Abstract
Purpose
To investigate potential target genes associated with the diabetic condition in mouse cavernous endothelial cells (MCECs) for the treatment of diabetes-induced erectile dysfunction (ED).
Materials and Methods
Mouse cavernous tissue was embedded into Matrigel, and sprouted cells were subcultivated for other studies. To mimic diabetic conditions, MCECs were exposed to normal-glucose (NG, 5 mmoL) or high-glucose (HG, 30 mmoL) conditions for 72 hours. An RNA-sequencing assay was performed to evaluate gene expression profiling, and RT-PCR was used to validate the sequencing data.
Results
We isolated MCECs exposed to the two glucose conditions. MCECs showed well-organized tubes and dynamic migration in the NG condition, whereas tube formation and migration were significantly decreased in the HG condition. RNA-sequencing analysis showed that MCECs had different gene profiles in the NG and HG conditions. Among the significantly changed genes, which we classified into 14 major gene categories, we identified that aging-related (9.22%) and angiogenesis-related (9.06%) genes were changed the most. Thirteen genes from the two gene categories showed consistent changes on the RNA-sequencing assay, and these findings were validated by RT-PCR.
Conclusions
Our gene expression profiling studies showed that Cyp1a1, Gclm, Igfbp5, Nqo1, Il6, Cxcl5, Olr1, Ctgf, Hbegf, Serpine1, Cyr61, Angptl4, and Loxl2 may play a critical role in diabetes-induced ED through aging and angiogenesis signaling. Additional research is necessary to help us understand the potential mechanisms by which these genes influence diabetes-induced ED.
Keywords: Diabetes mellitus, Erectile dysfunction, Gene expression, Penis, RNA sequencing
Graphical Abstract
INTRODUCTION
Erectile dysfunction (ED) is a highly age-dependent disorder, and recent epidemiologic studies suggest that approximately 5% to 35% of men aged 40 to 70 years have experienced different levels of ED [1]. The focus on diabetes mellitus (DM) as an important cause of ED has increased recently. About 50% to 75% of men with diabetes have ED [2]. Medications for diabetes, cardiovascular disease, and hypertension can also cause ED [3]. Proposed mechanisms include elevated advanced glycation end products, increased levels of oxygen free radicals, impaired nitric oxide cyclic guanosine monophosphate, and severe endothelial dysfunction or neuropathic damage [4]. The currently available drugs, which are phosphodiesterase-5 (PDE5) inhibitors, have shown poor responsiveness in patients with severe angiopathy. Many studies have shown that severe diabetes-induced endothelial dysfunction leads to insufficient bioavailable NO for PDE5 inhibitors to induce penile erection [5].
The vascular endothelium plays an important role in regulating blood flow and penile erection [6]. Endothelial dysfunction is considered one cause of ED, not only through the regulation of muscle tone but also through the coordination of signals from neural and other sources [7]. Our previous studies established the functional importance of endothelial cells (ECs) in mouse corpus cavernosum tissue and demonstrated their differential distribution [8]. In addition, we successfully isolated mouse and human corpus cavernosum ECs for in vitro cell-based models, which have been widely used in vascular biology research and have given us valuable insight for understanding potential angiogenesis mechanisms in various vascular diseases [9,10]. Many studies have already tried to identify the key genes associated with ED [11]. However, the ED-associated genetic mechanisms in mouse cavernous ECs from diabetes-induced ED mice remain unclear in vitro, and more new molecular targets responsible for diabetes-induced ED are needed.
Gene expression profiling in physiologic and pathologic conditions can give us a foundation for finding the molecular mechanisms of ED and global genetic alteration caused by diabetes. In the present study, we performed an RNA-sequencing assay on mouse cavernous ECs (MCECs) exposed to normal-glucose (NG) and high-glucose (HG) conditions, mimicking diabetes-induced angiopathy, to gain a systematic view of physiologic and pathologic processes and to suggest new therapeutic targets for diabetic-induced ED.
MATERIALS AND METHODS
1. Ethics statement and design of the animal study
In total, 33 adult male C57BL/6J mice (8 weeks old) were used in this study (5 for mouse cavernous EC characterization, 10 for in vitro function study, 8 for RNA-sequencing assay, and 10 for reverse transcription PCR validation). All animal experiments performed in this study were approved by the Institutional Animal Care and Use Committee of Inha University (approval number: 190813-661).
2. Primary culture of mouse cavernous endothelial cells
The primary culture of MCECs was performed as described previously [10]. Briefly, penis tissues were harvested and kept in sterile vials with Hank's balanced salt solution (HBSS; Gibco, Carlsbad, CA, USA). The tissues were then washed twice in phosphate-buffered saline, and the glans, dorsal nerve bundle, and urethra were removed, and only the corpus cavernosum tissues were used. The corpus cavernosum tissues were cut into small pieces and covered with Matrigel on 60-mm cell culture dishes at 37℃ for 10 minutes in a 5% CO2 atmosphere. Then, we added 3 mL of complement medium 199 (M199; Gibco) containing 20% fetal bovine serum (FBS), 1% penicillin/streptomycin, 0.5 mg/mL heparin (Sigma-Aldrich, St. Louis, MO, USA), and 5 ng/mL vascular endothelial growth factor (VEGF; R&D Systems Inc., Minneapolis, MN, USA). After 2 weeks of culture until the cells were more than 90% confluent and spread over the bottom of the dish, only sprouting cells were used for subcultivation, which were seeded onto dishes coated with 0.2% gelatin (Sigma-Aldrich). Cells from passages 2 to 4 were used for all experiments.
