Abstract
Sophora flavescens, with high medicinal value, is a traditional Chinese medical plant wildly distributed in China. In this study, the complete chloroplast (cp) genome of Sophora flavescens was determined through Illumina sequencing method. The complete chloroplast genome of S. flavescens was 154,378 bp in length and contained a pair of IR regions (28,876 bp) separated by a small single copy region (18,110 bp) and a large single copy region (84,516 bp). The cp genome of S. flavescens encoded 130 genes including 84 protein-coding genes, 37 tRNA genes and eight ribosomal RNA genes. The overall GC content of S. flavescens cp genome is 36.6%. By phylogenetic analysis using ML method, S. flavescens showed the closest relationship with Sophora alopecuroides.
Keywords: Sophora flavescens, chloroplast genome, Illumina sequencing, phylogenetic analysis
The shrubby sophora (Sophora flavescens) is a species of plant in the genus Sophora, which belongs to the Fabaceae family. It is a traditional Chinese medicine that has been used for anti-tumour, viral hepatitis, enteritis, viral myocarditis, arrhythmia, and skin diseases (Sun et al. 2012). Due to its important medicinal value, lots of scientific researches were reported to clarify corresponding functions of secondary metabolites in the root of Sophora flavescens. While the DNA information derived from genome and organelle genomes of S. flavescens is still limited. Here, we assembled and analyzed the chloroplast genome of S. flavescens based on the next-generation sequencing method. Our aim was to retrieve valuable cp molecular markers, indels and SSRs by comparative analyses with other valuable Sophora species.
Plant materials of Sophora flavescens Ait. sequenced in this study were acquired from medical plants garden in Guiyang University of Traditional Chinese Medicine. Total genomic DNA for genome sequencing was extracted exclusively from fresh young leaves using the cetyltrimethylammonium bromide (CTAB) method and was stored at -20 °C in the Key Laboratory of Miao Medicine, Guiyang University of Traditional Chinese Medicine. For high-throughput sequencing (NGS), the paired-end library from DNA extracts was prepared with a NEBNext Library building kits, following manufacturer’s protocol. Then, the library was sequenced on an Illumina HiSeq2500 platform. After reads quality filtration, the clean reads were assembled by SPAdes 3.6.1 (Bankevich et al. 2012) with default settings. We used the chloroplast genome of Sophora alopecuroides (MF156140.1) as a reference sequence to align the contigs and identify gaps. To fill the gap, Price (Ruby et al. 2013) and MITObim v1.8 (Hahn et al. 2013) were applied and Bandage (Wick et al. 2015) was used to identify the borders of the IR, LSC, and SSC regions. The complete sequence was primarily annotated by Plann (Huang and Cronk 2015) combined with manual correction. All tRNAs were confirmed using the tRNAscan-SE search server (Lowe et al. 1997). Other protein-coding genes were verified by BLAST search on the NCBI website (http://blast.ncbi.nlm.nih.gov/), and manual correction for start and stop codons was conducted. The circular cp genome map was drawn using OrganellarGenomeDRAW (Lohse et al. 2007). This complete chloroplast genome sequence together with gene annotations were submitted to GenBank under the accession numbers of MH748034.
The chloroplast genome of Sophora flavescens Ait. is a typical quadripartite structure with a length of 154,378 bp. The whole cp genome contains a large single-copy (LSC) region of 84,516 bp, a small single-copy (SSC) region of 18,110 bp, and two inverted repeats (IRs) regions of 28,876 bp (Figure 1). The cp genome possesses 130 genes, including 84 protein-coding genes (78 PCG species), eight ribosomal RNA genes (four rRNA species) and 37 tRNA genes (30 tRNA species). The overall GC content of the cp genome is 36.6%. The genome structure, gene order and GC content are similar to those of Sophora alopecuroides cp genome.
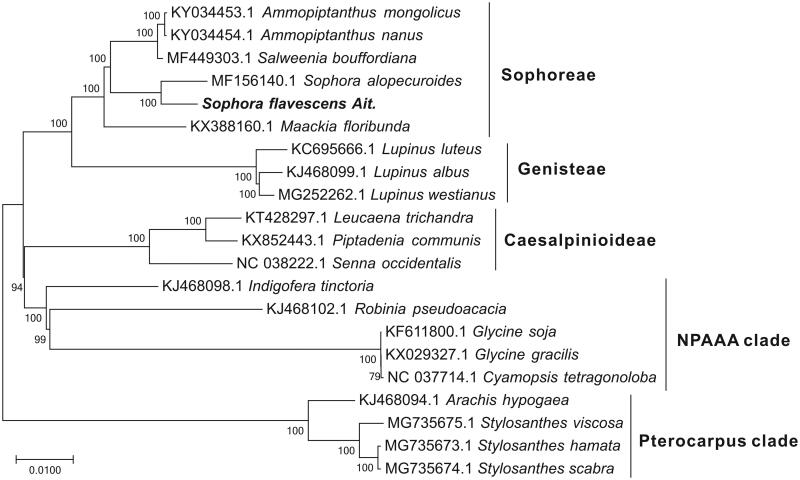
Figure 1.
Phylogenetic tree yielded by ML analysis of 21 higher plant cp genomes. ML consensus tree is shown with bootstrap supports indicated by numbers besides branches.
For phylogenetic analysis assessing the relationship of this plastid, we selected other 20 higher plant cp genomes from Sophoreae (five taxa), Genisteae (three taxa), Caesalpinioideae (three taxa), NPAAA clade (five taxa) and the Pterocarpus clade (four taxa) to construct a genome-wide alignment. We considered plastids of the Pterocarpus clade as the outgroup. The genome-wide alignment of all cp genomes was done by HomBlocks (Bi et al. 2018), resulting in 69,597 positions in total. The whole genome alignment was analyzed by IQ-TREE version 1.6.6 (Nguyen et al. 2014) under the TIM3 + F + R3 model. The tree topology was verified under both 1000 bootstrap and 1000 replicates of SH-aLRT test. As shown in Figure 1, the phylogenetic positions of these 21 cp genomes were successfully resolved with full bootstrap supports across almost all nodes. Sophora flavescens Ait. belongs to the Sophoreae clade as expected, and exhibited the closest relationship with Sophora alopecuroides.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22. [DOI] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genetics. 52:267–274. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Bellare P, Joseph L. DRisi. 2013. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3: Genes| Genomes| Genetics. 3:865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Cao H, Sun L, Dong S, Bian Y, Han J, Zhang L, Ren S, Hu Y, Liu C, et al. 2012. Antitumor activities of kushen: literature review. Evid Based Complement Altern Med. 2012. Article ID 373219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]