Abstract
Background
Hepatocellular carcinoma (HCC) lacks effective treatments and has a poor prognosis. Therefore it is needed to develop more effective drug targets. Kinesin family member 11 (KIF11) has been reported to affect the progression of several cancers, and its high expression associates with the prognosis of patients. However, the relevant mechanisms of KIF11 in HCC progression have not been studied.
Method
Through the cancer genome atlas (TCGA) database and immunohistochemical (IHC) staining of patients' specimens, we determined that KIF11 was highly expressed in HCC tissues and associated with prognosis. We established a KIF11 stably depleted hepatoma cell line, through cell-cloning experiments and cell counting kit-8 (CCK-8) assays to detect the effects on proliferation in vitro. The role of KIF11 in promoting cell proliferation was verified in mice.
Result
The expression of KIF11 was negatively correlated with the overall survival (OS) and disease-free survival (DFS) and positively correlated with tumor size of HCC patients. KIF11 depletion inhibits cell proliferation and tumor growth in vitro and in vivo. Conclusion. KIF11 can be used as a positive correlation marker for HCC prognosis and served as a potential therapeutic target.
1. Introduction
Hepatocellular carcinoma (HCC) is the fifth most frequently diagnosed cancer and the second most common cause of cancer death worldwide, especially in South Africa and East Asia [1–6]. In 2010, new cases of HCC were up to 24,120. Unfortunately, the number of HCC patients has been rising, increasing to 42,030 new cases in 2012 in the U.S. Most patients with HCC were diagnosed at the advanced stage, and a minority patients were diagnosed at an early stage [2, 7, 8]. So, the prognosis of HCC patients was still poor. The mortality rate of HCC has also obviously increased, with 18,910 deaths in 2010 and 31,780 deaths in 2019, accounting for 5.2% of all cancer deaths in 2019 [9, 10]. Hepatocellular carcinoma (HCC) accounts for 90% of primary hepatic malignancies and is the most common form of primary liver cancer [11, 12]. The surgical therapy of HCC is not effective. For HCC patients with high metastasis, chemotherapy is the most common treatment. However, traditional chemotherapy has poor efficacy with obvious side effects [13, 14]. Therefore, we need to develop new molecular biomarkers and targets that indicate prognosis and improve treatment.
Kinesin family member 11 (KIF11, also known as EG5) consists of 1056 amino acids encoding a motor protein that belongs to the kinesin-like protein family. The kinesin superfamily (KIF) is a highly conserved motor protein subdivided into 14 families (Kinesin 1–14A/B) [15–17]. Their domains bind and step across microtubules by transforming the chemical energy of ATP into mechanical forces [18, 19]. KIF proteins play vital roles in cellular functions, including mitosis and intracellular trafficking of vesicles and organelles [18, 20]. Due to the overexpression of KIF proteins, such as KIF11, additional forces are generated during mitosis, leading to premature separation of sister chromatids and unequal chromosome distribution, which may further result in aneuploidy of progeny cells [18]. The genetic instability due to KIF11 defects leads to cancer progression, for example by increasing the development of invasion and metastasis [21]. KIF11 is broadly expressed in lymph node, bone marrow, and other normal tissues. Recent researches reported that KIF11 was overexpressed in several tumors and associated with the poor prognosis, such as prostate cancer [22], gastric cancer [23], non-small cell lung cancer [24], oral cancer [25], meningioma [26], hepatic carcinoma [27], and pancreatic tumor [28].
In this study, firstly, we revealed the expression of KIF11 in hepatocellular carcinoma through analyzing TCGA database and IHC staining of patient specimens. And then, we clarified the relationship between the expression of KIF11 and the prognosis and clinicopathological characteristics of hepatocellular carcinoma. Finally, we use in vivo and in vitro experiments to verify the expression of hepatocellular carcinoma after knockdown KIF11. Our main purpose is to provide new targets for the diagnosis and treatment of HCC.
2. Materials and Methods
2.1. Bioinformatics and Statistical Analysis
To explore the different expressions of KIF11 in HCC and paired/unpaired adjacent normal liver tissues, we searched in TCGA database (https://www.cancer.gov/). For overall survival and disease-free survival analysis, we used the web tool of GEPIA (http://gepia.cancer-pku.cn/), median for group cutoff, 95% confidence interval.