3. Characterization of primary cultured mouse cavernous endothelial cells
To characterize cell types, the cells were cultured on sterile cover glasses (Marienfeld Laboratory, Lauda-Königshofen, Germany), which were placed on the bottom of 12-well plates and grown until nearly confluent. Then, the cells were stained with antibodies to CD31 (an EC marker; Chemicon, Temecula, CA, USA; 1:50), CD90 (a fibroblast marker; R&D Systems Inc.; 1:50), NG2 chondroitin sulfate proteoglycan (NG2; a pericyte marker, Millipore, San Francisco, CA, USA; 1:50), smooth muscle α-actin (α-SMA, a smooth muscle cell marker; Sigma-Aldrich; 1:100), or DAPI (a nucleus marker; Vector Laboratories Inc., Burlingame, CA, USA) as previously described [10]. Signals were visualized, and digital images were obtained with a confocal fluorescence microscope (K1-Fluo; Nanoscope Systems, Inc., Daejeon, Korea).
4. Establishment of in vitro model that mimics diabetes-induced angiopathy
To mimic diabetes-induced angiopathy conditions, primary cultured MCECs were serum-starved overnight and exposed to the NG (5 mmoL, Sigma-Aldrich) or HG (30 mmoL) condition for 72 hours at 37℃ in a 5% CO2 atmosphere [12].
5. Tube formation assay
An in vitro angiogenesis assay was performed to evaluate the angiogenic ability of the cultured MCECs in the NG and HG conditions. Approximately 100 µL of growth factor-reduced Matrigel (Becton Dickinson, Mountain View, CA, USA) was dispensed into 48-well cell culture plates at 4℃. After the plates were incubated at 37℃ for at least 10 to 15 minutes, the MCECs that were preconditioned under NG and HG conditions for 72 hours were seeded onto the Matrigel at 1×105 cells/well in 300 µL of M199 medium. After 16 hours, images were taken with a phase-contrast microscope, and the number of integrated tubes was counted at a screen magnification of ×40.
6. Cell migration assay
The MCEC migration assay was performed with the SPLScar™Block system (SPL Life Sciences, Pocheon, Korea) on 60-mm culture dishes. The conditioned MCECs (NG and HG condition) were seeded into the 3-well block at >95% confluence, and 5 hours later, the block was removed and the cells were further incubated in M199 medium with 2% FBS for 24 hours. The images were taken with a phase-contrast microscope, and migrated cells were analyzed by measuring the density of the cells that had moved into the frame line.
7. RNA-sequencing assay
For the RNA-sequencing study, MCECs were cultured and treated (n=2 per group) under NG and HG conditions. The RNA-sequencing assay was performed by E-Biogen Inc. (Seoul, Korea). Briefly, total RNA was isolated 72 hours after exposure to the glucose condition using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA quality was assessed by use of an Agilent 2100 Bioanalyzer (Agilent Technologies, Amstelveen, The Netherlands), and RNA quantification was performed by using an ND-2000 Spectrophotometer (Thermo Inc., Wilmington, DE, USA).
8. Library preparation and sequencing
Libraries were prepared from total RNA by using the SMARTer Stranded RNA-Seq Kit (Clontech Laboratories, Inc., Mountain View, CA, USA). The isolation of mRNA was performed by using the Poly(A) RNA Selection Kit (LEXOGEN, Inc., Vienna, Austria). The isolated mRNAs were used for cDNA synthesis and shearing. Indexing was performed with Illumina indices 1–12. The enrichment step was performed by PCR. Subsequently, libraries were checked by using the Agilent 2100 Bioanalyzer (DNA High Sensitivity Kit) to evaluate the mean fragment size. Quantification was performed by using the library quantification kit with a StepOne Real-Time PCR System (Life Technologies, Inc., Carlsbad, CA, USA). High-throughput sequencing was performed as paired-end 100 sequencing using HiSeq 2500 (Illumina, Inc., San Diego, CA, USA).
9. Data analysis
The quality control of raw sequencing data was performed by using FastQC (Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Adapter and low-quality reads (<Q20) were removed by using FASTX_Trimmer (Available at: http://hannonlab.cshl.edu/fastx_toolkit/) and BBMap (Available at: https://sourceforge.net/projects/bbmap/). Then, the trimmed reads were mapped to the reference genome by using TopHat [13]. Gene expression levels were estimated by using read count and fragments per kb per million reads values by BEDTools and Cufflinks [14]. The expression values were normalized with the Quantile normalization method by using EdgeR within R (Available at: https://www.r-project.org). Data mining and graphic visualization were performed with ExDEGA. The RNA-sequencing data have been deposited in the Gene Expression Omnibus database (Available at: www.ncbi.nlm.nih.gov/geo)accession no. GSE146078.
10. Validation of sequencing data by RT-PCR
Total RNA was extracted from cultured cells by using TRIzol (Invitrogen) following the manufacturer's protocols. Reverse transcription was performed with 1 µg of RNA in 20 µL of reaction buffer with oligo dT primer and AccuPower RT Premix (Bioneer Inc., Daejeon, Korea). The primers used in this study are shown in Table 1. The PCR reaction was performed with denaturation at 94℃ for 30 seconds, annealing at 60℃ for 30 seconds, and extension at 72℃ for 1 minute in a DNA Engine Tetrad Peltier Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA). For the analysis of PCR products, 10 µL of each PCR products was electrophoresed on a 1% agarose gel and detected under ultraviolet light. GAPDH was used as an internal control. All phase images and PCR bands from densitometry analysis were measured with an image analyzer system (ImageJ 1.34; National Institutes of Health, Bethesda, MD, USA; http://rsbweb.nih.gov/ij/).
