Abstract
Conus striatus is a kind of piscivorous cone snail. We have sequenced it by next generation sequencing method. We used de novo assembly and reference mapping methods to assemble mitogenome. The mitochondrial genome is 15,738 bp, containing 13 protein coding genes, 22 transfer RNAs and 2 ribosomal RNAs genes. The overall base composition of C. striatus is 25.9% for A, 16.3% for C, 20.8% for G and 38.6% for T. The phylogenetic analysis was conducted with 18 related species and confirmed the classification status. The complete mitogenome of the C. striatus provides an essential and important DNA molecular data for further phylogeography and evolutionary analysis for cone snail phylogeny.
Keywords: Conus striatus, mitogenome, next-generation sequencing
Conus is a kind of sea snails which are predators and venomous animals. They use their harpoon-like radular teeth to inject the venom to anesthetize the prey. There are about 600 different cone snails that are classified according to their feeding behavior into piscivorous, molluscivorous and vermivorous species (Le Gall et al. 1999). The cone snail C. striatus (Linnaeus 1758) belongs to a clade of piscivorous snails from the Indo–Pacific. They have been observed to elicit a spastic paralysis upon injection of venom into a fish during prey capture (Kelley et al. 2006).
Specimens of C. striatus (voucher no. 20150207-002) were collected from Penghu, Taiwan (23.564N, 119.693E) and deposited in Marine Toxins Lab., Department of Food Science, National Taiwan Ocean University. The total genomic DNA was extracted from muscle using magnetic bead technique with the KingFisher magnetic processors (ThermoFisher Scientific Inc., Worcester, MA). The raw next-generation sequencing reads generated from MiSeq sequencer (Illumina, San Diego, CA) were de novo assembled and reference mapping was conducted by commercial software (Geneious V9, Auckland, New Zealand) to produce a single circular form of complete mitogenome with about an average 40.8 coverage (2244 out of 6,214,252 reads, 0.036%). The complete mitochondrial genome of C. striatus is 15,738 bp in size (GenBank: KX156937), including 13 protein coding genes, 22 transfer RNA genes and 2 ribosomal RNA genes. The overall base composition of C. striatus is 25.9% for A, 16.3% for C, 20.8% for G and 38.6% for T. The protein coding rRNA and tRNA genes of C. striatus mitogenome were predicted using MITOS (Bernt et al. 2013) and tRNAscan-SE (Schattner et al. 2005).
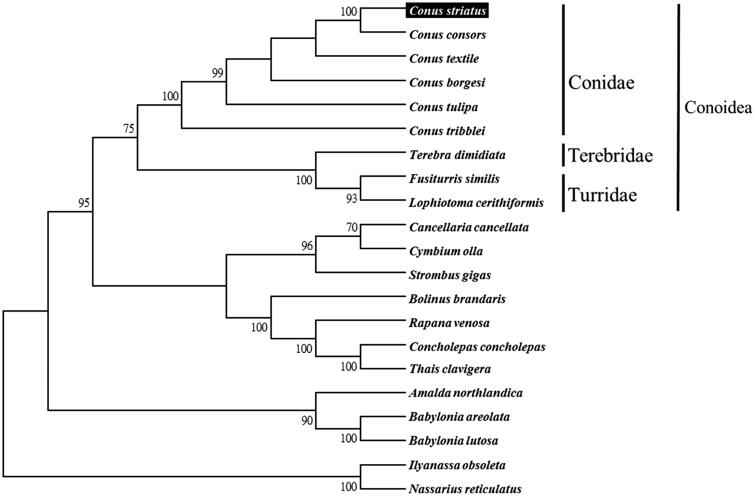
We used MEGA 6 (Tamura et al. 2013) to construct the phylogenetic relationships of the C. striatus and related families by Neighbour-joining method with 1000 bootstrap replicates based on the 13 protein-coding genes and 2 ribosomal RNA genes of the other 20 complete mitochondrial genomes of Neogastropoda marine mollusks, which are reported in Genbank of NCBI database. Bootstrap support values were relatively high, with 13 nodes having values >95% and 8 nodes demonstrating 100% bootstrap support (Figure 1). C. striatus was grouped together with six other conus species from the family Conidae. The lineages of Conidea are strongly supported in this report and are agreed with previous studies (Bouchet et al. 2011; Puillandre et al. 2014).
Figure 1.
Phylogenetic tree generated using the Neighbour-joining method with 1000 bootstrap replicates based on complete mitochondrial genomes. C. striatus (KX156937), C. consors (KF887950), C. textile (DQ862058), C. borgesi (EU827198), C. tulipa (KR006970), C. tribblei (KT199301), Terebra dimidiate (EU827196), Fusiturris similis (EU827197), Lophiotoma cerithiformis (DQ284754), Cancellaria cancellata (EU827195), Cymbium olla (EU827199), Strombus gigas (KM245630), Bolinus brandaris (EU827194), Rapana venosa (KM213962), Concholepas concholepas (JQ446041), Thais clavigera (DQ159954), Amalda northlandica (GU196685), Babylonia areolata (HQ416443), B. lutosa (KF897830), Ilyanassa obsoleta (DQ238598) and Nassarius reticulatus (EU827201).
Acknowledgments
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation . Mol Phylogenet Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Bouchet P, Kantor Y, Sysoev A, Puillandre N.. 2011. A new operational classification of the Conidea (Gastropoda). J Molluscan Stud. 77:273–308. [Google Scholar]
- Kelley WP, Schulz JR, Jakubowski JA, Gilly WF, Sweedler JV.. 2006. Two toxins from Conus striatus that individually induce tetanic paralysis. Biochemistry. 45:14212–14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall F, Favreau P, Richard G, Letourneux Y, Molgo J.. 1999. The strategy used by some piscivorous cone snails to capture their prey: the effects of their venoms on vertebrates and on isolated neuromuscular preparations. Toxicon. 37:985–998. [DOI] [PubMed] [Google Scholar]
- Linnaeus C. 1758. Systema Naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis. Editio decima, reformata. Laurentius Salvius: Holmiae: ii, p. 176. [Google Scholar]
- Puillandre N, Bouchet P, Duda TF, Kauferstein S Jr., Kohn AJ, Olivera BM, Watkins M, Meter C.. 2014. Molecular phylogeny and evolution of the cone snails. Mol Phylogenet E. 78:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM.. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0 . Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]