2.2. Cell Lines and Tumor Samples
Hep 3B cells were cultured in Eagle's Minimum Essential Medium (30-2003, ATCC, Manassas, USA), and SNU-475 cells were culture in RPMI-1640 medium (30-2001, ATCC, USA), respectively. All cells were maintained with 10% fetal bovine serum (FBS, BioLegend, Beit HaEmek, Israel) at 37°C with 5% CO2. Liver tumor samples and adjacent normal tissues were collected from patients undergoing surgeries at our hospital. All samples in this research were approved by the patients and Ethics Committee of Hospital.
2.3. Immunohistochemical (IHC) Staining
All tissues were formalin-fixed and paraffin-embedded, and then, the tissues were cut into 4 μm sections. After being deparaffinized and rehydrated, antigen retrieval was performed in citrate buffer, blocked with goat serum. After being washed, the tissues were incubated with primary antibodies (KIF11 1 : 100 dilution, ab61199) overnight at 4°C. Then, the sections were washed with PBS. Then, the sections were incubated with secondary antibody-HRP conjugate at room temperature. Finally, the sections were stained with DAB for 5 min and counterstained with hematoxylin.
2.4. Lentivirus-Mediated shRNA Interference
The sequence of siRNA-KIF11 (sense 5′-CCACGTACCCTTCATCAA-3′) and negative control siRNA (sense 5′-UUCUCCGAGCGUGUCACGUTT-3′) were cloned into the PLKO.1 vector. Then, the lentivirus vectors cotransfected into HEK-293T cells with the packing vector. After 72 hours, the lentivirus was collected from the supernatant and added to target tumor cells. After 3 days, 2 μg/mL puromycin (P8230, Solarbio) was used to screen positive cells.
2.5. Quantitative RT-PCR
Total RNA was extracted using the TRIzol reagent and made into cDNA using reverse transcription kit. Then, quantitative PCR was performed using the SYBR MasterMix (4913850001, Roche, Basel, Switzerland), ABI 7900HT, according to the manufacturer's instructions, and then the difference was calculated using the 2–ΔΔCT. KIF11 primer is as follows: forward 5′-GAA CAATCATTAGCAGCAGAA-3′ and reverse 5′-TCAGTATAGACA CCACAGTTG-3′ [27]. β-Actin primer is as follows: forward primer 5′-TAATCTTCGCCTTAA TACTT-3′ and reverse primer 5′-AGCCTTCATACATCTCAA-3′ [27].
2.6. Western Blot
All proteins from cells or tumors were resolved by SDS-PAGE gel (P1200, Solarbio, Beijing, China) and transferred onto PVDF membranes (LC2002, Thermo, Waltham, US). Then, the membranes were blocked in 5% fat-free milk in TBST for 1 hour at room temperature and then washed, and primary antibodies were incubated at 4°C overnight. After being washed, the PVDF was incubated with horseradish peroxidase-conjugated secondary antibodies. The bands were visualized by ECL reagent (32106, Thermo). Antibodies are as follows: KIF11 (1 : 1000 dilution, ab61199, Abcam, Cambridge, UK); anti-β-actin (1 : 1000 dilution, ab5694, Abcam, Cambridge, UK).
2.7. Colony Formation Assay
Cells were seeded into 6-wells with 2 mL complete medium, containing 500 cells. After 2 weeks of culture in a cell culture incubator, 4% paraformaldehyde was used for the fixation of the cells and then stained with 1% crystal violet. Then, the ImageJ software was used to calculate the number of cell clones.
2.8. CCK-8 Assay
1 × 104 control tumor cells and sh-KIF11 tumor cells with 100 μL complete medium were seeded into 96-wells. After 3 days incubation, cell viability was quantified using CCK-8 reagent (B34302, Bimake, China). Briefly, 10 μL of CCK-8 reagent was added to each well according to the instructions and cultured at 37° C for 3 hours, and the absorbance was measured at 490 nm wavelength using a microplate reader.