Table 1. Primer list for RT-PCR.
| Gene | Primer sequence | Product size (bp) |
|---|---|---|
| Ctgf | F: CCAGGAAGTAAGGGACACG | 370 |
| R: TAATTTCCCTCCCCGGTTAC | ||
| Hbegf | F: TGTGTTCAAGTAGCCGCAAG | 448 |
| R: GATCCCTGCACTCTGACCAT | ||
| Serpine1 | F: GTAGCACAGGCACTGCAAAA | 417 |
| R: TGAGACCTTTGTGGGGTAGG | ||
| Cyr61 | F: GCACCTCGAGAGAAGGACAC | 332 |
| R: AGTTTTGCTGCAGTCCTCGT | ||
| Angptl4 | F: CTACAGCCTGCAGCTCACTG | 450 |
| R: CTTTGTCCACAAGACGCAGA | ||
| Loxl2 | F: GGATGACCTTGGACCTCTGA | 453 |
| R: AGGCCTGGTACCTGAGGTTT | ||
| Cyp1a1 | F: AAGTGCAGATGCGGTCTTCT | 454 |
| R: CCATTTGGGAAGGCTGTTTA | ||
| Gclm | F: TGTTTTGGAATGCACCATGT | 461 |
| R: AGAGCAGTTCTTTCGGGTCA | ||
| Igfbp5 | F: GAGCAACACAAAGGGAGAGC | 390 |
| R: TAGGCAGTTCCTGGCTCAGT | ||
| Nqo1 | F: TAGCCTGTAGCCAGCCCTAA | 401 |
| R: GTCTGCAGCTTCCAGCTTCT | ||
| Il6 | F: CCGGAGAGGAGACTTCACAG | 421 |
| R: GGAAATTGGGGTAGGAAGGA | ||
| Cxcl5 | F: TAGAGCCCCAATCTCCACAC | 439 |
| R: GTGCATTCCGCTTAGCTTTC | ||
| Olr1 | F: TGGCTATGGGAGAATGGAAC | 444 |
| R: GTTGGTTGGGAGACTTTGGA | ||
| GAPDH | F: CCACTGGCGTCTTCACCAC | 501 |
| R: CCTGCTTCACCACCTTCTTG |
11. Statistical analysis
All data were expressed as mean±standard error. Statistical analysis was performed by using Mann–Whitney U tests. All p-values less than 0.05 were considered significant.
RESULTS
1. Isolation and characterization of MCECs
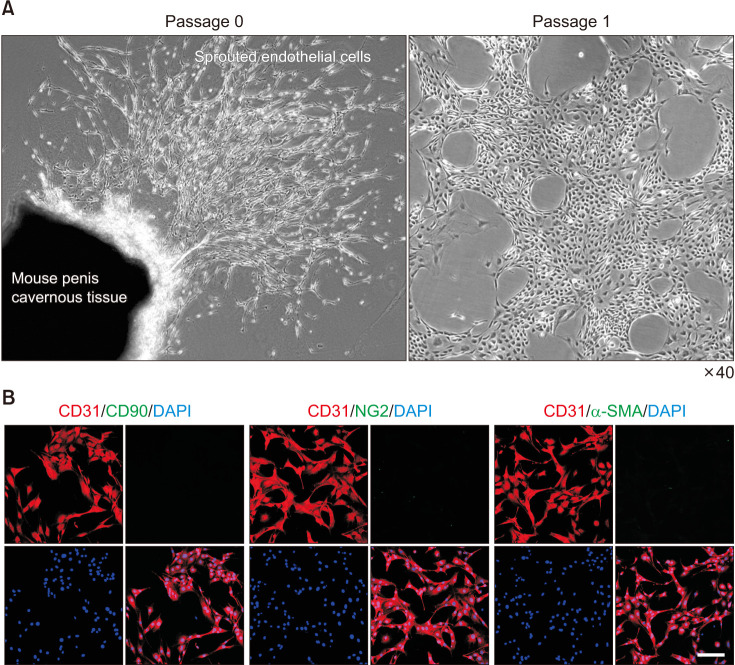
Representative images of sprouted cells from mouse cavernous tissue at passage 0 and passage 1 are shown in Fig. 1A. After the cells spread over the entire bottom (about 2 weeks), only sprouting cells were used for other studies. Immunofluorescent staining of sprouted cells showed high positive staining for CD31 (an EC marker) but not for CD90 (a fibroblast marker), NG2 (a pericytes marker), or α-SMA (a smooth muscle cell marker) (Fig. 1B).
Fig. 1. Isolation and characterization of primary cultured mouse cavernous endothelial cells (MCECs). (A) Representative phage images (screen magnification, ×40) of sprouted cells from mouse penis cavernous tissues at passage 0, and sprouted cells subcultured at passage 1. (B) Immunofluorescent staining of MCECs with antibodies against CD31 (an EC-positive marker), CD90 (a fibroblast marker), NG2 (a pericyte marker), and α-SMA (a smooth muscle cell marker). Nuclei were labeled with the DNA dye DAPI (4,6-diamidino-2-phenylindole). Scale bar indicates 100 µm.
2. Reduced tube formation and migration in MCECs exposed to the high-glucose condition
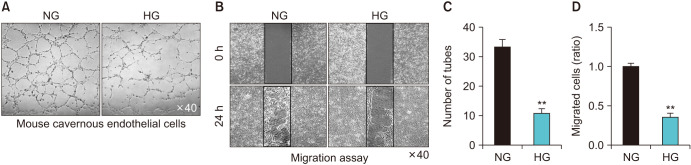
To evaluate whether primary cultured MCECs can form tube-like structures in NG and HG conditions, we performed a tube formation assay on Matrigel in vitro. After 16 hours of incubation, MCECs formed well-organized capillary-like structures in the NG condition, whereas significantly decreased tube formation was detected in MCECs exposed to the HG condition (Fig. 2A, B). In addition, MCEC migration was also reduced under the HG condition (Fig. 2C, D).
