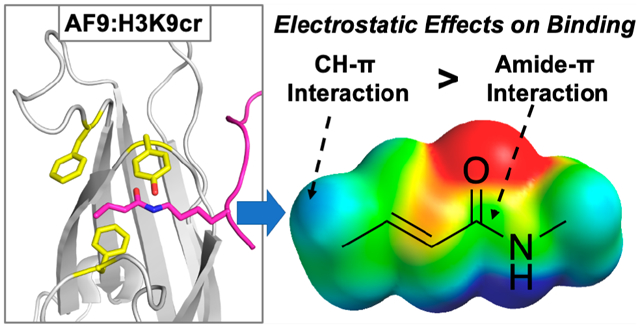
Abstract
Lysine crotonylation (Kcr) is a histone post-translational modification that is implicated in numerous epigenetic pathways and diseases. Recognition of Kcr by YEATS domains has been proposed to occur through intermolecular amide–π and alkene–π interactions, but little is known about the driving force of these key interactions. Herein, we probed the recognition of lysine crotonylation and acetylation by the AF9 YEATS domain through incorporation of noncanonical Phe analogs with distinct electrostatics at two positions. We found that amide–π interactions between AF9 and acyllysines are electrostatically tunable, with electron-rich rings providing more favorable interactions. This differs from trends in amide–heteroarene interactions and provides insightful information for therapeutic design. Additionally, we report for the first time that CH–π interactions at Phe28 directly contribute to AF9’s recognition of acyllysines, illuminating differences among YEATS domains, as this residue is not highly conserved but has been shown to impart selectivity for specific post-translational modification.
Graphical Abstract
INTRODUCTION
Critical cellular processes in eukaryotes, including DNA replication, DNA repair, and gene expression, are facilitated by the epigenetic regulation of chromatin.1,2 In particular, the patterns of post-translational modifications (PTMs) on histones, such as methylation, acylation, and phosphorylation, are critical to epigenetic regulation, with factors including PTM identity and chromosomal location dictating cellular responses.3,4 Although lysine acetylation was first reported in the 1960s, numerous larger acyl groups with functionally distinct roles have only been discovered in the past decade.5
Among the recently discovered histone acyl PTMs, lysine crotonylation (Kcr) has been shown to influence a number of biological processes and disease states.6–13 Studies have indicated that lysine crotonylation is widespread in the proteome, including 28 Kcr sites on histone tails.6,14 Data suggests that histone lysine crotonylation is a stronger activator of transcription than lysine acetylation (Kac) and is congruently enriched at active gene promoters and enhancers in mammalian epigenomes.8 Recently, Wan et al. reported that lysine crotonylation is downregulated in hepatocellular carcinoma and demonstrated that increased lysine crotonylation decreases tumor proliferation, suggesting that Kcr regulation is an attractive strategy for therapeutic campaigns.11
As an increasing number of biological roles of histone lysine crotonylation are discovered, gaining insight into the fundamental mechanisms for Kcr recognition is required for the development of chemical probes for crotonyl reader proteins. Reader proteins and their associated histone-modification interaction domains are central to the recognition of histone PTMs, and numerous disease states have been linked to reader protein dysfunction and dysregulation.15–19 YEATS-domain-containing proteins have emerged as a key family of reader proteins that are capable of recognizing Kcr on histones.20,21 Notably, YEATS domains preferentially bind Kcr over other acyllysine modifications.22–24
AF9, a human reader protein that is part of the super elongation complex and is implicated in numerous epigenetic regulatory pathways, has a YEATS domain shown to recognize acyllysine modifications at multiple positions on histone 3 (H3), including K9, K18, and K27.22,23 AF9’s recognition of Kcr has been directly linked to active transcription.22 The interactions of the YEATS domain of ENL, a closely related homologue to AF9, with Kac have been implicated in the expression of oncogenic genes in leukemia.25,26 Further, gain of function mutations in the YEATS domain of ENL in Wilms tumors were shown to promote oncogenesis.27 Accordingly, YEATS-domain-containing reader proteins have emerged as promising drug targets, and a deeper understanding of the YEATS:Kcr binding interaction would advance inhibitor development.28–33
AF9 binds Kcr via what has been described as a π–π–π sandwich, in which the amide and alkene of the crotonyl group are bound between two highly conserved aromatic side chains.23 The crotonyl group also makes contacts with a less conserved Phe28 via its terminal methyl group. While several campaigns have successfully developed inhibitors for AF9 with binding affinities in the nanomolar range,28–33 precise analyses of intermolecular interactions in YEATS binding pockets are needed to guide inhibitor design. In particular, several factors contribute to favorable interactions with aromatic groups in water, including not only dispersion forces and the hydrophobic effect but also electrostatic interactions arising from the quadrupole moment of aromatic rings, which results in an electron-rich face and electron-poor edge, as can be visualized with electrostatic potential (ESP) maps.34–43 These electrostatic interactions are particularly relevant in cation–π interactions but may also contribute to amide–π and CH–π interactions, both of which are prevalent in proteins.44–47 Indeed, the electrostatic tunability of CH–π interactions between proline and aromatic groups has been demonstrated in several systems, as the CH groups in proline are polarized by the neighboring amide bonds,48–50 and carbohydrate–π interactions have also been shown to be electrostatically tunable in a number of cases.51–54 Studies of amide–π interactions have primarily focused on dipole–dipole interactions in amide–heteroarene interactions,44,45,55 with one example of an amide–arene interaction that is more favorable for electron-poor aromatic residues.56 We hypothesized that, in addition to alkene–π interactions,28 amide–π and CH–π interactions also play a role in AF9’s recognition of Kcr and are influenced by electrostatic effects, and optimizing electrostatic contacts is an attractive strategy for maximizing inhibitor binding to YEATS domains. However, the magnitude and significance of the tunability of these noncovalent interactions on crotonyl recognition have not been reported.
Previously, our group reported a novel approach to study the contributions of cation–π interactions in the binding mechanism of the HP1 chromodomain, a reader domain that recognizes histone trimethyllysine (Kme3). The methodology utilized genetic code expansion (GCE) to tune the electrostatics of individual aromatic residues in the binding pocket and thus determine their electrostatic contribution to binding Kme3.57 Herein, we apply this approach to analyze the binding contributions of two key amino acids that we hypothesize to contribute to acyllysine recognition: Phe28 and Tyr78. Given AF9’s biological relevance in epigenetic regulation and the reported π–π–π stacking binding mode, our studies of the mechanism for selectivity of Kcr readers improve the fundamental understanding of this reader protein family by precisely evaluating noncovalent binding interactions between AF9 and acylated lysine. These studies demonstrate that both amide–π and CH–π interactions consist of electrostatic contributions to binding rather than hydrophobic or dispersion forces alone. Notably, this is the first report that Phe28 has significant interactions with bound acyllysine. Our findings shed light onto the unique recognition of lysine crotonylation by AF9 and also represents a novel approach to study the general PTM promiscuity of reader proteins that recognize lysine acylation. Further, this work affords valuable insight into AF9–PTM interactions that may inform probe or inhibitor development for this promising drug target. Lastly, these studies provide new insight into the driving forces that contribute to amide–π and CH–π interactions, which are ubiquitous in protein structures and protein–ligand inter actions.
RESULTS AND DISCUSSION
AF9 Binds Kcr via Amide–π, Alkene–π, and CH–π Interactions.
The AF9 YEATS domain takes on an immunoglobulin-like global fold with eight antiparallel beta sheets.23 The AF9 PTM-binding motif features an aromatic cage containing three aromatic amino acids [Figures 1a and S1 (Supporting Information, SI)].22,58 Two of these positions, Tyr78 and Phe59 in AF9, are highly conserved as aromatic side chains among YEATS family members (Figure 1b).20–22 Mutations of each residue to alanine in AF9 result in a substantial loss of binding to acyllysine modifications, highlighting the importance of both aromatic residues for PTM recognition.22 A third aromatic residue, Phe28, is conserved in AF9 and ENL but is not present in other YEATS domains (Figure 1b).20–22,59,60 Mutating Phe28 to alanine in AF9 affords 5-fold attenuated binding, but not nearly to the extent of the other two aromatic residues (200-fold for Phe59 and 80-fold for Tyr78) in the cage.22 Thus, we sought to investigate whether the three aromatic cage residues contribute differently to binding of Kcr and Kac.
Figure 1.