2.9. Xenograft Mouse Tumors
A total of ten 6-8-week-old nude-BALB/c mice were randomly divided into sh-KIF11 group and control group. 5 × 106 SNU-475 cells (sh-KIF11 or control cell) embedded into 100 mL Matrigel were injected into the flank of mice. The tumors volume was recorded every 3 days (volume = length × width2 × 0.5). After 30 days, the mice were sacrificed, and the tumors were removed for analysis.
2.10. Statistics
GraphPad 6.0 was used for statistical analysis. All results in this study were represented as mean ± SEM. The Kaplan-Meier (KM) method was used to detect the relationship between the expression and postoperative survival time. The correlations between clinical features and KIF11 expression were analyzed through the χ2 analysis. Student's t-test was used for statistical comparisons. ∗ indicated p < 0.05, which was also considered a statistically significant difference, and also, ∗∗ indicated p < 0.01, and ∗∗∗ indicated p < 0.001.
3. Results
3.1. KIF11 Is Associated with the Poor Prognosis of HCC Patients
To explore the relationship between KIF11 expression and HCC progression, we first analyzed the data in TCGA database, showing that KIF11 expression in HCC tissues was significantly higher than normal liver tissue (Figure 1(a)). Patients with high KIF11 expression had poor overall survival (OS, p = 0.00072) and disease-free survival (DFS, p = 0.00027, Figure 1(b)). We collected the clinical case information from 70 patients with HCC and used IHC staining to detect KIF11 expression in the patient's tissues and found that KIF11 was positively correlated with tumor size (p = 0.016, Table 1, Figures 2(a) and 2(b)). In conclusion, we thought KIF11 was associated with poor prognosis in HCC patients.
Figure 1.
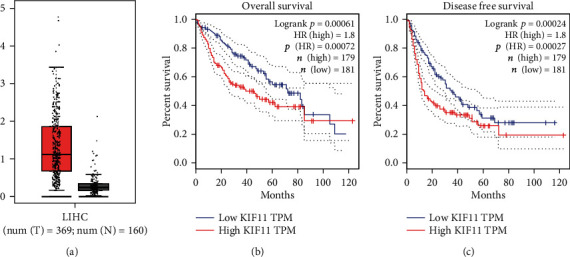
KIF11 expression and the correlation with the prognosis in TCGA database. (a) KIF11 expression levels between HCC tissues (n = 369) and normal liver tissues (n = 160) in TCGA database. (b) Overall survival (OS) and disease-free survival (DFS) of patients with different expression levels of KIF11. The OS and DFS were analyzed using the GEPIA web tool.
Table 1.
Relationships of KIF11 expression and clinicopathological characteristics in 70 patients with HCC.
| Feature | All n = 70 | KIF11 expression | χ 2 | p | |
|---|---|---|---|---|---|
| Low | High | ||||
| n = 36 | n = 34 | ||||
| Age (year) | 0.489 | 0.484 | |||
| <55 | 38 | 21 | 17 | ||
| ≥55 | 32 | 15 | 17 | ||
| Gender | 0.477 | 0.490 | |||
| Male | 40 | 22 | 18 | ||
| Female | 30 | 14 | 16 | ||
| Tumor grade | 2.979 | 0.084 | |||
| Low | 30 | 19 | 11 | ||
| High | 40 | 17 | 23 | ||
| Tumor size | 5.826 | 0.016∗ | |||
| ≥5 cm | 48 | 20 | 28 | ||
| <5 cm | 22 | 16 | 6 | ||
| AFP (ng/mL) | 0.278 | 0.598 | |||
| <50 | 41 | 20 | 21 | ||
| ≥50 | 29 | 16 | 13 | ||
Figure 2.
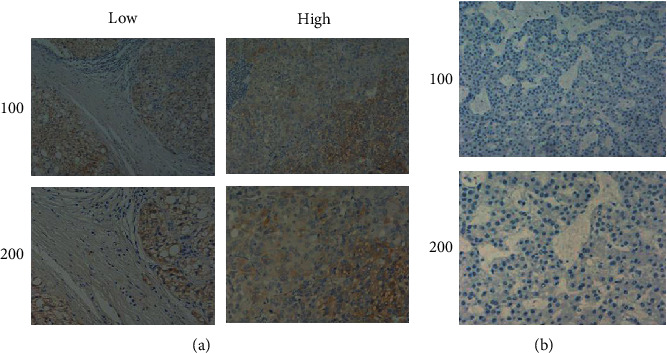
IHC staining for KIF11 in HCC and normal liver tissues. (a) KIF11 was high expression in HCC tissues compared with normal liver tissues (b) and mainly expressed in the cytoplasm.