Fig. 2. Decreased tube formation and migration in mouse cavernous endothelial cells (MCECs) exposed to the high-glucose (HG) condition. Phase images of MCECs (16 hours, screen magnification, ×40). MCECs were incubated in normal-glucose (NG) and HG conditions for 72 hours. (A) Tube formation assay was performed on Matrigel in 48-well dishes. Scale bar indicates 500 µm. (B) Migration assay in MCECs exposed to NG and HG conditions for same time with tube formation. After 24 hours, the images were taken by microscopy. Scale bar indicates 500 µm. (C) Number of tubes and (D) migrated cells per field. Each bar depicts the mean values (±standard error) from four separate experiments. **p<0.01 compared with the NG group.
3. Overview of the RNA-sequencing data
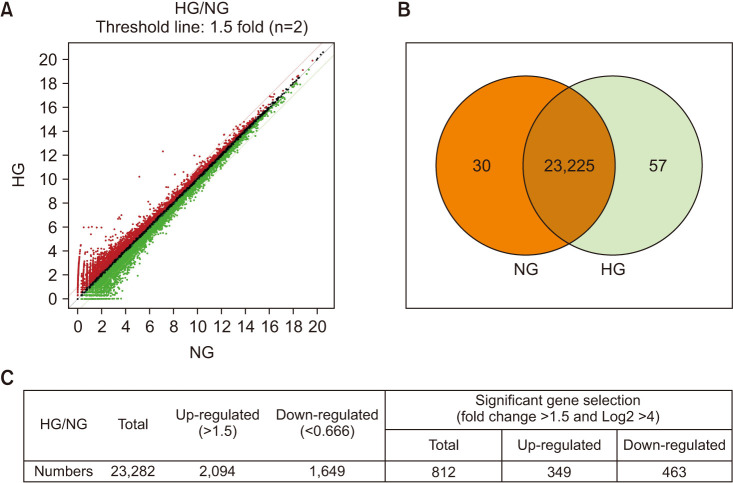
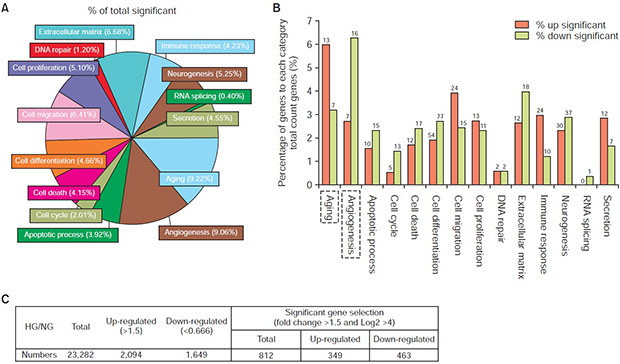
For this study, two gene libraries for the NG group and the HG group (n=2 for each group) were constructed for the RNA-sequencing assay. In total, 23,282 genes were detected in both libraries, whereas 30 and 57 genes were detected in only the NG and HG groups, respectively (Fig. 3A, B). Among all the detected genes, 2,094 genes were up-regulated and 1,649 genes were down-regulated. Differentially expressed gene analysis was performed with three conditions: fold change >1.5, and log2 >4. Consequently, among the 812 identified genes, 349 genes were up-regulated and 463 genes were down-regulated (Fig. 3C).
Fig. 3. Analysis of two libraries of differentially expressed genes. (A) Total gene expression of mouse cavernous endothelial cells in normal-glucose (NG) and high-glucose (HG) conditions. (B) Differentially expressed genes in two libraries. (C) Differentially expressed gene analysis for significantly changed gene selection following the conditions set in advance. Two separate samples for each group were subject to analysis.
4. Gene ontology category analysis of the RNAsequencing data
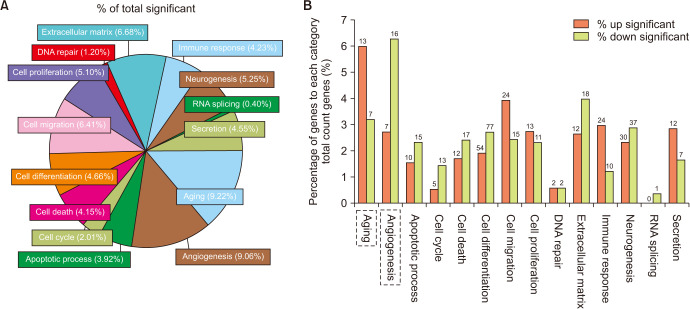
To evaluate the gene ontology (GO) category of the selected genes, a total of 812 identified genes were analyzed in 14 GO categories by use of ExDEGA. In these GO categories, aging (9.22%) and angiogenesis (9.06%) had highest proportions (Fig. 4A). In the former, 13 up-regulated (Table 2) and 7 down-regulated aging-related genes were detected. In addition, 7 up-regulated and 16 down-regulated angiogenesisrelated genes (Table 3) were detected (Fig. 4B).
Fig. 4. The significantly altered genes of the RNA-sequencing data were allocated to gene ontology (GO) categories. (A) The percentage of total significantly changed genes allocated to the top 14 GO categories. (B) The percentage of genes in each category of total counted genes with detailed up-regulated and down-regulated gene numbers. The significantly altered genes were enriched in Aging and Angiogenesis GO categories, as showed with dotted frame.
