Abstract
Coelomactra antiquata is a famous delicacy and a promising new candidate for aquaculture, which belongs to the family Mactridae (Mollusca: Veneroida). The complete mitochondrial genome of C. antiquata (Liao Ning province, in China, LN) was finished, which is the first representative from this province. The results showed that the total length of LN-mtDNA sequence is 16,797 bp and the content of A + T is 65.01%. It encodes 35 genes, including 12 protein-coding genes, 21 transfer RNA genes and two ribosomal RNA genes. All coding genes are encoded on the heavy strand. Compared with the typical gene content of animal mitochondrial genomes, atp8 and trnSer(UCN) genes are missing in the mitochondrial genome. The complete mitochondrial genome contains 26 non-coding regions (1598 bp), one major non-coding region consists of 1046 bp in which 4.9 tandem repeat sequences (99bp per sequence) was observed. The phylogenetic tree showed that Liaoning population was clustered into one clade with Shandong (Rizhao, Jiaonan and Jimo) and Guangxi (Beihai) populations, meanwhile all of them are far from the Fujian populations (Pingtan, Zhangzhou and Changle). So, Liaoning, Shandong and Guangxi populations have a close relationship. Actually, Fujian is located between Liaoning, Shandong and Guangxi. So, the result challenges the previously assumed relevance between geographic distance and genetic distance. The genetic distance of Liaoning C. antiquata and Fujian (Changle, Zhangzhou and Pingtan) C. antiquata (0.176–0.177) is greater than the genetic distance between Mytilus galloprovincialis and Mytilus trossulus (0.160). The genetic difference of Liaoning population and Fujian populations has reached species level.
Keywords: Cryptic species, mactridae, mollusca, phylogeny, veneroida
Researches on cryptic species have increased quickly over the past two decades, which is in large part resulted from the increasing availability of molecular data (Bickford et al. 2007). In recent years, detection of cryptic species based on complete mitochondrial genomes has become increasingly popular (Iannelli et al. 2007; Krzywinski et al. 2011; Gasser et al. 2012; Burger et al. 2014; Griggio et al. 2014; Shen et al. 2014). Generally, animal mitochondrial genomes are circular molecules, containing 37 genes: 2 ribosomal RNA genes (lrRNA and srRNA), 13 PCGs (protein-coding genes) and 22 tRNA genes (Shen et al. 2012; Schierwater et al. 2013). Coelomactra antiquata is a famous Chinese delicacy and a promising new candidate for aquaculture, which belongs to the family Mactridae (Mollusca: Veneroida) (Liu et al. 2006; Meng et al. 2012; Yuan et al. 2013; Meng et al. 2013).
A specimen of C. antiquata was obtained from the Dalian city, Liaoning province in China (N: 38.889, E: 121.707), which is stored at Marine Museum of Huaihai Institute of Technology (Accession number: Can-003). The mtDNA of C. antiquata was extracted from muscle tissues using Phenol-chloroform method. Seven pairs of specific primers were designed, including cox1-nad6F: TGG TGA AAG AGT GTT GGA CG, cox1-nad6R: CTA CAA ATA CGA GCA GCA GCA A, nad6-srRNAF: CTG GGC CTG TTC TTG GTG TAT, nad6-srRNAR: GGC GGT TTG TAC GCA TTT CA, srRNA-cytbF: AAA ACT CAA AAA GGC TGG CGG, srRNA-cytbR: GCC TCC GCA TCA GAA TCA AC, cytb-nad4F: CAT GGT TGG TGG GGT GTA CT, cytb-nad4R: CCT GCC AAA GGT GCC TCT AC, nad4-nad3F: TGA GGG TAC CAA CCT GAA CG, nad4-nad3R: TGC ATT CCC CCT CCA AAA CG, nad3-cox2F: TGA AGA GGG TCG AAC GGT TT, nad3-cox2R: TGA CCA TGC GCT TCT ACA CT, cox2-cox1F: GGT TGC TGG ACC TTT GTA CG and cox2-cox1R: CTA TTG CCG GAC CAG GAT GT. The DNA fragments were obtained by polymerase chain reaction amplification and sequenced by primer-walking strategy. Finally, the complete mitochondrial genome of C. antiquata (Liaoning province, in China, LN) was finished, which is the first representative from this province. The locations of the protein-coding genes (PCGs) and rRNA genes were determined with DOGMA (Wyman et al. 2004). Transfer RNA genes were identified by using tRNAscan-SE 1.21 (Schattner et al. 2005). Tandem repeats finder was used to search for tandem repeats of the mitochondrial genome (Benson 1999).