3.2. Establish Stable Cell Lines of Knockdown KIF11 Using shRNA Plasmids
To further investigate the regulatory mechanism and effects of KIF11 on HCC cells, we selected two HCC cell lines, Hep3B and SNU-475, using lentivirus-mediated shRNA to interfere with KIF11 expression. A stably KIF11-depleted cell line was established by screening for puromycin to study the mechanism by which KIF11 affected tumor progression. Knockdown efficiency was verified at mRNA (Figure 3(a)) and protein levels (Figure 3(b)) using QPCR and western blot, respectively.
Figure 3.
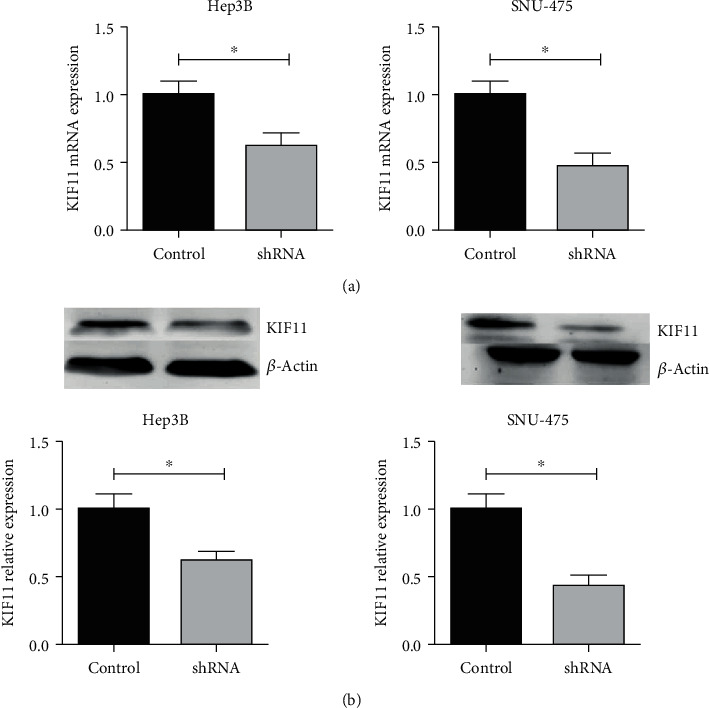
Lentiviral-mediated shRNA interference with KIF11 expression. After adding virus-mediated shRNA for 3 days, the total mRNAs and proteins of the cells were extracted, and the interference effect was detected by qPCR (a) and western blot (b). Adding puromycin to cells for screening positive cells.
3.3. Interfering with KIF11 Inhibits Cell Proliferation in Hep3B and SNU-475 Cells
Then, we investigated whether KIF11 modulated cancer cell proliferation in HCC cells. The colony formation assays were performed to detect cell proliferation with normal cancer cells and KIF11 depletion cells, suggesting that KIF11 depletion suppressed cell colony formation in Hep3B and SNU-475 cell lines (Figure 4(a)). Similar results were shown in CCK-8 assays (Figure 4(b)). Ki-67 and PCNA are two proliferation-related proteins, and their expression levels can reflect cell proliferation degree. Western blot assays showed that Ki-67 and PCNA expression was decreased after knockdown of KIF11 (Figures 4(c) and 4(d)).
Figure 4.