Table 2. Summary of selected aging-related genes that increased in the high-glucose (HG) group compared with the normal-glucose (NG) group.
| Gene symbol | Fold change | Raw data (RC) | KEGG input | Transcript_id | Description | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HG/NG | NG1 | NG2 | NG3 | NG4 | HG1 | HG2 | HG3 | HG4 | Entrez ID | |||
| Cyp1a1 | 1.525 | 3,387 | 2,152 | 3,387 | 3,387 | 4,382 | 4,455 | 5,903 | 5,714 | 13076 | NM_009992 | Cytochrome P450, family 1, subfamily a, polypeptide 1 |
| Pde4d | 1.545 | 2,775 | 1,986 | 2,775 | 2,774 | 3,960 | 3,917 | 4,605 | 4,985 | 238871 | NM_011056 | Phosphodiesterase 4D, cAMP specific |
| Igfbp2 | 1.573 | 43 | 58 | 43 | 43 | 65 | 83 | 89 | 105 | 16008 | NM_008342 | Insulin-like growth factor binding protein 2 |
| Ifi27l2a | 1.597 | 118 | 46 | 118 | 118 | 145 | 189 | 193 | 144 | 76933 | NM_001281830 | Interferon, alpha-inducible protein 27 like 2A |
| Agtr1a | 1.613 | 39 | 28 | 39 | 39 | 82 | 61 | 48 | 64 | 11607 | NM_177322 | Angiotensin II receptor, type 1a |
| Gclm | 1.620 | 11,802 | 8,054 | 11,802 | 11,802 | 18,786 | 16,625 | 19,134 | 22,199 | 14630 | NM_008129 | Glutamate-cysteine ligase, modifier subunit |
| Igfbp5 | 1.858 | 66,035 | 55,527 | 66,035 | 66,035 | 120,144 | 110,948 | 136,878 | 157,159 | 16011 | NM_010518 | Insulin-like growth factor binding protein 5 |
| Sod2 | 1.869 | 1,742 | 746 | 1,744 | 1,743 | 2,487 | 2,762 | 3,361 | 3,211 | 20656 | NM_013671 | Superoxide dismutase 2, mitochondrial |
| Aldh3a1 | 1.875 | 1,926 | 1,255 | 1,926 | 1,926 | 3,120 | 3,482 | 3,817 | 3,912 | 11670 | NM_001112725 | Aldehyde dehydrogenase family 3, subfamily A1 |
| Nqo1 | 1.934 | 11,826 | 5,971 | 11,826 | 11,826 | 19,154 | 22,142 | 20,839 | 23,020 | 18104 | NM_008706 | NAD(P)H dehydrogenase, quinone 1 |
| Gclc | 2.008 | 12,979 | 7,884 | 12,979 | 12,979 | 22,042 | 24,161 | 27,450 | 28,034 | 14629 | NM_010295 | Glutamate-cysteine ligase, catalytic subunit |
| Il6 | 2.498 | 1,078 | 635 | 1,078 | 1,078 | 2,369 | 2,613 | 2,453 | 2,972 | 16193 | NM_031168 | Interleukin 6 |
| Olr1 | 1.802 | 1,166 | 856 | 1,164 | 1,160 | 2,150 | 1,982 | 2,107 | 2,323 | 108078 | NM_138648 | Oxidized low density lipoprotein (lectin-like) receptor 1 |
Table 3. Summary of selected angiogenesis-related genes that decreased in the high-glucose (HG) group compared with the normal-glucose (NG) group.
| Gene symbol | Fold change | Raw data (RC) | KEGG input | Transcript_id | Description | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HG/NG | NG1 | NG2 | NG3 | NG4 | HG1 | HG2 | HG3 | HG4 | Entrez ID | |||
| Wnt7b | 0.383 | 25 | 8 | 25 | 25 | 6 | 4 | 4 | 18 | 22422 | NM_009528 | Wingless-type MMTV integration site family, member 7B |
| Ctgf | 0.432 | 26,261 | 28,683 | 26,265 | 26,262 | 12,606 | 12,690 | 12,153 | 15,186 | 14219 | NM_010217 | Connective tissue growth factor |
| Ramp1 | 0.523 | 191 | 108 | 191 | 191 | 124 | 72 | 93 | 88 | 51801 | NM_016894 | Receptor (calcitonin) activity modifying protein 1 |
| Nox1 | 0.542 | 47 | 18 | 47 | 47 | 23 | 15 | 23 | 29 | 237038 | NM_172203 | NADPH oxidase 1 |
| Cxcl12 | 0.545 | 732 | 691 | 730 | 731 | 380 | 446 | 410 | 523 | 20315 | NM_013655 | Chemokine (C-X-C motif) ligand 12 |
| Hbegf | 0.564 | 20,479 | 11,774 | 20,479 | 20,479 | 10,103 | 9,102 | 12,227 | 13,205 | 15200 | NM_010415 | Heparin-binding EGF-like growth factor |
| Adm2 | 0.565 | 1,074 | 1,139 | 1,074 | 1,074 | 620 | 666 | 713 | 791 | 223780 | NM_182928 | Adrenomedullin 2 |
| Serpine1 | 0.581 | 88,771 | 67,443 | 88,771 | 88,771 | 50,387 | 52,935 | 52,574 | 56,240 | 18787 | NM_008871 | Serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| Cyr61 | 0.598 | 32,600 | 27,400 | 32,600 | 32,600 | 20,205 | 18,159 | 21,347 | 23,270 | 16007 | NM_010516 | Cysteine rich protein 61 |
| Loxl2 | 0.607 | 27,739 | 17,667 | 27,738 | 27,733 | 14,919 | 14,002 | 18,232 | 19,561 | 94352 | NM_033325 | Lysyl oxidase-like 2 |
| Sox18 | 0.630 | 31 | 24 | 31 | 31 | 17 | 23 | 15 | 24 | 20672 | NM_009236 | SRY (sex determining region Y)-box 18 |
| Angptl4 | 0.637 | 29,720 | 26,036 | 29,720 | 29,720 | 18,692 | 20,439 | 20,586 | 21,674 | 57875 | NM_020581 | Angiopoietin-like 4 |
| Thbs1 | 0.638 | 418,163 | 252,972 | 418,164 | 418,164 | 234,104 | 246,596 | 267,973 | 289,271 | 21825 | NM_011580 | Thrombospondin 1 |
| Shb | 0.647 | 850 | 619 | 850 | 850 | 472 | 425 | 659 | 713 | 230126 | NM_001033306 | Src homology 2 domain-containing transforming protein B |