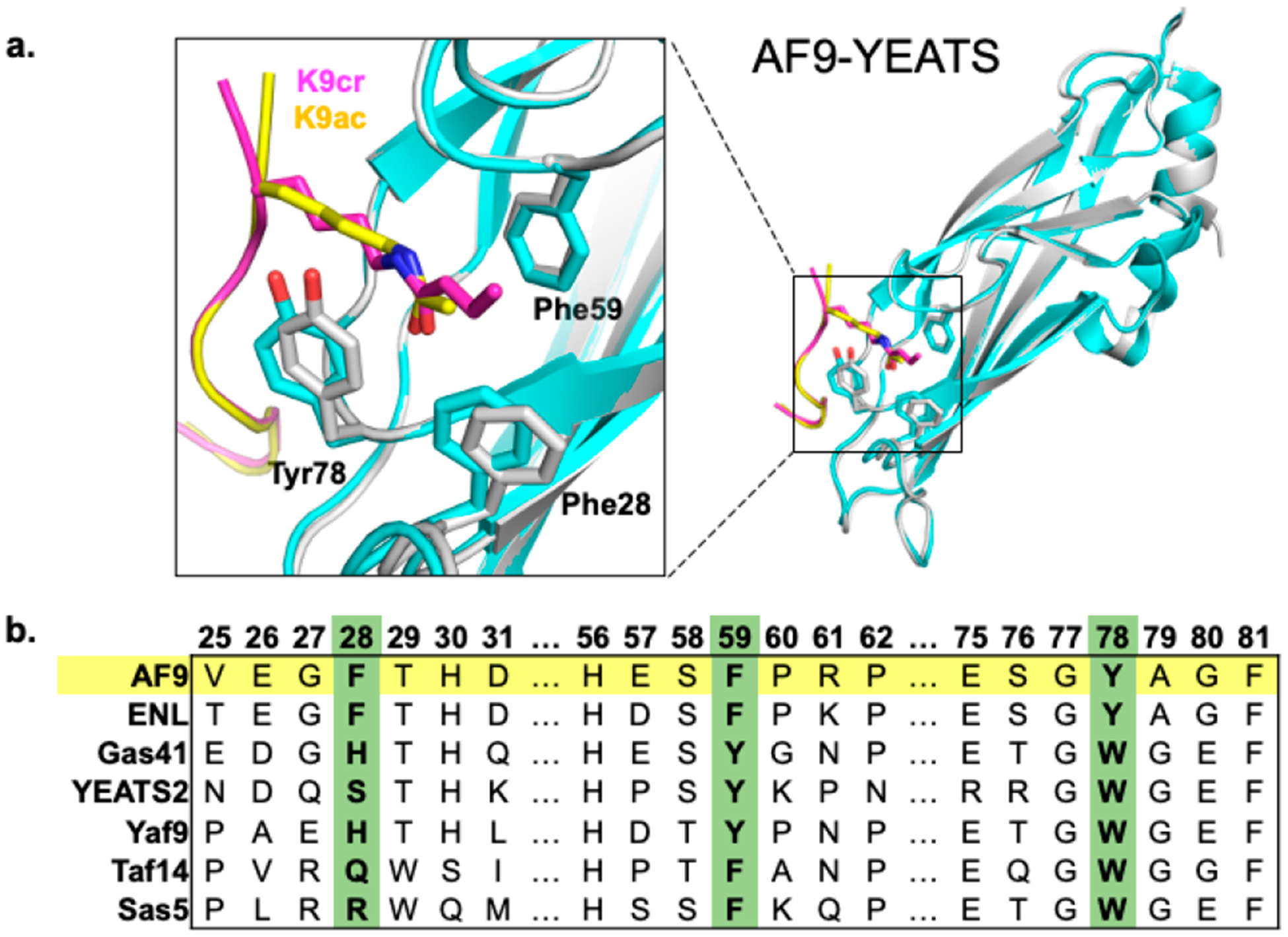
(a) Reported structures of the AF9 YEATS domain in complex with Kcr (gray, purple) overlaid with the AF9 YEATS domain in complex with Kac (cyan, yellow) (PDB: 5HJB and 4TMP).22,58 (b) Sequence alignment of members of the YEATS domain family prepared using Clustal Omega.
Akin to other lysine acyl PTMs, crotonylation introduces an amide bond and neutralizes the positive charge on the lysine side chain. However, the alkene in the crotonyl group provides unique functionality among histone PTMs, since the π bond can participate in intermolecular π–π interactions. Structural studies have suggested that in addition to hydrogen bonding to the NH and CO of the crotonyl group, AF9 and other YEATS domains bind Kcr via an unusual π–π–π stacking mechanism, in which two aromatic residues sandwich the crotonyl group.22–24 Specifically, Tyr78 is located 3.8–3.9 Å above the amide functionality on the acyl group and has been hypothesized to engage in amide–π interactions (Figure 2). Phe59 is located 3.6–3.7 Å above the alkene moiety on the crotonyl group and has been hypothesized to engage in π–π interactions (Figure 2). The role of Phe28 in binding Kcr, which is 4.5 Å away from the γ-carbon of the crotonyl group (Figure S1, SI), has not previously been described, although it has been hypothesized to play a role in selectivity for Kcr and Kac.22
Figure 2.
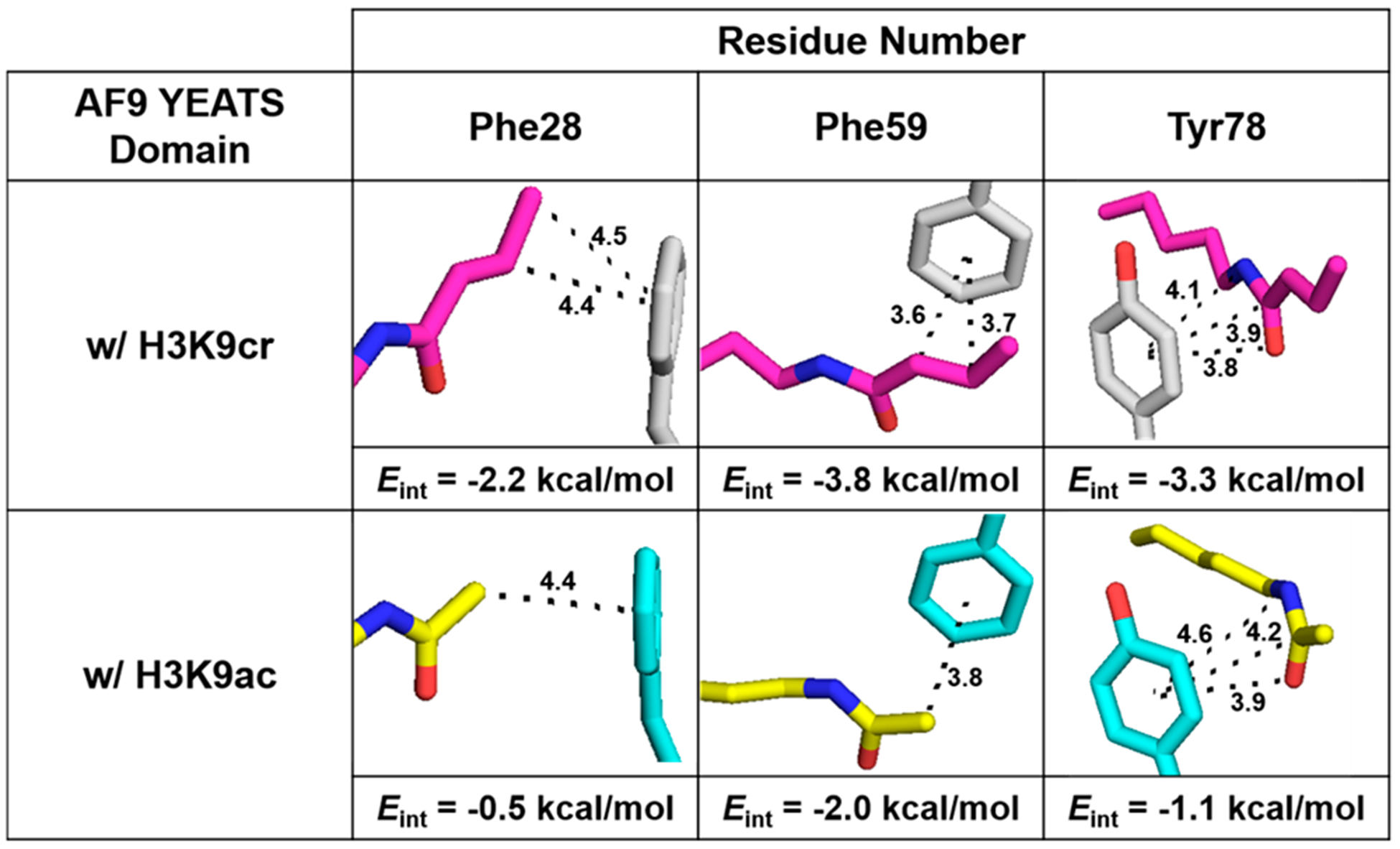
Images of Kcr (top) and Kac (bottom) contacts with residues Phe28, Phe59, and Tyr78 of AF9. Distances displayed are in angstroms. Interaction energies (Eint) for Kcr or Kac with each residue were calculated at the M06–2X/6–311+G(d,p) level of theory using the SMD implicit solvent model (see the SI). Previously reported structures (PDB: 5HJB and 4TMP) were used.22,58
AF9 also binds other acyllysine modifications, including Kac. While AF9 preferentially recognizes Kcr, the degree of selectivity and overall binding affinity are dependent on the position of the modified lysine within the histone tail. Studies have demonstrated that AF9 is 2.5–3.5-fold selective for Kcr over Kac at K9, K18, and K27 in histone 3.23 Crystal and NMR structures with AF9 indicate that the binding pocket takes on a similar, but not identical, conformation, depending on which acyllysine is bound (Figure 1a).22–24 Notably, Tyr78 is slightly rotated in orientation relative to the amide bond when bound to Kac as opposed to Kcr (Figures 1 and 2a). Additionally, Phe28 moves closer inward to the binding pocket, residing 4.4 Å away from the α-carbon of the acetyl group (Figures 1 and 2a). This structural analysis indicates that all three of these aromatic residues, Phe28, Phe59, and Tyr78, are in van der Waals contact with both of the crotonyl and acetyl groups and thus likely contribute to binding. On the basis of reported crystallographic data, we hypothesized that three different types of interactions contribute to different degrees to AF9’s recognition of Kcr and Kac: amide–π interactions at Tyr78, alkene–π interactions at Phe59, and CH–π interactions at Phe28.