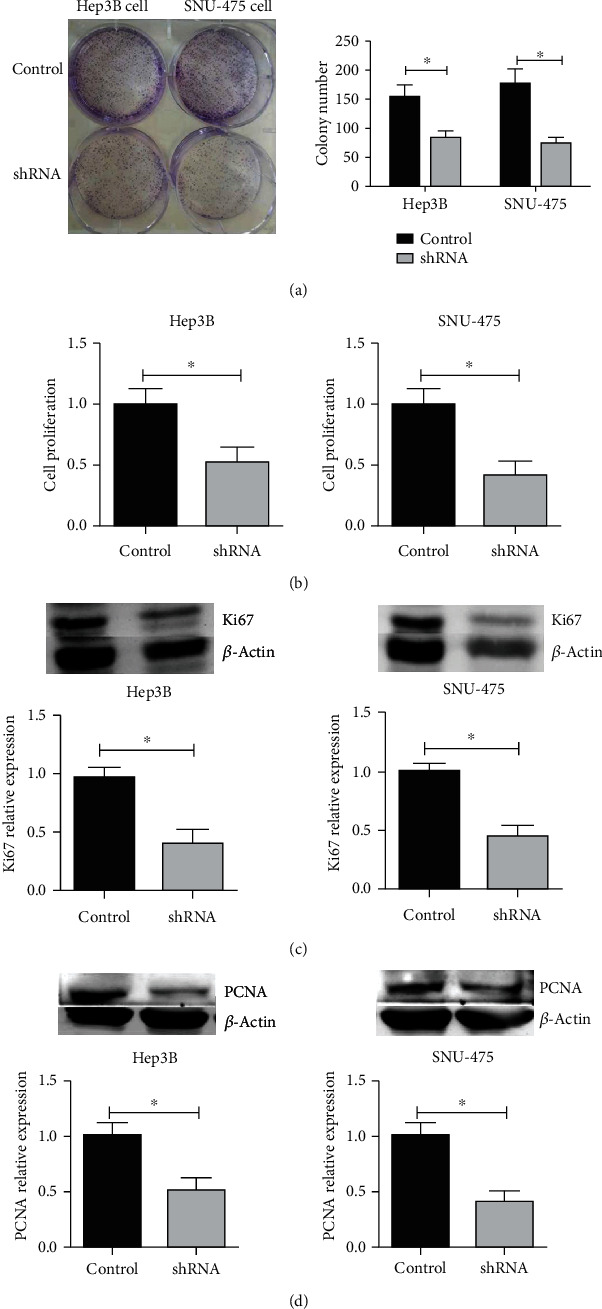
Interfering with KIF11 inhibits the proliferation of Hep3B and SNU-475 cells. (a) The cell colony formation assays and CCK-8 assay (b) were performed to detect cell proliferation after knockdown KIF11 in Hep3B and SNU-475 cells. Western blot showed that after knocking down KIF11, Ki-67, and PCNA expressions were decreased (c, d).
3.4. Knockdown KIF11 Decrease HCC Proliferation and Tumor Growth In Vivo
To confirm the functions of KIF11 in vivo, 5 × 106 SNU-475 cells embedded into Matrigel were injected in the flank of 6-8-week-old nude-BALBc mice. The tumor volume reached a level of macroscopicity after 15 days of implantation. The size of the tumor volume was measured every 3 days, and after 2 addition weeks, the control tumor volume reached approximately 200 mm3, whereas sh-KIF11 only reached 110 mm3 (Figure 5(a)). To further characterize the control cells and sh-KIF11 cells, the KIF11 and proliferation marker, Ki-67, were detected using western blot in tumors, showing that the expression of Ki-67 and KIF11 were lower than control cells (Figures 5(b) and 5(c)). In short, knockdown KIF11 also resulted in reduced cell proliferation in vivo.
Figure 5.
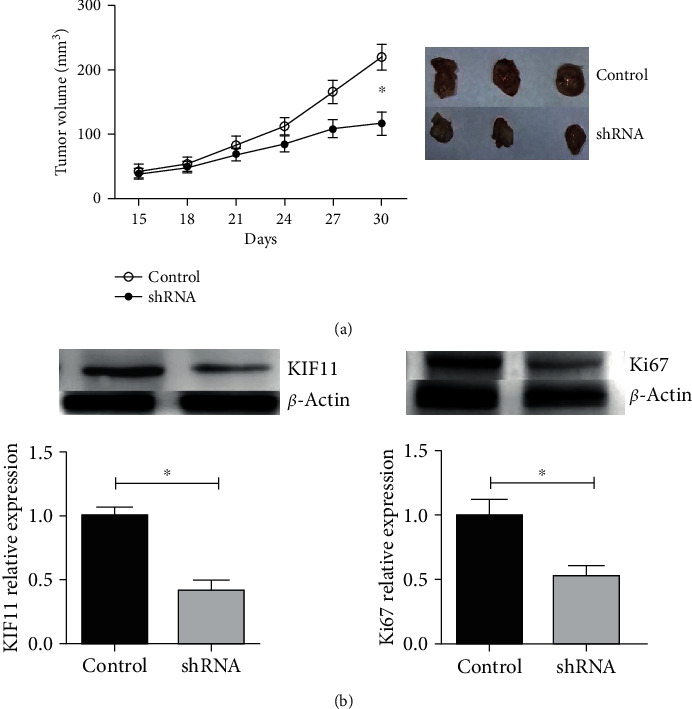
Knockdown of KIF11 decreases HCC proliferation in vivo. (a) After tumor inoculation, the volume of the tumor was observed and recorded every 3 days. After 30 days, the mice were sacrificed, and the tumors were taken out. (b) The protein expression of KIF11 and Ki-67 were decreased in tumor tissues from the KIF11 depletion group.