| Arhgap22 | 0.651 | 3,455 | 2,224 | 3,455 | 3,455 | 2,080 | 1,759 | 2,470 | 2,624 | 239027 | NM_153800 | Rho GTPase activating protein 22 |
| Pgf | 0.657 | 978 | 531 | 978 | 979 | 384 | 529 | 712 | 860 | 18654 | NM_008827 | Placental growth factor |
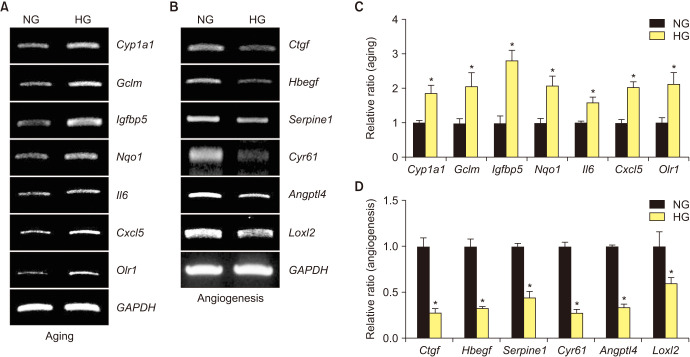
5. Validation of RNA-sequencing results by RT-PCR
To validate the RNA-sequencing results, we selected 13 significantly changed genes, comprising 7 up-regulated aging-related genes and 6 down-regulated angiogenesis-related genes for validation study. After exposure to the NG and HG conditions, total RNA was extracted from MCECs. The RT-PCR results showed that Cyp1a1, Gclm, Igfbp5, Nqo1, Il6, Cxcl5, and Olr1 were increased (Fig. 5A, C), whereas Ctgf, Hbegf, Serpine1, Cyr61, Angptl4, and Loxl2 were decreased in the HG condition (Fig. 5B, D).
Fig. 5. RT-PCR validation of differentially expressed genes in RNA sequencing. (A) Seven aging-related genes were evaluated in mouse cavernous endothelial cells (MCECs) exposed to normal-glucose (NG) and high-glucose (HG) conditions. (B) Six angiogenesis-related genes were evaluated in MCECs exposed to NG and HG conditions. (C, D) Each bar depicts the mean values (±standard error) from three separate experiments. *p<0.05 compared with the NG group.
DISCUSSION
Dysfunction of ECs in corpus cavernosum tissue is one of the most common reasons for the low effect of PDE5 inhibitors in the treatment of diabetes-induced ED [15]. However, most genome studies have focused on corpus cavernosum at a tissue level in vivo [16] and not at a cellular level in vitro. Here, we isolated MCECs and performed an RNA-sequencing assay to investigate potential target genes for ED.
After exposure to the HG condition for 3 days, MCECs showed a decreased ability for tube formation and migration, which provided an ideal model to mimic diabetesinduced angiopathy [9,17]. In addition, many studies have already shown that hyperglycemia promotes endothelial dysfunction through oxidative stress and the AGE pathway [18]. However, there remains no accepted gold criterion for evaluating glucose variability. In this study, MCECs exposed to NG and HG conditions were used to investigate the genes important for diabetes-induced ED, and we found some oxidative stress markers. For example, NQO1, GPX2, SOD2, and SOD3 were significantly increased in the HG condition in MCECs. Consistent with a previous study, we also found that the ratio of collagen to elastin was also significantly increased in the HG compared with the NG condition, which also may be attributed to AGE-induced crosslinking and results in a decreased coronary flow reserve in the diabetic condition [19,20]. These findings suggest that the HG condition is ideal for simulating diabetes-induced MCEC dysfunction.
The RNA-sequencing results and analysis of GO categories detected numerous differentially expressed genes, and many top GO categories were evaluated in this study. The GO analysis showed that the significantly altered genes were enriched in aging and angiogenesis. Although age-related ED is known to be associated with endothelial dysfunction in the penis, the molecular basis of this mechanism remains largely unknown [21]. In addition, previous studies have shown that the expression of many angiogenic mediators, such as angiopoietin (Ang)-1 [22], Ang-4 [23], and VEGF [24], is decreased in the HG condition at the protein level but not at the RNA level and that intracavernous administration of these angiogenic mediators improves erectile function in diabetes-induced ED. However, the most effective molecular mediator in regulating angiogenesis remains unknown; furthermore, ways to overcome the limited regenerative capacity for mature ECs remain unknown [25]. In this study, we used a more global method to evaluate the molecular changes in cavernous EC under varied glucose conditions. From the significantly altered genes, we selected seven aging-related genes that were up-regulated and six angiogenesis-related genes that were down-regulated in the HG condition as the targets for diabetic-induced ED, because ED is an age-dependent disorder and diabetes causes severe endothelial dysfunction. In addition, ED may be an important predictor of cardiovascular disease [26]. These genes may be the key to improving local EC regeneration by reducing the aging of cells and inducing angiogenic mechanisms in diabetes-induced ED. These findings may have clinical implications for early diagnosis of vascular disease caused by aging and angiopathy.