Computational Investigations of AF9 Binding to Acyllysine.
We undertook computational studies to gain a deeper insight into the relative contribution of each position in the aromatic cage. Interaction energies (Eint) between Kcr and Tyr78, Phe59, or Phe28 were calculated at the M06–2X/6–311+G(d,p) level of theory using the SMD implicit solvent model (see the computational methods in the SI). Geometries of each dimer and monomer were obtained from a previously reported crystal structure of AF9 in complex with H3K9cr (PDB: 5HJB).22 Computed Eint values predict interactions between Kcr and Phe59 (Eint = −3.8 kcal/mol) to be stronger than those between Kcr and Tyr78 (Eint = −3.3 kcal/mol) (Figure 2, top panel). Additionally, calculations demonstrate potentially significant interactions between Kcr and Phe28 (Eint = −2.2 kcal/mol).
Calculations were also completed to determine interaction energies (Eint) between Kac and the same three aromatic cage residues using a previously reported crystal structure of AF9 in complex with H3K9ac (PDB: 4TMP).58 The trend was analogous to the Kcr results, with Phe59 having the strongest interactions (Eint = −2.0 kcal/mol), followed by Tyr78 (Eint = −1.1 kcal/mol) and Phe28 (Eint = −0.5 kcal/mol). However, the magnitudes were lower in every case due to fewer van der Waals contacts (Figure 2, bottom panel).
At the 78 position, slightly differing conformations of Tyr are likely responsible for differences in calculated Eint between Kcr and Kac, as Tyr78 has a more optimal stacking geometry with Kcr (Figure 2). Differences in the interaction energies for Phe59 with Kcr and Kac may be due to the alkene–π stacking for Kcr versus the CH–π interaction in Kac, as well as the larger surface area of crotonyl compared to acetyl, which leads to a greater number of contacts to Phe59. Although both Kcr and Kac are positioned 4.4 Å from Phe28 in their respective crystal structures, a larger interaction energy for Phe28 with Kcr was calculated compared to that of Kac due to the presence of both the sp2 β-carbon and the sp3 γ-carbon of Kcr making contact with Phe28, compared to just the α-carbon of Kac (Figure 2). Analysis of the geometry of the Phe28 CH–π interaction suggests it may not be ideal based on the parameters defined by Brandl et al. (Figure S1, SI).61 However, the crystal structures may not adequately represent the accessible geometries of interaction, as Phe28 is found in a loop that may be somewhat flexible in solution. Overall, calculations of intermolecular contacts between AF9 and two of its acyllysine binding partners suggest that three aromatic residues contribute to different degrees to PTM recognition, with contributions of each residue differing depending on the identity of the bound acyllysine.
Tuning AF9–PTM Interactions with Genetic Code Expansion.
To determine the contribution of electrostatics arising from the π-systems versus van der Waals or hydrophobic interactions, we measured the sensitivity to electrostatic effects of the aromatic interactions in the recognition of acyllysine PTMs, as an insight into the driving force for binding influences on strategies for inhibitor design. We employed genetic code expansion to modulate the electrostatic potential (ESP) of Phe28, Phe59, and Tyr78 within the aromatic cage binding motif of the AF9 YEATS domain. Specifically, we utilized amber codon suppression to site-specifically incorporate para-substituted phenylalanine derivatives into the aromatic cage of AF9 (Figure 3). An evolved orthogonal tRNA/aminoacyl tRNA synthetase pair, referred to as pCNF-RS, enables a wide range of para-substituted phenylalanine analogs to be encoded into the Escherichia coli genome.62 With this technology, the desired amino acids are selectively and orthogonally installed at positions within the recombinant gene that have been mutated to a TAG codon.
Figure 3.
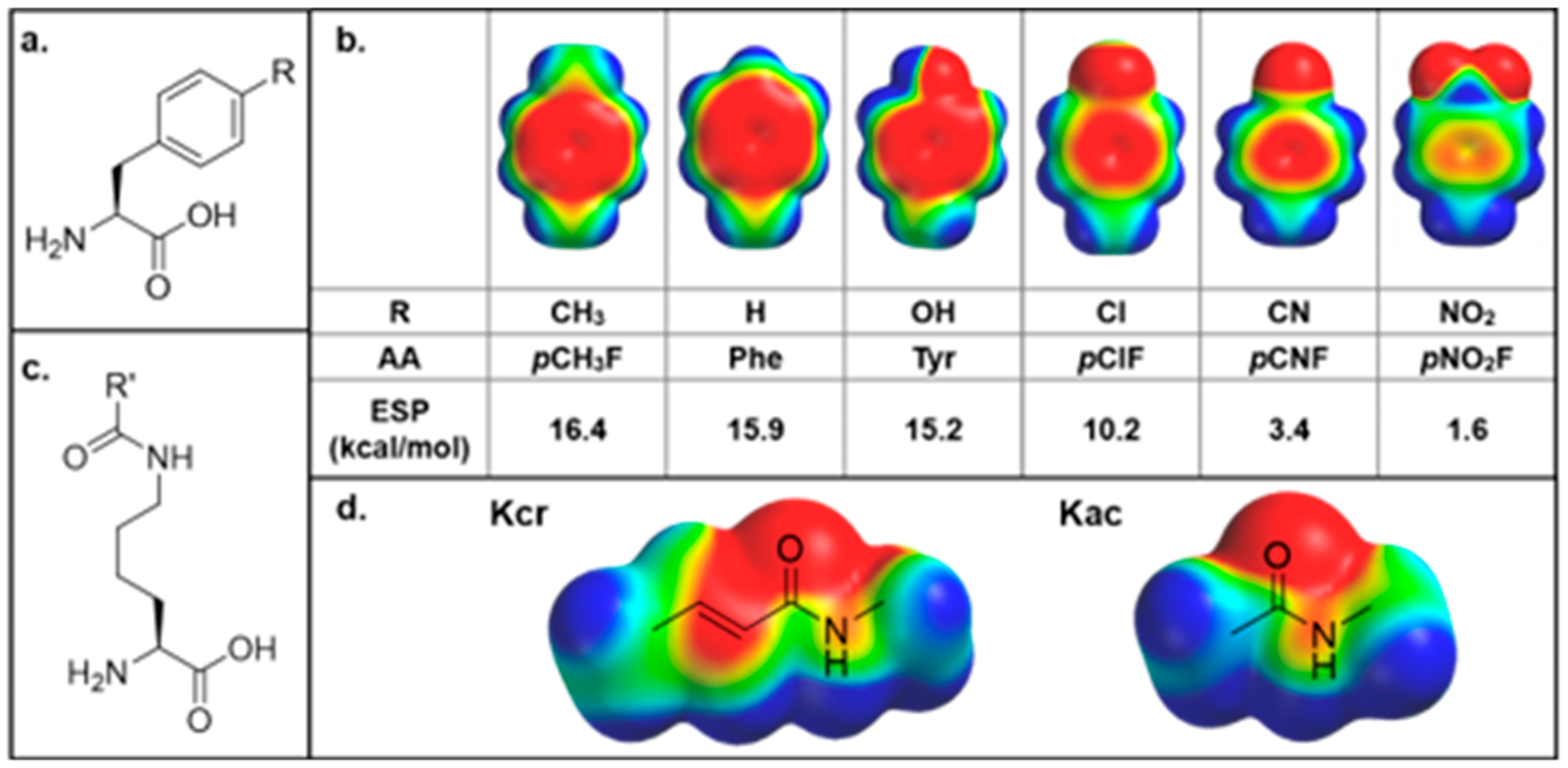
(a) General structure of para-substituted aromatic noncanonical amino acids incorporated at positions of interest. (b) Electrostatic potential (ESP) maps and calculated ESP values of amino acids tested within the aromatic cage. (c) General structure of acyllysine. (d) ESP maps of crotonyllysine and acetyllysine. ESP maps were all calculated in Spartan using the DFT wB97X-D 6–31G* level of theory with an energetic range from −50 to +50 kcal/mol, where red indicates negative electrostatic potential, blue indicates positive electrostatic potential, and green is neutral.