4. Discussion
HCC is a devastating disease with a poor prognosis in all major malignancies. There is an urgent need to identify molecular markers associated with the pathogenesis of the disease for more timely and effective diagnosis and treatment. Importantly, we found KIF11 is a highly expressed protein by searching the TCGA database and IHC stain of clinical HCC samples. In addition, we also uncovered a strong correlation between KIF11 expression and clinical pathological parameters, such as OS, DFS, tumor size, and malignancy of HCC. Liu et al. previously evaluated mRNA and protein expression levels of KIF11 in 26 freshly frozen HCC tissues and normal samples, which showed that mRNA and protein of KIF11 were significantly higher than matched normal live tissues. Moreover, the KIF11 expression level was strongly associated with TNM stage and liver cirrhosis, and Cox regression analysis showed that KIF11 was an independent factor for overall survival [27]. IHC stain showed that KIF11 protein was mainly expressed in cell cytoplasm, consistent with previous studies [29]. Due to poor patient compliance and since we have not established a complete follow-up system, we only did a single variable analysis, and we have not done a follow-up for survival analysis, etc. We strive to make multifactor analysis in future research to make the results more diversified and more credible.
In other types of cancers, KIF11 also acts as an oncogene, and knockdown of KIF11 inhibits tumor growth [29–31]. Our experiments implicated knockdown of KIF11 by lentiviral-mediated shRNA inhibits tumor cell growth in vitro and in vivo.
As we know, KIF11 plays an essential role in the formation and maintenance of bipolar spindles during mitosis as a microtubule-based motor. In addition to its role in mitosis, more studies have shown other biological functions. In the immune system, KIF11 acts with Tat to promote apoptosis in CD4+ cells [32]. In pancreatic cancer cells, Liu et al. showed that KIF11 promoted cell proliferation dependence ATPase, and ATPase-dead mutant inhibited cell proliferation [28]. Higher expression of KIF11 disturbed bipolar spindle formation, triggering multipole spindle assembly and weakening chromosome segregation. Additionally, the overexpression of KIF11 causes genomic instability in cells and prompts abnormal proliferation in mice [33].
Research and development of small molecules targeting KIF11 are undergoing clinical trials [34]. We found that KIF11 is overexpressed in HCC tissues, and KIF11 shRNA significantly inhibits proliferation and tumor formation of HCC cells, suggesting that KIF11 may serve as an effective target for the treatment of HCC.
In this study, we showed that KIF11 was highly expressed in HCC tissues and associated with the prognosis. Our results in the present study have uncovered underlying mechanisms how the expression of KIF11 is involved in the progression of HCC. The exact role of KIF11 in the development of liver cancer needs to be clarified in further research.
5. Conclusions
In short, we firstly showed that elevated KIF11 expression was significantly correlated with the poor clinical outcome in HCC. Then, our in vitro experiments proved that interfering with KIF11 inhibits cell proliferation in Hep3B and SNU-475 cells. Similarly, knockdown KIF11 inhibited the proliferation of HCC cells and tumor growth in vivo. Due to limited energy and funds, we will conduct external validation to verify the above conclusion in future research. And we speculate that KIF11 may be a potential target for the treatment of HCC in the future.
