goodyera fumata (NC026773), Goodyera velutina (NC029365), Habenaria radiata (KX871237), Habenaria pantlingiana (NC026775), Iris gatesii (NC024936), Iris sanguinea (NC029227), Kuhlhasseltia nakaiana (KY354041), Listera fugongensis (NC030711), Ludisia discolor (NC030540), Masdevallia coccinea (NC026541), Neottia ovata (NC030712), Neottia pinetorum (NC030710), Neuwiedia zollingeri var. singapureana (KM244735), Oncidium sphacelatum (NC028148), Paphiopedilum armeniacum (NC026779), Paphiopedilum niveum (NC026776), Phalaenopsis equestris (NC017609), Phragmipedium longifolium (NC028149), Sobralia callosa (NC028147), Vanilla planifolia (NC026778).
Acknowledgements
The endangered plant material was collected under the proper permit from the environmental protection authority of the Korean government. The voucher specimen of Habenaria radiata used in this study was deposited in Korea University herbarium (KUS acc. no. 2015-1271). The extracted DNA material was deposited in Plant DNA Bank of Korea (PDBK acc. no. 2015-1271).
Disclosure statement
The authors report no conflicts of interest, and are independently responsible for the content and writing of the paper.
References
- APG IV 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20. [Google Scholar]
- Chang CC, Lin HC, Lin IP, Chow TY, Chen HH, Chen WH, Cheng CH, Lin CY, Liu SM, Chang CC, et al. 2006. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 23:279–291. [DOI] [PubMed] [Google Scholar]
- Chase M, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Berg C, Schuiteman A.. 2015. An updated classification of Orchidaceae. Bot J Linn Soc. 177:151–174. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-J, Lee H-L.. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11:247–261. [DOI] [PubMed] [Google Scholar]
- Lee NS. 2011. Illustrated flora of Korean orchids. Seoul (South Korea): Ewha Womans University Press; (in Korean). [Google Scholar]
- Lin CS, Chen JJ, Huang YT, Chan MT, Daniell H, Chang WJ, Hsu CT, Liao DC, Wu FH, Lin SY, et al. 2015. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci Rep. 5:9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K.. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5:2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J.. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57:758–771. [DOI] [PubMed] [Google Scholar]
- Wu FH, Chan MT, Liao DC, Hsu CT, Lee YW, Daniell H, Duvall MR, Lin CS.. 2010. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 10:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi DK, Kim KJ.. 2012. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS One. 7:e35872. [DOI] [PMC free article] [PubMed] [Google Scholar]