A few studies have been conducted on aging-related genes such as Nqo1, Il6, and Cxcl5 in diabetes-induced ED [25,26,27]. There are, however, no studies on Cyp1a1, Gclm, Olr1, or Igfbp5 in diabetes-induced ED. Igfbp5 acts as a switch to regulate insulin-like growth factor (IGF) signaling [27]; therefore, it seems have high relevance for diabetes-induced ED. Concerning the angiogenesis-related genes, there are no studies on any of the six genes in diabetes-induced ED. However, Calenda et al. [28] showed that Serpine1 may be related to nerve-injury induced ED, and Angptl4 is a predictive marker for diabetic nephropathy [29]. Therefore, Igfbp5, Serpine1, and Angptl4 would be preferred candidates for the mechanism study of diabetes-induced ED.
To our knowledge, this is the first study to perform systematic profiling of gene alterations in MCECs on exposure to NG and HG conditions. However, our study has some limitations. First, the glucose conditions could not completely mimic diabetes-induced ED. Second, we could not perform the gene set enrichment analysis or hierarchical clustering analysis with our RNA-sequencing results. Third, there are several other significantly altered genes that were not validated by RT-PCR.
In this study, we profiled the differentially expressed genes in MCECs under NG and HG conditions. Further protein expression and functional study in a DM mouse model of these validated genes and significantly changed genes, such as Igfbp5, Serpine1, and Angptl4, will be important and helpful for us to understand the detailed mechanisms of aging and angiogenesis in diabetes-induced ED.
CONCLUSIONS
We profiled differentially expressed genes in MCECs under NG and HG conditions. In the analysis of GO categories, we found that aging and angiogenesis had the highest proportion of genes. Seven aging-related genes and six angiogenesis-related genes were significantly changed in the HG condition compared with the NG condition. Our suggested genes may play a critical role in aging and angiogenesis, and additional research is necessary to help us understand the potential mechanisms for diabetes-induced ED.
ACKNOWLEDGMENTS
This research was supported by Inha University Research Grant (Guo Nan Yin).
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
- Research conception and design: Guo Nan Yin and Jiyeon Ock.
- Performing the research and data analysis: Guo Nan Yin, Jiyeon Ock, Min-Ji Choi, Anita Limanjaya, Kang-Moon Song, Kalyan Ghatak, and Mi-Hye Kwon.
- Drafting of the manuscript: Guo Nan Yin and Jiyeon Ock.
- Critical revisions to the manuscript: Ji-Kan Ryu and Jun-Kyu Suh.
References
- 1.Kubin M, Wagner G, Fugl-Meyer AR. Epidemiology of erectile dysfunction. Int J Impot Res. 2003;15:63–71. doi: 10.1038/sj.ijir.3900949. [DOI] [PubMed] [Google Scholar]
- 2.Malavige LS, Levy JC. Erectile dysfunction in diabetes mellitus. J Sex Med. 2009;6:1232–1247. doi: 10.1111/j.1743-6109.2008.01168.x. [DOI] [PubMed] [Google Scholar]
- 3.Yafi FA, Jenkins L, Albersen M, Corona G, Isidori AM, Goldfarb S, et al. Erectile dysfunction. Nat Rev Dis Primers. 2016;2:16003. doi: 10.1038/nrdp.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorve VS, Kshirsagar AD, Vyawahare NS, Joshi VS, Ingale KG, Mohite RJ. Diabetes-induced erectile dysfunction: epidemiology, pathophysiology and management. J Diabetes Complications. 2011;25:129–136. doi: 10.1016/j.jdiacomp.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Angulo J, González-Corrochano R, Cuevas P, Fernández A, La Fuente JM, Rolo F, et al. Diabetes exacerbates the functional deficiency of NO/cGMP pathway associated with erectile dysfunction in human corpus cavernosum and penile arteries. J Sex Med. 2010;7(2 Pt 1):758–768. doi: 10.1111/j.1743-6109.2009.01587.x. [DOI] [PubMed] [Google Scholar]
- 6.Verma S, Buchanan MR, Anderson TJ. Endothelial function testing as a biomarker of vascular disease. Circulation. 2003;108:2054–2059. doi: 10.1161/01.CIR.0000089191.72957.ED. [DOI] [PubMed] [Google Scholar]
- 7.Gerber RE, Vita JA, Ganz P, Wager CG, Araujo AB, Rosen RC, et al. Association of peripheral microvascular dysfunction and erectile dysfunction. J Urol. 2015;193:612–617. doi: 10.1016/j.juro.2014.08.108. [DOI] [PubMed] [Google Scholar]
- 8.Yin GN, Park SH, Choi MJ, Limanjaya A, Ghatak K, Minh NN, et al. Penile neurovascular structure revisited: immunohistochemical studies with three-dimensional reconstruction. Andrology. 2017;5:964–970. doi: 10.1111/andr.12387. [DOI] [PubMed] [Google Scholar]