Abbreviations
- KIF11:
Kinesin family member 11
- IHC:
Immunohistochemistry
- DAB:
3,3-Diaminobenzidin
- HRP:
Horseradish peroxidase
- PCNA:
Proliferating cell nuclear antigen
- PBS:
Phosphate-buffered saline
- PAGE:
Polyacrylamide gel electrophoresis
- QRT-PCR:
Quantificational real-time polymerase chain reaction
- shRNA:
Short hairpin RNA.
Data Availability
The dataset supporting the conclusions of this article is included within the article.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of human specimens and animals were followed. In our hospital, we have achieved the informed consents of patients before the surgeries in the surgical informed consents and agreement. We cannot provide these materials because of the Chinese language and privacy protection. The animal study was carried out in accordance with the guidelines approved by the Animal Experimentation Ethics Committee of Department of Pathology in Tianjin First Center Hospital. The protocol was approved by the Committee, all surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering.
Consent
All of the authors have agreed to publish this article in your journal if it is accepted.
Conflicts of Interest
The authors declare that they have no competing interests.
Authors' Contributions
Zhan-Dong Hu, Ying Jiang, and Li-Li Zhai carried out the experiment of molecular biology and drafted the manuscript. Hong-Mei Sun and Zhi-Qi Yin carried out the animal experiment. Zhan-Dong Hu and Jing-Wen Wang participated in the design of the study and performed the statistical analysis. Jun Yan conceived of the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in Globocan 2012. International Journal of Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Haider C., Hnat J., Wagner R., et al. Transforming growth factor-β and axl induce cxcl5 and neutrophil recruitment in hepatocellular carcinoma. Hepatology. 2019;69(1):222–236. doi: 10.1002/hep.30166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA: A Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Kim E., Kim D., Lee J.-S., et al. Capicua suppresses hepatocellular carcinoma progression by controlling the etv4–mmp1 axis. Hepatology. 2018;67(6):2287–2301. doi: 10.1002/hep.29738. [DOI] [PubMed] [Google Scholar]
- 5.Wang H., Zhang C. Z., Lu S.-X., et al. A coiled-coil domain containing 50 splice variant is modulated by serine/arginine-rich splicing factor 3 and promotes hepatocellular carcinoma in mice by the ras signaling pathway. Hepatology. 2019;69(1):179–195. doi: 10.1002/hep.30147. [DOI] [PubMed] [Google Scholar]
- 6.Mittal S., El-Serag H. B. Epidemiology of hcc: Consider the population. Journal of Clinical Gastroenterology. 2013;47:S2–S6. doi: 10.1097/mcg.0b013e3182872f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan J.-H., Yang F., Wang F., et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 8.L’Hermitte A., Pham S., Cadoux M., et al. Lect2 controls inflammatory monocytes to constrain the growth and progression of hepatocellular carcinoma. Hepatology. 2019;69(1):160–178. doi: 10.1002/hep.30140. [DOI] [PubMed] [Google Scholar]
- 9.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 10.Jemal A., Siegel R., Xu J., Ward E. Cancer statistics, 2010. CA: A Cancer Journal for Clinicians. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 11.Venook A. P., Papandreou C., Furuse J., de Guevara L. L. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. The Oncologist. 2010;15(Supplement 4):5–13. doi: 10.1634/theoncologist.2010-s4-05. [DOI] [PubMed] [Google Scholar]
- 12.Bosch F. X., Ribes J., Cléries R., Díaz M. Epidemiology of hepatocellular carcinoma. Clinics in Liver Disease. 2005;9(2):191–211. doi: 10.1016/j.cld.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Rampone B., Schiavone B., Martino A., Viviano C., Confuorto G. Current management strategy of hepatocellular carcinoma. World Journal of Gastroenterology: WJG. 2009;15(26, article 3210) doi: 10.3748/wjg.15.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S., Hoffmann K., Schemmer P. Treatment of hepatocellular carcinoma: a systematic review. Liver Cancer. 2012;1(3-4):144–158. doi: 10.1159/000343828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miki H., Setou M., RIKEN GER Group, GSL Members, Hirokawa N. Kinesin superfamily proteins (kifs) in the mouse transcriptome. Genome Research. 