At both the Phe28 and Tyr78 positions, we successfully prepared a library of AF9 mutants, containing either Tyr, Phe, or one of the four screened p-Phe analogs at the desired position (R = CH3, Cl, CN, or NO2, Figure 3a). By modulating the para-position on the phenyl ring, the ESP of the aromatic ring can be tuned, thereby offering precise control over the strength of noncovalent interactions involving the side chain (Figure 3b). Notably, the negative ESP, represented by red on the face of the aromatic ring, is nearly completely neutralized by the nitro group. Successful noncanonical amino acid (ncAA) incorporation and protein purity were confirmed via LC–MS and SDS–PAGE (Figures S2 and S3, SI). However, we found that para-substitutions at position 59 appear to be structurally perturbing; we hypothesize that the para-group sterically clashes with a conserved, ordered water molecule that is present in reported AF9 structures (Figure S4, SI).
Since the epitope recognized by AF9 is within the unstructured N-terminal region of histone 3 (H3), synthetic peptides are commonly used as ligands to evaluate the binding of reader proteins. To this end, we synthesized residues 4–15 of H3 with either a crotonyl or acetyl modification at K9 via Fmoc-based solid-phase peptide synthesis for binding studies. Peptide identity and purity were confirmed via LC–MS (Figure S5, SI). Histone tail peptides containing K9cr and K9ac have been shown to have approximately a 3-fold difference in binding affinity for AF9, with Kd values of 4.6 μM for K9cr and 16.6 μM for K9ac.23
To ensure that protein structure and fold were not perturbed by the introduction of canonical or noncanonical mutations at positions 28 and 78, we performed HSQC NMR experiments on wild-type (WT) AF9, AF9 F28Y, and AF9 Y78pClF to compare the structures of both unbound and ligand-bound AF9 mutants to the wild-type protein [Figures 4 and S6 and S7 (SI)]. Peaks were assigned corresponding to the previously reported NMR structure of wild-type AF9 bound to H3K18ac and H3K18cr (BMRB ID: 26059 and 26060).23 Experiments confirm that the global protein fold is unperturbed by these mutations; both mutants overlay well with the wild-type in the bound and unbound states [Figures 4 and S6 and S7 (SI)].
Figure 4.
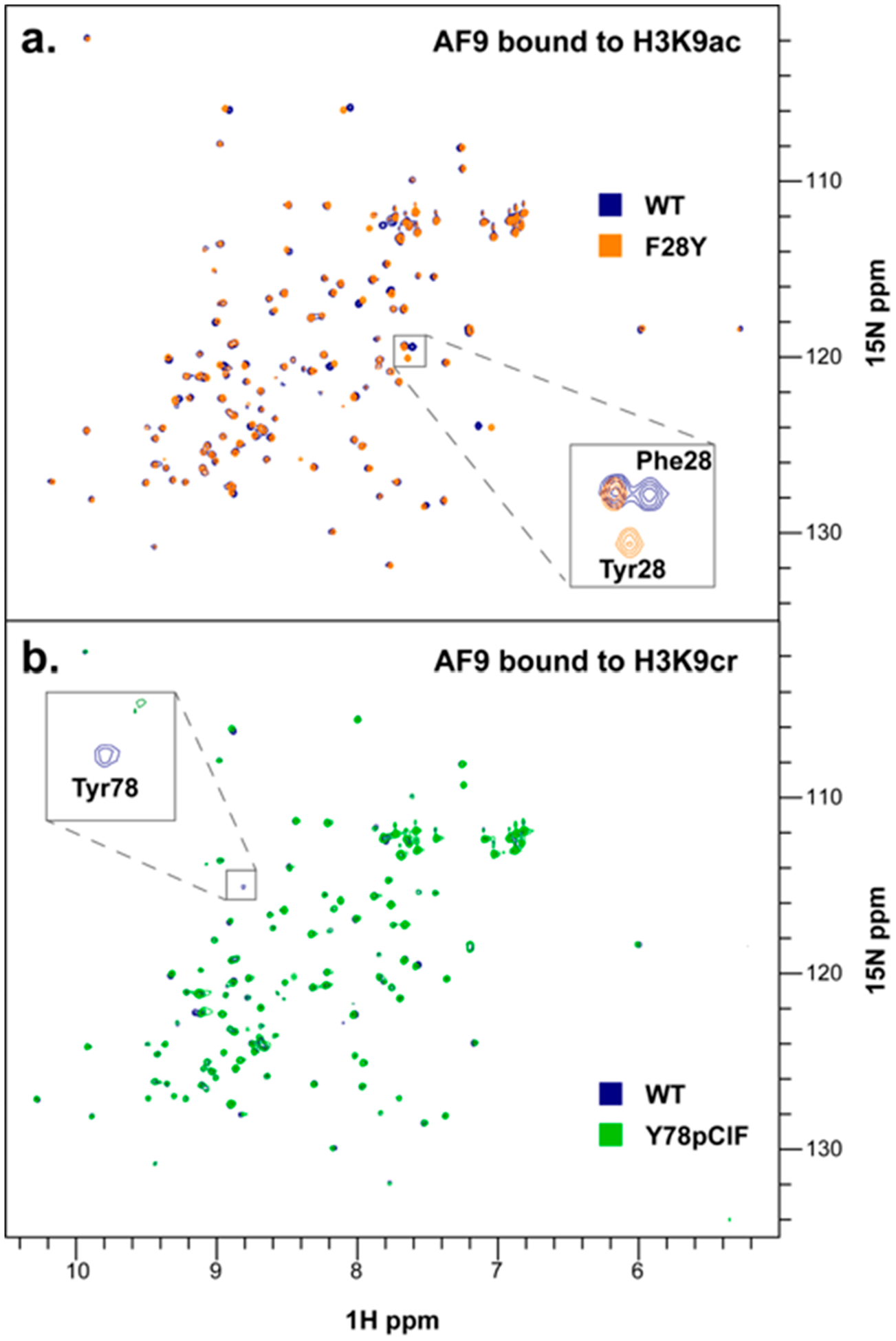
(a) Overlay of HSQC NMR spectra for AF9 WT (blue) and AF9 F28Y (orange), each bound to H3K9ac. (b) Overlay of HSQC NMR spectra for AF9 WT (blue) and AF9 Y78pClF (green), each bound to H3K9cr.
An expected peak shift is observed for Phe28 when mutated to Tyr due to the small change in chemical environment (Figure 4a, inset), and the Tyr78 cross-peak is absent in the 78pClF spectra since 78pClF was not15N-labeled (Figure 4b, inset). Thermal melting data obtained from circular dichroism (CD) measurements also indicate that the fold and stability of the Phe28 and Tyr78 mutant proteins are comparable to those of wild-type AF9 and are not perturbed by the incorporation of these ncAAs (Figure S8, SI).
Analysis of the Amide–π Interaction at the Tyr78 Position.
Changes to the binding affinity as a result of electrostatically tuning aromatic residues were quantified using isothermal titration calorimetry (ITC) [Figures 5 and S9 (SI)]. Analysis of the amide–π interaction between Tyr78 and the acyl amide indicated measurable changes in binding affinity between the AF9 mutants and H3K9cr/ac that correlate with the identity of the phenyl para-substituent, in which electron-withdrawing groups weaken the K9cr and K9ac association. Plotting the free energy of binding (ΔGbinding) as a function of the computationally determined ESP for the unnatural amino acids incorporated at position 78 resulted in a linear free energy relationship (LFER), confirming the presence of electrostatically tunable intermolecular contacts between Tyr78 and H3K9cr (Figure 6a). We also evaluated a LFER between ΔGbinding and Hammett σpara values, an experimental measure of substituent electronic effects.63 Plotting ΔGbinding as a function of σpara also showed strong correlation (Figure 6b). In contrast, plotting ΔGbinding as a function of polarizability (a measure of dispersion forces) or log P (a measure of hydrophobicity) did not yield a significant correlation, demonstrating that the tunability observed at this position is indeed electrostatic in nature and not driven by dispersion forces or the hydrophobic effect (Figure S10, SI).
Figure 5.