The results showed that the total length of LN-mtDNA sequence is 16797 bp (GenBank accession number: KR362246) and the content of A + T is 65.01% (AT skew = −0.1955, GC skew= 0.2592). It encodes 35 genes, including 12 protein-coding genes, 21 transfer RNA genes and two ribosomal RNA genes. All coding genes are encoded on the heavy strand. Compared with the typical gene content of animal mitochondrial genomes, atp8 and trnSer(UCN) genes are missing in the mitochondrial genome. The complete mitochondrial genome contains 26 non-coding regions (1598 bp), one major non-coding region consists of 1046 bp, in which 4.9 tandem repeat sequences (AAA GAG TTT ACC CTT TGT CCT AGT ACA CCT CCT GAA GCC CCA AGA GAT TAC CCT TGG GAT TCG GAG GTG TAC TGG TCT AGG GGG TGC GGG AAA CCC AAC, 99 bp in length) was observed.
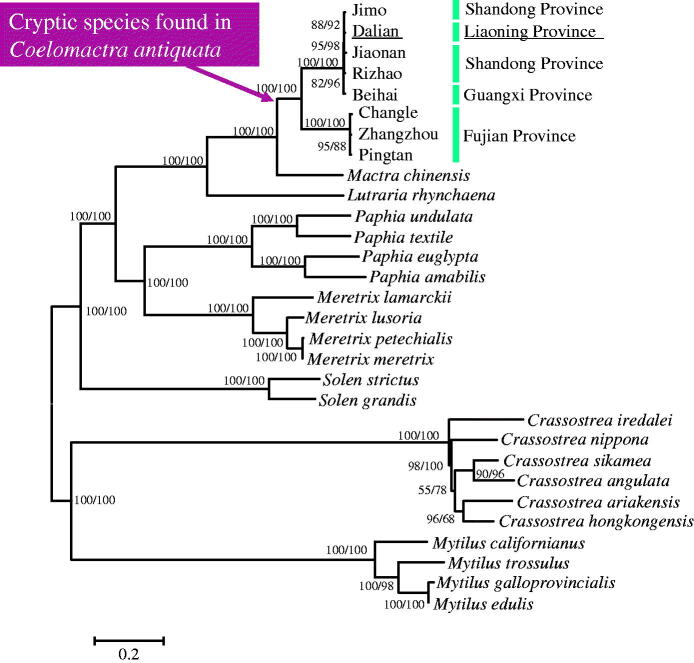
Topologies constructed based on neighbor-joining (NJ) and maximum-likelihood (ML) methods of 12 PCGs (nucleotide sequences) are identical (Figure 1). The phylogenetic tree showed that Liaoning population was clustered into one clade with Shandong (Rizhao, Jiaonan and Jimo) and Guangxi (Beihai) populations, meanwhile all of them are far from the Fujian populations (Pingtan, Zhangzhou and Changle). So, Liaoning, Shandong and Guangxi populations have a close relationship. Actually, Fujian is located between Liaoning, Shandong and Guangxi. So, the result challenges the previously assumed relevance between geographic distance and genetic distance. The genetic differences among different species or populations were explored based on mitochondrial genomic sequences. The genetic distance between Mytilus galloprovincialis and Mytilus trossulus is 0.160. However, the genetic distance of Liaoning and Fujian (Changle, Zhangzhou and Pingtan) C. antiquata is ranging between 0.176 and 0.177, which is larger than that of two Mytilus species. Therefore, the genetic difference of Liaoning and Fujian populations (Changle, Zhangzhou and Pingtan) has reached interspecies level.
Figure 1.
Phylogenetic trees constructed from neighbor-joining (NJ) and maximum-likelihood (ML) methods of 12 PCGs (nucleotide sequences).
The numbers at the nodes indicate the support values from NJ and ML methods.
GenBank accession numbers: Coelomactra antiquata (Jimo: KC503288, Dalian: KR362246, Rizhao: JN692486, Beihai: KR362245, Jiaonan: KC503289, Changle: KC503290, Zhangzhou: NC_021375, Pingtan: KC503291), Mactra chinensis: NC_025510, Lutraria rhynchaena: NC_023384, Solen grandis: NC_016665, Solen strictus: NC_017616, Paphia undulata: NC_016891, Paphia textile: NC_016890, Paphia euglypta: NC_014579, Paphia amabilis: NC_016889, Mytilus galloprovincialis: NC_006886, Mytilus edulis: NC_006161, Mytilus trossulus: NC_007687, Mytilus californianus: NC_015993, Meretrix petechialis: NC_012767, Meretrix meretrix: NC_013188, Meretrix lusoria: NC_014809, Meretrix lamarckii: NC_016174, Crassostrea sikamea: NC_012649, Crassostrea nippona: NC_015248, Crassostrea iredalei: NC_013997, Crassostrea hongkongensis: NC_011518, Crassostrea ariakensis: NC_012650, Crassostrea angulata: NC_012648.