2003;13(6b):1455–1465. doi: 10.1101/gr.984503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirokawa N., Tanaka Y. Kinesin superfamily proteins (kifs): various functions and their relevance for important phenomena in life and diseases. Experimental Cell Research. 2015;334(1):16–25. doi: 10.1016/j.yexcr.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence C. J., Dawe R. K., Christie K. R., et al. A standardized kinesin nomenclature. The Journal of Cell Biology. 2004;167(1):19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wordeman L. How kinesin motor proteins drive mitotic spindle function: lessons from molecular assays; proceedings of the Seminars in cell & developmental biology. Elsevier; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vale R. D., Reese T. S., Sheetz M. P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42(1):39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279(5350):519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 21.Liu X., Gong H., Huang K. Oncogenic role of kinesin proteins and targeting kinesin therapy. Cancer Science. 2013;104(6):651–656. doi: 10.1111/cas.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wissing M. D., De Morrée E. S., Dezentjé V. O., et al. Nuclear eg5 (kinesin spindle protein) expression predicts docetaxel response and prostate cancer aggressiveness. Oncotarget. 2014;5(17, article 7357) doi: 10.18632/oncotarget.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan G.-R., Zou F.-Y., Dang B.-L., et al. Genistein-induced mitotic arrest of gastric cancer cells by downregulating KIF 20 a, a proteomics study. Proteomics. 2012;12(14):2391–2399. doi: 10.1002/pmic.201100652. [DOI] [PubMed] [Google Scholar]
- 24.Saijo T., Ishii G., Ochiai A., et al. Eg5 expression is closely correlated with the response of advanced non-small cell lung cancer to antimitotic agents combined with platinum chemotherapy. Lung Cancer. 2006;54(2):217–225. doi: 10.1016/j.lungcan.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 25.Daigo K., Takano A., Manh T., et al. Characterization of kif11 as a novel prognostic biomarker and therapeutic target for oral cancer. International Journal of Oncology. 2018;52(1):155–165. doi: 10.3892/ijo.2017.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J., Rapp M., Jungk W., et al. Identification of kif11 as a novel target in meningioma. Cancers. 2019;11(4):p. 545. doi: 10.3390/cancers11040545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C., Zhou N., Li J., Kong J., Guan X., Wang X. Eg5 overexpression is predictive of poor prognosis in hepatocellular carcinoma patients. Disease Markers. 2017;2017:9. doi: 10.1155/2017/2176460.2176460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M., Wang X., Yang Y., et al. Ectopic expression of the microtubule-dependent motor protein eg5 promotes pancreatic tumourigenesis. The Journal of Pathology. 2010;221(2):221–228. doi: 10.1002/path.2706. [DOI] [PubMed] [Google Scholar]
- 29.Lu M., Zhu H., Wang X., et al. The prognostic role of eg5 expression in laryngeal squamous cell carcinoma. Pathology. 2016;48(3):214–218. doi: 10.1016/j.pathol.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Sun D., Lu J., Ding K., et al. The expression of eg5 predicts a poor outcome for patients with renal cell carcinoma. Medical Oncology. 2013;30(1):p. 476. doi: 10.1007/s12032-013-0476-0. [DOI] [PubMed] [Google Scholar]
- 31.Ding S., Xing N., Lu J., et al. Overexpression of eg5 predicts unfavorable prognosis in non-muscle invasive bladder urothelial carcinoma. International Journal of Urology. 2011;18(6):432–438. doi: 10.1111/j.1442-2042.2011.02751.x. [DOI] [PubMed] [Google Scholar]
- 32.Liu M., Li D., Sun L., et al. Modulation of eg5 activity contributes to mitotic spindle checkpoint activation and tat-mediated apoptosis in cd4-positive t-lymphocytes. The Journal of Pathology. 2014;233(2):138–147. doi: 10.1002/path.4333. [DOI] [PubMed] [Google Scholar]
- 33.Castillo A., Morse H. C., Godfrey V. L., Naeem R., Justice M. J. Overexpression of eg5 causes genomic instability and tumor formation in mice. Cancer Research. 2007;67(21):10138–10147. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- 34.Duhl D. M., Renhowe P. A. Inhibitors of kinesin motor proteins--research and clinical progress. Current Opinion in Drug Discovery & Development. 2005;8(4):431–436. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article.