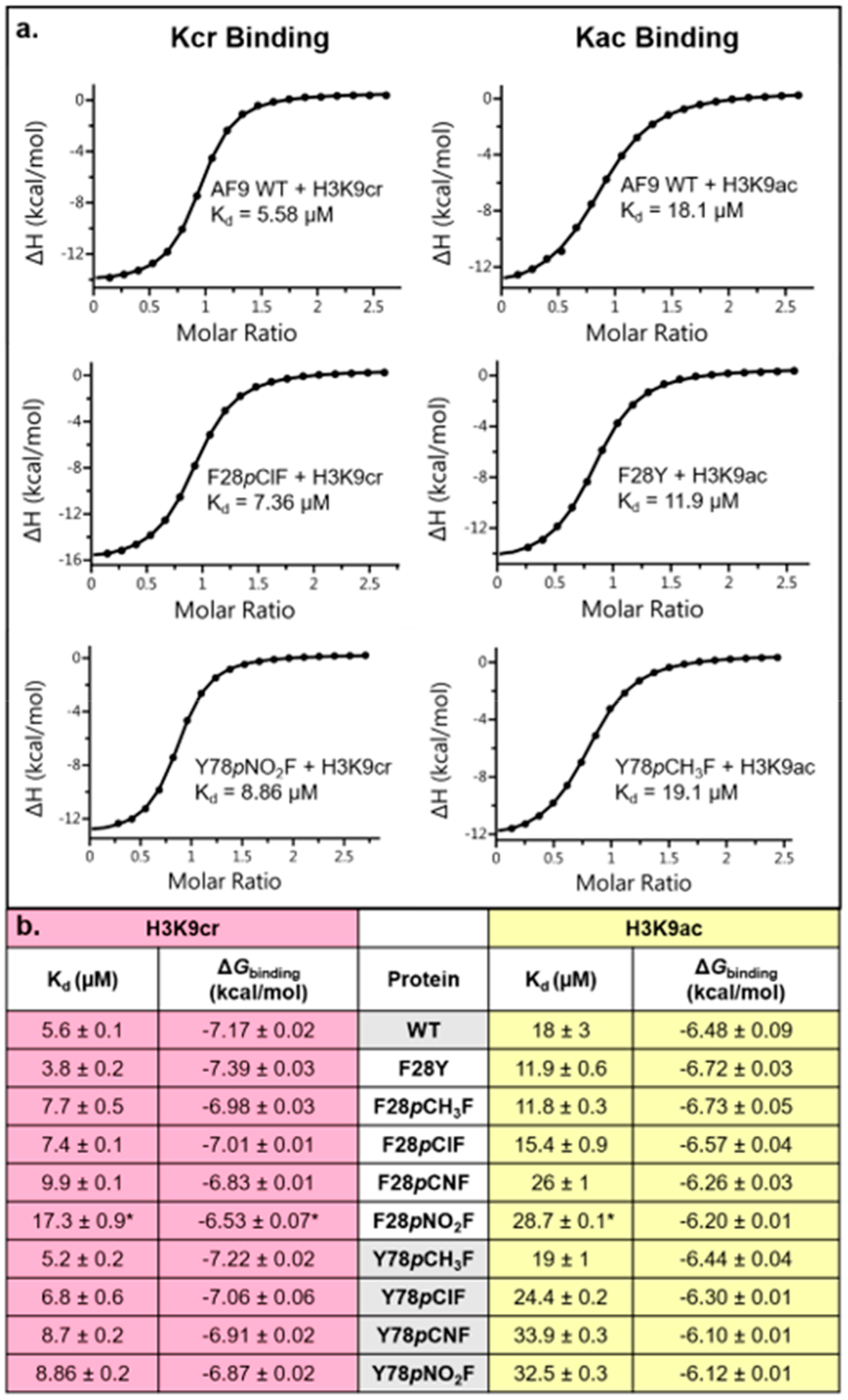
(a) Representative ITC traces from experiments measuring binding of H3K9cr or H3K9ac to AF9 variants. (b) Collective ITC data with average Kd and ΔGbinding values for mutants binding both peptides. Values shown are the average ± standard deviation of three independent experiments (*for F28pNO2F, values shown are the average of two independent experiments).
Figure 6.
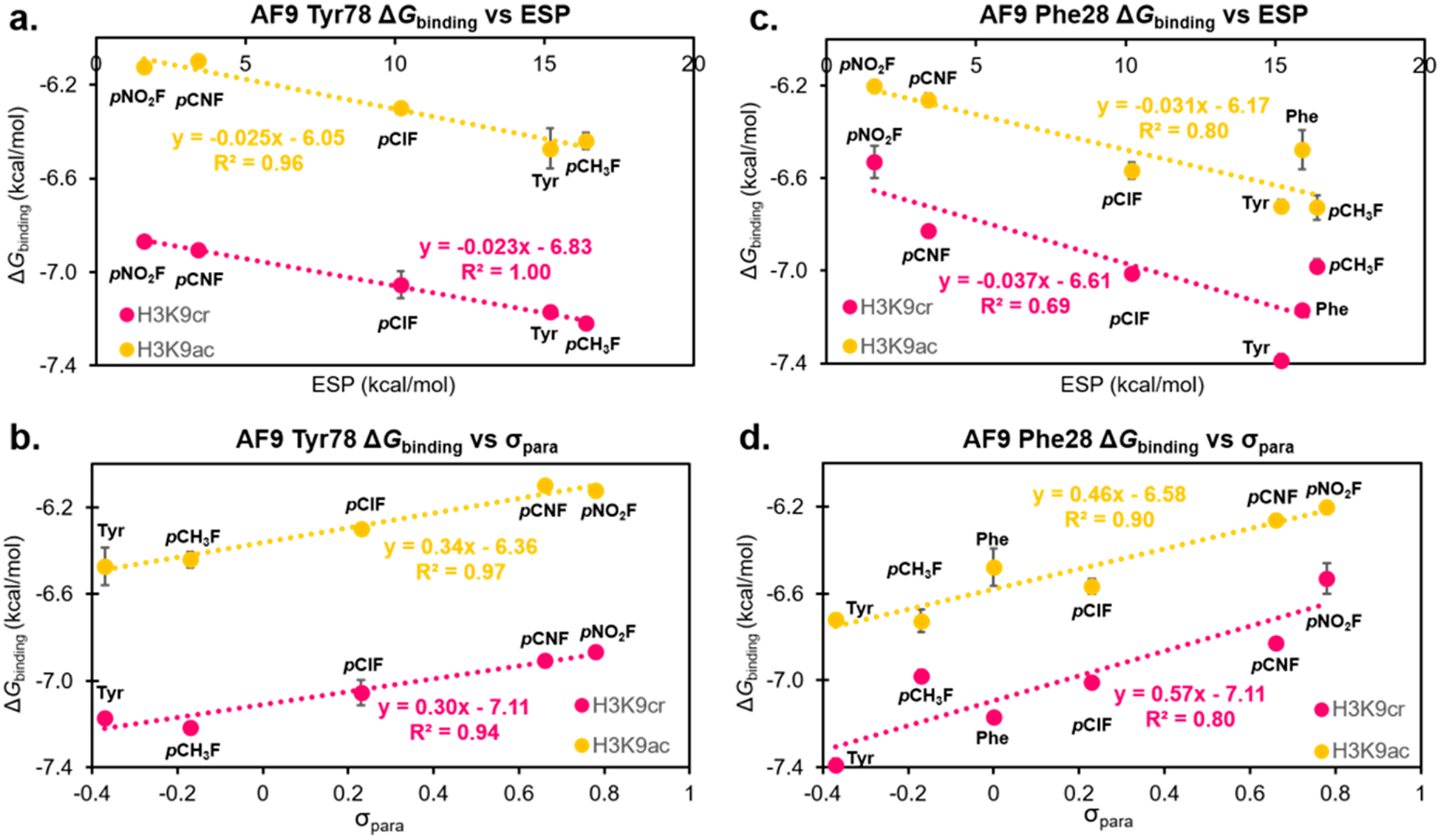
LFER plots analyzing the correlation of ΔGbinding with calculated ESP values (a) and σpara values (b) for a range of AF9 position 78 mutants binding H3K9cr and H3K9ac. LFER plots analyzing the correlation of ΔGbinding and calculated ESP values (c) and σpara values (d) for a range of AF9 position 28 mutants binding H3K9cr and H3K9ac. Error bars reflect the standard deviation of three independent ITC experiments for each data point.
Surprisingly, we also found that the ΔGbinding is anticorrelated with the magnitude of the dipole moment of the aromatic ring, using para-substituted toluene as a model for the aromatic amino acids, with larger dipole moments resulting in weaker binding (Figure 7). This is unexpected, since the dipole moment of the Tyr78 and the acyl amide are oriented antiparallel to each other in AF9, such that the dipole–dipole interaction is expected to be increasingly favorable as the para-substituent becomes more electron-withdrawing. Indeed, previous studies of amide–heteroarene interactions have found that such favorable dipole–dipole interactions dominate the interaction,44 which is not observed here. Our results also contrast with an experimental study on amide–arene interactions with fluorinated arenes, which found that electron-poor arenes generally interacted more strongly with the amide, depending on a combined influence of dipole–dipole interactions with neighboring groups and quadrupolar interactions.45,56 The opposite trend of electrostatic effects observed here thus may be due to differences in the local environment in AF9 that influences both dipolar and quadrupolar interactions, such as differences in hydrogen bonding to the amide.
Figure 7.