Acknowledgments
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This work was sponsored by the Natural Science Foundation of the Jiangsu Higher Education Institutions (15KJA170001), Jiangsu Natural Science Funds (BK20131210), Open-end Fund of Jiangsu Key Laboratory of Marine Biotechnology (2011HS009 and 2014HS003), projects funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions, Huaihai Institute of Technology Natural Science Fund (Z2014019), Open-end Fund of Jiangsu Institute of Marine Resources (JSIMR201411), Project supported by The Jiangsu Provincial Platform for Conservation and Utilization of Agricultural Germplasm, Qing Lan Project, Jiangsu 333 and Lianyungang 521 Talent Projects.
References
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I.. 2007. Cryptic species as a window on diversity and conservation. Trends Ecol Evol (Amst.). 22:148–155. [DOI] [PubMed] [Google Scholar]
- Burger TD, Shao RF, Barker SC.. 2014. Phylogenetic analysis of mitochondrial genome sequences indicates that the cattle tick, Rhipicephalus (Boophilus) microplus, contains a cryptic species. Mol Phylogenet E. 76:241–253. [DOI] [PubMed] [Google Scholar]
- Gasser RB, Jabbar A, Mohandas N, Schnyder M, Deplazes P, Littlewood DTJ, Jex AR.. 2012. Mitochondrial genome of Angiostrongylus vasorum: comparison with congeners and implications for studying the population genetics and epidemiology of this parasite . Infect Genet Evol. 12:1884–1891. [DOI] [PubMed] [Google Scholar]
- Griggio F, Voskoboynik A, Iannelli F, Justy F, Tilak MK, Turon X, Pesole G, Douzery EJP, Mastrototaro F, Gissi C.. 2014. Ascidian mitogenomics: comparison of evolutionary rates in closely related taxa provides evidence of ongoing speciation events. Genome Biol E. 6:591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannelli F, Pesole G, Sordino P, Gissi C.. 2007. Mitogenomics reveals two cryptic species in Ciona intestinalis. Trends Genet. 23:419–422. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Li C, Morris M, Conn JE, Lima JB, Povoa MM, Wilkerson RC.. 2011. Analysis of the evolutionary forces shaping mitochondrial genomes of a Neotropical malaria vector complex. Mol Phylogenet Evol. 58:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhu JX, Sun HL, Fang JG, Gao RC, Dong SL.. 2006. The clam, Xishi tongue Coelomactra antiquata (Spengler), a promising new candidate for aquaculture in China. Aquaculture. 255:402–409. [Google Scholar]
- Meng X, Shen X, Zhao N, Tian M, Liang M, Hao J, Cheng H, Yan BL, Dong Z, Zhu X.. 2013. Mitogenomics reveals two subspecies in Coelomactra antiquata (Mollusca: Bivalvia). Mitochondrial DNA. 24:102–104. [DOI] [PubMed] [Google Scholar]
- Meng X, Zhao N, Shen X, Hao J, Liang M, Zhu X, Cheng H, Yan B, Liu Z.. 2012. Complete mitochondrial genome of Coelomactra antiquata (Mollusca: Bivalvia): the first representative from the family Mactridae with novel gene order and unusual tandem repeats. Comp Biochem Physiol D. 7:175–179. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM.. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierwater B, Stadler P, Desalle R, Podsiadlowski L.. 2013. Mitogenomics and metazoan evolution . Mol Phylogenet Evol. 69:311–312. [DOI] [PubMed] [Google Scholar]
- Shen X, Li X, Sha ZL, Yan BL, Xu QH.. 2012. Complete mitochondrial genome of the Japanese snapping shrimp Alpheus japonicus (Crustacea: Decapoda: Caridea): Gene rearrangement and phylogeny within Caridea. Sci China-Life Sci. 55:591–598. [DOI] [PubMed] [Google Scholar]
- Shen X, Meng XP, Chu KH, Zhao NN, Tian M, Liang M, Hao J.. 2014. Comparative mitogenomic analysis reveals cryptic species: a case study in Mactridae (Mollusca: Bivalvia). Comp. Biochem Physiol D. 12:1–9. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Kong L, Li Q.. 2013. Mitogenome evidence for the existence of cryptic species in Coelomactra antiquata. Genes Genomics. 35:693–701. [Google Scholar]