- 9.Yin GN, Ock J, Choi MJ, Song KM, Ghatak K, Minh NN, et al. A simple and nonenzymatic method to isolate human corpus cavernosum endothelial cells and pericytes for the study of erectile dysfunction. World J Mens Health. 2020;38:123–131. doi: 10.5534/wjmh.180091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin GN, Ryu JK, Kwon MH, Shin SH, Jin HR, Song KM, et al. Matrigel-based sprouting endothelial cell culture system from mouse corpus cavernosum is potentially useful for the study of endothelial and erectile dysfunction related to high-glucose exposure. J Sex Med. 2012;9:1760–1772. doi: 10.1111/j.1743-6109.2012.02752.x. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan CJ, Teal TH, Luttrell IP, Tran KB, Peters MA, Wessells H. Microarray analysis reveals novel gene expression changes associated with erectile dysfunction in diabetic rats. Physiol Genomics. 2005;23:192–205. doi: 10.1152/physiolgenomics.00112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Detaille D, Guigas B, Chauvin C, Batandier C, Fontaine E, Wiernsperger N, et al. Metformin prevents high-glucose-induced endothelial cell death through a mitochondrial permeability transition-dependent process. Diabetes. 2005;54:2179–2187. doi: 10.2337/diabetes.54.7.2179. [DOI] [PubMed] [Google Scholar]
- 13.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musicki B, Burnett AL. Endothelial dysfunction in diabetic erectile dysfunction. Int J Impot Res. 2007;19:129–138. doi: 10.1038/sj.ijir.3901494. [DOI] [PubMed] [Google Scholar]
- 16.Andrade E, Andrade PM, Borra RC, Claro J, Srougi M. cDNA microarray analysis of differentially expressed genes in penile tissue after treatment with tadalafil. BJU Int. 2008;101:508–512. doi: 10.1111/j.1464-410X.2007.07285.x. [DOI] [PubMed] [Google Scholar]
- 17.Yin GN, Jin HR, Choi MJ, Limanjaya A, Ghatak K, Minh NN, et al. Pericyte-derived Dickkopf2 regenerates damaged penile neurovasculature through an angiopoietin-1-Tie2 pathway. Diabetes. 2018;67:1149–1161. doi: 10.2337/db17-0833. [DOI] [PubMed] [Google Scholar]
- 18.Castela Â, Costa C. Molecular mechanisms associated with diabetic endothelial-erectile dysfunction. Nat Rev Urol. 2016;13:266–274. doi: 10.1038/nrurol.2016.23. [DOI] [PubMed] [Google Scholar]
- 19.Katz PS, Trask AJ, Souza-Smith FM, Hutchinson KR, Galantowicz ML, Lord KC, et al. Coronary arterioles in type 2 diabetic (db/db) mice undergo a distinct pattern of remodeling associated with decreased vessel stiffness. Basic Res Cardiol. 2011;106:1123–1134. doi: 10.1007/s00395-011-0201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sims TJ, Rasmussen LM, Oxlund H, Bailey AJ. The role of glycation cross-links in diabetic vascular stiffening. Diabetologia. 1996;39:946–951. doi: 10.1007/BF00403914. [DOI] [PubMed] [Google Scholar]
- 21.Andersson KE. Mechanisms of penile erection and basis for pharmacological treatment of erectile dysfunction. Pharmacol Rev. 2011;63:811–859. doi: 10.1124/pr.111.004515. [DOI] [PubMed] [Google Scholar]
- 22.Jin HR, Kim WJ, Song JS, Piao S, Choi MJ, Tumurbaatar M, et al. Intracavernous delivery of a designed angiopoietin-1 variant rescues erectile function by enhancing endothelial regeneration in the streptozotocin-induced diabetic mouse. Diabetes. 2011;60:969–980. doi: 10.2337/db10-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon MH, Ryu JK, Kim WJ, Jin HR, Song KM, Kwon KD, et al. Effect of intracavernous administration of angiopoietin-4 on erectile function in the streptozotocin-induced diabetic mouse. J Sex Med. 2013;10:2912–2927. doi: 10.1111/jsm.12278. [DOI] [PubMed] [Google Scholar]
- 24.Jesmin S, Sakuma I, Salah-Eldin A, Nonomura K, Hattori Y, Kitabatake A. Diminished penile expression of vascular endothelial growth factor and its receptors at the insulin-resistant stage of a type II diabetic rat model: a possible cause for erectile dysfunction in diabetes. J Mol Endocrinol. 2003;31:401–418. doi: 10.1677/jme.0.0310401. [DOI] [PubMed] [Google Scholar]
- 25.Costa C, Soares R, Schmitt F. Angiogenesis: now and then. APMIS. 2004;112:402–412. doi: 10.1111/j.1600-0463.2004.apm11207-0802.x. [DOI] [PubMed] [Google Scholar]
- 26.Kloner RA. Erectile dysfunction as a predictor of cardiovascular disease. Int J Impot Res. 2008;20:460–465. doi: 10.1038/ijir.2008.20. [DOI] [PubMed] [Google Scholar]
- 27.Duan C, Allard JB. Insulin-like growth factor binding protein-5 in physiology and disease. Front Endocrinol (Lausanne) 2020;11:100. doi: 10.3389/fendo.2020.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calenda G, Strong TD, Pavlovich CP, Schaeffer EM, Burnett AL, Yu W, et al. Whole genome microarray of the major pelvic ganglion after cavernous nerve injury: new insights into molecular profile changes after nerve injury. BJU Int. 2012;109:1552–1564. doi: 10.1111/j.1464-410X.2011.10705.x. [DOI] [PubMed] [Google Scholar]
- 29.Al Shawaf E, Abu-Farha M, Devarajan S, Alsairafi Z, Al-Khairi I, Cherian P, et al. ANGPTL4: a predictive marker for diabetic nephropathy. J Diabetes Res. 2019;2019:4943191. doi: 10.1155/2019/4943191. [DOI] [PMC free article] [PubMed] [Google Scholar]