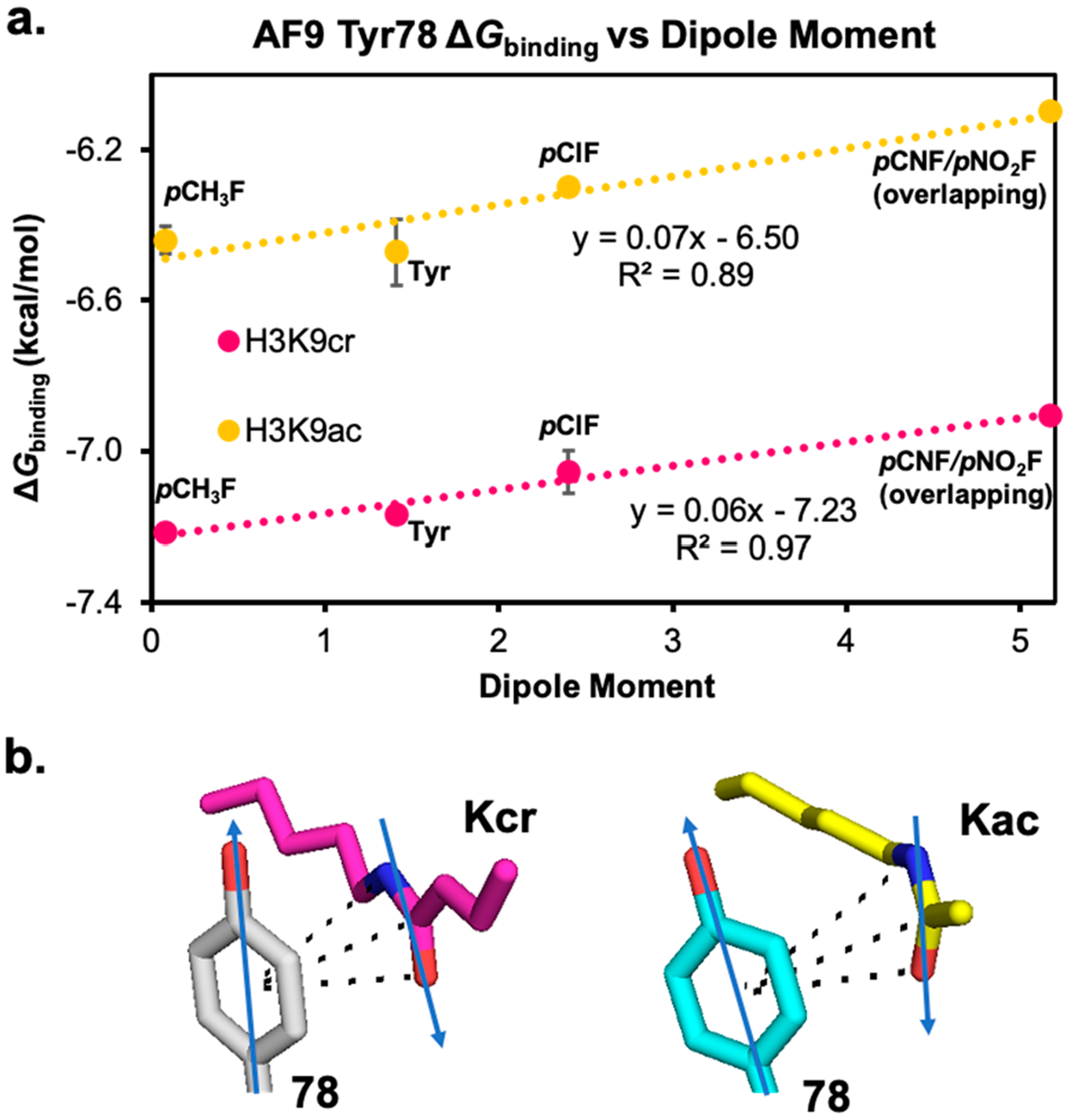
(a) LFER plot showing the anticorrelation of ΔGbinding with the dipole moment of the aromatic ring at residue 78, calculated using para-substituted toluenes as models for aromatic residues. (b) Arrows representing the orientation of the dipole moments of Tyr78 and the amide group in crotonyl- and acetyllysine.22,58
We observed a similar magnitude of attenuation for AF9 binding of both H3K9cr and H3K9ac, as seen in the similar slopes on the LFER plots. This suggests that the identity of the acyl group does not significantly influence the sensitivity of the amide–π binding to variation in electrostatics, at least for these two acyl groups. This is consistent with the similar ESPs of the amide bonds of the crotonyl and acetyl groups (Figure 3d), which indicate that the presence of the alkene in the crotonyl group does not significantly perturb the ESP relative to acetyl.
Because we used the same method to measure electrostatic effects in cation–π interactions in the binding of Kme3 to the HP1 chromodomain reader protein,57 we can compare the magnitude of the electrostatic effects in the amide–π interactions found here to the previously reported cation–π interactions. It is important to note that these interactions are distance-dependent. Thus, we compared cation–π interaction at Tyr24 in HP1, with distances from 3.6 to 4.2 Å, which is similar to the 3.8–4.2 Å distances for Tyr78–Kcr. Interestingly, varying the ESP of Tyr78 in AF9 has a 3-fold smaller influence on the amide–π interaction than the same changes in ESP have on cation–π interactions in HP1 recognizing H3K9me3, demonstrating experimentally that electrostatic interactions contribute less to amide–π interactions than to cation–π interactions.57 The presence of a tunable electrostatic interaction that is anticorrelated with the dipole moment at this position informs inhibitor design efforts that there exists significant room for optimization of π–π interactions at Tyr78. This represents a previously underexplored area of research in the development of a probe or inhibitor of this key drug target; exploiting this interaction could afford improved peptide or small molecule inhibitors.
Analysis of the CH3–π Interaction at the Phe28 Position.
Using the same approach, we evaluated potential contributions of Phe28 to AF9 binding H3K9cr and H3K9ac. Previously reported crystal structures indicate that this residue takes on different conformations depending on which acyl group is bound such that the γ-carbon of H3K9cr and the α-carbon of H3K9ac are similarly distanced from the Phe28 residue (Figure 2b).23,24 Analysis of ESP maps of these two acyl groups reveals that the atoms in closest contact with Phe28 are electro-positive, suggesting that CH–π interactions may be favorable and influenced by the electrostatics of the aromatic ring (Figure 3d). We utilized AF9 mutants at the 28 position with varying ESP to investigate the potential role of Phe28 in recognition of acyllysine substrates. Like at Tyr78, we observed measurable changes in binding affinity between the AF9 Phe28 mutants and both acyllysines. Plotting the ΔGbinding as a function of the electrostatic potential of the ncAA incorporated at position 28 resulted in a LFER, demonstrating the presence of a tunable CH–π interaction at this position, which has not previously been reported (Figure 6c). Interestingly, plotting ΔGbinding as a function of σpara values results in stronger correlations (Figure 6d). Similar to the 78 position, plotting ΔGbinding as a function of polarizability or log P shows no correlation, indicating that the tunability at Phe28 is not due to dispersion forces or the hydrophobic effect (Figure S10, SI). The magnitude of this binding attenuation with increasing electron-withdrawing ability is similar for H3K9cr and H3K9ac, as seen in the similar slopes of the two lines (Figures 6c,d), consistent with the similar ESP for the γ-CH3 of the crotonyl group and the α-CH3 of the acetyl group.
Such electrostatic tunability of CH–π interactions has been observed in several cases for prolyl–π interactions48–50 and carbohydrate–π interactions,51–53 but not in all model systems.64,65 Some reports have categorized CH–π interactions as “normal” or “activated”, in which the CH is not polarized in the normal interactions but carries a δ+ due to neighboring electron-withdrawing groups in the activated interactions.46,64 Prolyl–π and carbohydrate–π interactions fall into the category of activated CH–π interactions. Comparison of the ESPs for the crotonyl γ-methyl, the acetyl α-methyl, and the axial CH-groups in proline indicates that the methyls in the crotonyl and acetyl are polarized similarly to the prolyl CH groups (Figure S11), suggesting that the CH–π interaction in AF9 is of the activated variety. We compared the slope of the LFER plots of the AF9 Phe28 position with σpara to published values for two different systems with CH–π interactions: an intramolecular prolyl–π interaction within a peptide (slope = 0.29)50 and a designed miniprotein containing multiple prolyl–π and α-CH–π interactions (slope = 0.52).48 The LFER plots (Figure 6d) give slopes of 0.46 and 0.57 for acetyl and crotonyl, respectively, indicating that the electrostatic tunability of these CH–π interactions in an intermolecular protein–ligand interaction is comparable to previously reported interactions.
We also compared the magnitude of the electrostatic tunability of the CH–π interaction to our previously reported cation–π interactions.57 In this case, the CH–π interaction is at a greater distance (~4.5 Å) than is the cation–π interaction (3.6–4.2 Å). Nonetheless, the magnitude of the tunabilty as determined from the slope of the ESP LFER is only about 2–2.4-fold less than the cation–π interaction. Perhaps more surprisingly, our results indicate that the CH–π interaction at Phe28 in AF9 is more sensitive to electrostatic effects than the amide–π interaction at Tyr78, as seen from the larger slopes of the LFER plots for the Phe28 interactions with both H3K9cr and H3K9ac (Figure 6). This is potentially due to the larger positive electrostatic potential on the terminal methyl groups of both Kcr and Kac that interact with Phe28 compared to that of the amide carbonyl that interacts with Tyr78 (Figure 3d).
The discovery that Phe28 contributes to binding both H3K9cr and H3K9ac and furthermore exhibits significant electrostatic tunability shows its versatile role in the recognition of acyllysines. This finding emphasizes the value of our experimental strategy to study AF9 through direct analysis of the dynamic binding process rather than relying solely on static, structural data. This study expands on previous reports that “π–π–π” stacking interactions are the key binding forces involved in AF9’s recognition of Kcr, since CH–π interactions involving Phe28 also appear to hold a significant role.
CONCLUSION
In summary, we have elucidated the contributions of two key aromatic residues, Tyr78 and Phe28, to the recognition of two biologically relevant acyllysine PTMs by the AF9-YEATS domain. Calculations indicate that differences in the number of contacts between the aromatic cage and the crotonyl versus acetyl group account for the 3-fold difference in binding affinity. Incorporation of structurally similar ncAAs with differing electrostatic potentials demonstrated the electrostatic tunability of the amide–π interaction with Tyr78 that differs from previous reports on amide–heteroarene stacking. To the best of our knowledge, this is the first study demonstrating the electrostatic tunability of amide–π interactions with para-substituted benzene rings, which provides a measure of their sensitivity and suggests that the local environment, such as differences in hydrogen bonding to the amide, may influence this interaction. Analysis of electrostatic effects at position 28 also enabled the discovery of a previously unrecognized CH–π interaction involved in AF9 binding to both H3K9cr and H3K9ac that is also electrostatically tunable, thus representing an activated CH–π interaction. Surprisingly, the γ-CH3 of the crotonyl group was found to be just as sensitive to electrostatic effects as the α-CH3 of acetyl, despite the greater distance from the electron-withdrawing carbonyl. The discovery of the contribution of this CH–π interaction to binding enhances the understanding of the AF9 aromatic binding cage and offers insight into the differences in acyllysine recognition between AF9 and other YEATS-domain-containing proteins, as Phe28 is not conserved in all members of this histone-reader family.20–22 Previously, Phe28 has been proposed to inhibit binding of hydrophilic acyl groups such as hydroxyisobutyrate and succinate.20,23 Our studies show that it also actively contributes to binding of crotonyl- and acetyllysine. Comparison of the amide–π and CH–π tunability reported here to that of a cation–π interaction allows for the ordering of sensitivity to electrostatic effects as cation–π > CH–π > amide–π.66 While the sensitivity of CH–π interactions to electrostatic effects will no doubt vary from system to system, depending on neighboring groups, it is nonetheless an important benchmark for these interactions. In summary, our discovery that both the amide–π and CH–π interactions between AF9 and its substrates are electrostatically tunable informs medicinal chemistry efforts to target this protein and others through exploitation of interactions with aromatic residues. In a broader sense, these findings also have significance in the areas of protein folding and design, where a detailed knowledge of the noncovalent interactions that contribute to protein structure is required.36 Lastly, this work provides new experimental data on the nature of aromatic interactions within the context of a biologically relevant protein–protein interaction, which should help advance the development of theoretical models of these interactions.37–39,45
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge funding from an NIH grant (GM118499 to M.L.W.) and an NSF grant (GM124480 to K.N.H.) We thank Dr. Ashutosh Tripathy for assistance with ITC and CD, as well as Dr. Stu Parnham and Dr. David Williams for assistance with protein NMR.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c06568.
General and computational methods; genetic code expansion; protein expression, purification, and characterization; peptide synthesis, purification, and characterization; circular dichroism; NMR; binding studies; and Figures S1–S11 (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.0c06568
The authors declare no competing financial interest.
REFERENCES
- (1).Luger K; Mäder AW; Richmond RK; Sargent DF; Richmond TJ Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389 (6648), 251–260. [DOI] [PubMed] [Google Scholar]
- (2).Ramakrishnan V Histone structure and the organization of the nucleosome. Annu. Rev. Biophys. Biomol. Struct 1997, 26 (1), 83–112. [DOI] [PubMed] [Google Scholar]
- (3).Campos EI; Reinberg D Histones: annotating chromatin. Annu. Rev. Genet 2009, 43, 559–599. [DOI] [PubMed] [Google Scholar]
- (4).Peterson CL; Laniel M-A Histones and histone modifications. Curr. Biol 2004, 14 (14), R546–R551. [DOI] [PubMed] [Google Scholar]
- (5).Lin H; Su X; He B Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol 2012, 7 (6), 947–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Tan M; Luo H; Lee S; Jin F; Yang JS; Montellier E; Buchou T; Cheng Z; Rousseaux S; Rajagopal N; et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146 (6), 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Andrews FH; Strahl BD; Kutateladze TG Insights into newly discovered marks and readers of epigenetic information. Nat. Chem. Biol 2016, 12 (9), 662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sabari BR; Tang Z; Huang H; Yong-Gonzalez V; Molina H; Kong HE; Dai L; Shimada M; Cross JR; Zhao Y; et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 2015, 58 (2), 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Montellier E; Rousseaux S; Zhao Y; Khochbin S Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression. BioEssays 2012, 34 (3), 187–193. [DOI] [PubMed] [Google Scholar]
- (10).Ruiz-Andres O; Sanchez-Niño MD; Cannata-Ortiz P; Ruiz-Ortega M; Egido J; Ortiz A; Sanz AB Histone lysine crotonylation during acute kidney injury in mice. Dis. Models & Mech 2016, 9 (6), 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wan J; Liu H; Ming L Lysine crotonylation is involved in hepatocellular carcinoma progression. Biomed. Pharmacother 2019, 111, 976–982. [DOI] [PubMed] [Google Scholar]
- (12).Fellows R; Denizot J; Stellato C; Cuomo A; Jain P; Stoyanova E; Balázsi S; Hajnády Z; Liebert A; Kazakevych J; et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun 2018, 9 (1), 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jiang G; Nguyen D; Archin NM; Yukl SA; Méndez-Lagares G; Tang Y; Elsheikh MM; Thompson GR; Hartigan-O’Connor DJ; Margolis DM; et al. HIV latency is reversed by ACSS2-driven histone crotonylation. J. Clin. Invest 2018, 128 (3), 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wu Q; Li W; Wang C; Fan P; Cao L; Wu Z; Wang F Ultradeep lysine Crotonylome reveals the Crotonylation enhancement on both histones and nonhistone proteins by SAHA treatment. J. Proteome Res 2017, 16 (10), 3664–3671. [DOI] [PubMed] [Google Scholar]
- (15).Strahl BD; Allis CD The language of covalent histone modifications. Nature 2000, 403 (6765), 41–45. [DOI] [PubMed] [Google Scholar]
- (16).Musselman CA; Lalonde M-E; Côté J; Kutateladze TG Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol 2012, 19 (12), 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chervona Y; Costa M Histone modifications and cancer: biomarkers of prognosis? Am. J. Cancer Res 2012, 2 (5), 589. [PMC free article] [PubMed] [Google Scholar]
- (18).Waldmann T; Schneider R Targeting histone modifications—epigenetics in cancer. Curr. Opin. Cell Biol 2013, 25 (2), 184–189. [DOI] [PubMed] [Google Scholar]
- (19).Arrowsmith CH; Bountra C; Fish PV; Lee K; Schapira M Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discovery 2012, 11 (5), 384–400. [DOI] [PubMed] [Google Scholar]
- (20).Zhao D; Li Y; Xiong X; Chen Z; Li H YEATS domain—A histone acylation reader in health and disease. J. Mol. Biol 2017, 429 (13), 1994–2002. [DOI] [PubMed] [Google Scholar]
- (21).Li Y; Zhao D; Chen Z; Li H YEATS domain: Linking histone crotonylation to gene regulation. Transcription 2017, 8 (1), 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Li Y; Sabari BR; Panchenko T; Wen H; Zhao D; Guan H; Wan L; Huang H; Tang Z; Zhao Y; et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell 2016, 62 (2), 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang Q; Zeng L; Zhao C; Ju Y; Konuma T; Zhou M-M Structural insights into histone crotonyl-lysine recognition by the AF9 YEATS domain. Structure 2016, 24 (9), 1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Klein BJ; Vann KR; Andrews FH; Wang WW; Zhang J; Zhang Y; Beloglazkina AA; Mi W; Li Y; Li H; et al. Structural insights into the π–π–π stacking mechanism and DNA-binding activity of the YEATS domain. Nat. Commun 2018, 9 (1), 4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wan L; Wen H; Li Y; Lyu J; Xi Y; Hoshii T; Joseph JK; Wang X; Loh Y-HE; Erb MA; et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 2017, 543 (7644), 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Erb MA; Scott TG; Li BE; Xie H; Paulk J; Seo H-S; Souza A; Roberts JM; Dastjerdi S; Buckley DL; et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 2017, 543 (7644), 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wan L; Chong S; Xuan F; Liang A; Cui X; Gates L; Carroll TS; Li Y; Feng L; Chen G; et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 2020, 577 (7788), 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Li X; Li X-M; Jiang Y; Liu Z; Cui Y; Fung KY; van der Beelen SH; Tian G; Wan L; Shi X; et al. Structure-guided development of YEATS domain inhibitors by targeting π-π-π stacking. Nat. Chem. Biol 2018, 14 (12), 1140–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Heidenreich D; Moustakim M; Schmidt J; Merk D; Brennan PE; Fedorov O; Chaikuad A; Knapp S Structure-based approach toward identification of inhibitory fragments for eleven-nineteen-leukemia protein (ENL). J. Med. Chem 2018, 61 (23), 10929–10934. [DOI] [PubMed] [Google Scholar]
- (30).Moustakim M; Christott T; Monteiro OP; Bennett J; Giroud C; Ward J; Rogers CM; Smith P; Panagakou I; Díaz-Sáez L; et al. Discovery of an MLLT1/3 YEATS domain chemical probe. Angew. Chem., Int. Ed 2018, 57 (50), 16302–16307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Christott T; Bennett J; Coxon C; Monteiro O; Giroud C; Beke V; Felce SL; Gamble V; Gileadi C; Poda G; et al. Discovery of a selective inhibitor for the YEATS domains of ENL/AF9. SLAS DISCOVERY: Advancing Life Sciences R&D 2019, 24 (2), 133–141. [DOI] [PubMed] [Google Scholar]
- (32).Ni X; Heidenreich D; Christott T; Bennett J; Moustakim M; Brennan PE; Fedorov O; Knapp S; Chaikuad A Structural insights into interaction mechanisms of alternative piperazine-urea YEATS domain binders in MLLT1. ACS Med. Chem. Lett 2019, 10 (12), 1661–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Asiaban JN; Milosevich N; Chen E; Bishop TR; Wang J; Zhang Y; Ackerman CJ; Hampton EN; Young TS; Hull MV; et al. Cell-based ligand discovery for the ENL YEATS domain. ACS Chem. Biol 2020, 15 (4), 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ma JC; Dougherty DA The cation–π interaction. Chem. Rev 1997, 97 (5), 1303–1324. [DOI] [PubMed] [Google Scholar]
- (35).Dougherty DA The cation–π interaction. Acc. Chem. Res 2013, 46 (4), 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Newberry RW; Raines RT Secondary Forces in Protein Folding. ACS Chem. Biol 2019, 14 (8), 1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wheeler SE; Houk K Through-space effects of substituents dominate molecular electrostatic potentials of substituted arenes. J. Chem. Theory Comput 2009, 5 (9), 2301–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wheeler SE; Houk K Substituent effects in cation/π interactions and electrostatic potentials above the centers of substituted benzenes are due primarily to through-space effects of the substituents. J. Am. Chem. Soc 2009, 131 (9), 3126–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wheeler SE; Houk K Substituent effects in the benzene dimer are due to direct interactions of the substituents with the unsubstituted benzene. J. Am. Chem. Soc 2008, 130 (33), 10854–10855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Wheeler SE; McNeil AJ; Müller P; Swager TM; Houk K Probing substituent effects in aryl–aryl interactions using stereoselective Diels–Alder cycloadditions. J. Am. Chem. Soc 2010, 132 (10), 3304–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Cozzi F; Annunziata R; Benaglia M; Cinquini M; Raimondi L; Baldridge KK; Siegel JS Through-space interactions between face-to-face, center-to-edge oriented arenes: importance of polar–π effects. Org. Biomol. Chem 2003, 1 (1), 157–162. [DOI] [PubMed] [Google Scholar]
- (42).Cockroft SL; Hunter CA; Lawson KR; Perkins J; Urch CJ Electrostatic control of aromatic stacking interactions. J. Am. Chem. Soc 2005, 127 (24), 8594–8595. [DOI] [PubMed] [Google Scholar]
- (43).Gung BW; Xue X; Reich HJ The strength of parallel-displaced arene–arene interactions in chloroform. J. Org. Chem 2005, 70 (9), 3641–3644. [DOI] [PubMed] [Google Scholar]
- (44).Persch E; Dumele O; Diederich F Molecular recognition in chemical and biological systems. Angew. Chem., Int. Ed 2015, 54 (11), 3290–3327. [DOI] [PubMed] [Google Scholar]
- (45).Bootsma AN; Wheeler SE Stacking Interactions of Heterocyclic Drug Fragments with Protein Amide Backbones. ChemMedChem 2018, 13 (8), 835–841. [DOI] [PubMed] [Google Scholar]
- (46).Nishio M; Umezawa Y; Fantini J; Weiss MS; Chakrabarti P CH–π hydrogen bonds in biological macromolecules. Phys. Chem. Chem. Phys 2014, 16 (25), 12648–12683. [DOI] [PubMed] [Google Scholar]
- (47).Ferreira de Freitas R; Schapira M A systematic analysis of atomic protein–ligand interactions in the PDB. MedChemComm 2017, 8 (10), 1970–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Baker EG; Williams C; Hudson KL; Bartlett GJ; Heal JW; Porter Goff KL; Sessions RB; Crump MP; Woolfson DN Engineering protein stability with atomic precision in a monomeric miniprotein. Nat. Chem. Biol 2017, 13 (7), 764. [DOI] [PMC free article] [PubMed] [Google Scholar]; (Note: prior to comparison of slopes, the units in this work were converted to kcal/mol.)
- (49).Zondlo NJ Aromatic–proline interactions: electronically tunable CH/π interactions. Acc. Chem. Res 2013, 46 (4), 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Thomas KM; Naduthambi D; Zondlo NJ Electronic control of amide cis–trans isomerism via the aromatic–prolyl interaction. J. Am. Chem. Soc 2006, 128 (7), 2216–2217. [DOI] [PubMed] [Google Scholar]
- (51).Santana A.s. G.; Jiménez-Moreno E; Gómez AM; Corzana F; González C; Jiménez-Oses G; Jiménez-Barbero J; Asensio JL A dynamic combinatorial approach for the analysis of weak carbohydrate/aromatic complexes: Dissecting facial selectivity in CH/π stacking interactions. J. Am. Chem. Soc 2013, 135 (9), 3347–3350. [DOI] [PubMed] [Google Scholar]
- (52).Jiménez-Moreno E; Gómez AM; Bastida A; Corzana F; Jiménez-Oses G; Jiménez-Barbero J; Asensio JL Modulating Weak Interactions for Molecular Recognition: A Dynamic Combinatorial Analysis for Assessing the Contribution of Electrostatics to the Stability of CH–π Bonds in Water. Angew. Chem., Int. Ed 2015, 54 (14), 4344–4348. [DOI] [PubMed] [Google Scholar]
- (53).Laughrey ZR; Kiehna SE; Riemen AJ; Waters ML Carbohydrate–π interactions: What are they worth? J. Am. Chem. Soc 2008, 130 (44), 14625–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hudson KL; Bartlett GJ; Diehl RC; Agirre J; Gallagher T; Kiessling LL; Woolfson DN Carbohydrate–aromatic interactions in proteins. J. Am. Chem. Soc 2015, 137 (48), 15152–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Harder M; Kuhn B; Diederich F Efficient stacking on protein amide fragments. ChemMedChem 2013, 8 (3), 397–404. [DOI] [PubMed] [Google Scholar]
- (56).Giroud M; Harder M; Kuhn B; Haap W; Trapp N; Schweizer WB; Schirmeister T; Diederich F Fluorine Scan of Inhibitors of the Cysteine Protease Human Cathepsin L: Dipolar and Quadrupolar Effects in the π-Stacking of Fluorinated Phenyl Rings on Peptide Amide Bonds. ChemMedChem 2016, 11 (10), 1042–1047. [DOI] [PubMed] [Google Scholar]
- (57).Baril SA; Koenig AL; Krone MW; Albanese KI; He CQ; Lee GY; Houk KN; Waters ML; Brustad EM Investigation of trimethyllysine binding by the HP1 chromodomain via unnatural amino acid mutagenesis. J. Am. Chem. Soc 2017, 139 (48), 17253–17256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Li Y; Wen H; Xi Y; Tanaka K; Wang H; Peng D; Ren Y; Jin Q; Dent SY; Li W; et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 2014, 159 (3), 558–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Dobson C; Warren A; Pannell R; Forster A; Lavenir I; Corral J; Smith A; Rabbitts T The Mll–AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemo-genesis. EMBO journal 1999, 18 (13), 3564–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Corral J; Lavenir I; Impey H; Warren AJ; Forster A; Larson TA; Bell S; McKenzie AN; King G; Rabbitts TH An Mll–AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell 1996, 85 (6), 853–861. [DOI] [PubMed] [Google Scholar]
- (61).Brandl M; Weiss MS; Jabs A; Sühnel J; Hilgenfeld R CH⋯ π-interactions in proteins. J. Mol. Biol 2001, 307 (1), 357–377. [DOI] [PubMed] [Google Scholar]
- (62).Young DD; Young TS; Jahnz M; Ahmad I; Spraggon G; Schultz PG An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry 2011, 50 (11), 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Anslyn EV; Dougherty DA Modern Physical Organic Chemistry; University Science Books, 2006. [Google Scholar]
- (64).Aragay G; Hernández D; Verdejo B; Escudero-Adán EC; Martínez M; Ballester P Quantification of CH-π Interactions Using Calix [4] pyrrole Receptors as Model Systems. Molecules 2015, 20 (9), 16672–16686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Kim E-I; Paliwal S; Wilcox CS Measurements of molecular electrostatic field effects in edge-to-face aromatic interactions and CH-π interactions with implications for protein folding and molecular recognition. J. Am. Chem. Soc 1998, 120 (43), 11192–11193. [Google Scholar]
- (66).Hughes RM; Waters ML Effects of Lysine Acetylation in a β-Hairpin Peptide: Comparison of an Amide–π and a Cation–π Interaction. J. Am. Chem. Soc 2006, 128 (41), 13586–13591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


