Abstract
CD276 (also known as B7–H3, an immune checkpoint molecule) is aberrantly overexpressed in many cancers. However, the upregulation mechanism and in particular, whether oncogenic signaling has a role, is unclear. Here we demonstrate that a pro-oncogenic kinase PBK, the expression of which is associated with immune infiltration in nasopharyngeal carcinoma (NPC), stimulates the expression of CD276 epigenetically. Mechanistically, PBK phosphorylates MSL1 and enhances the interaction between MSL1 and MSL2, MSL3, and KAT8, the components of the MSL complex. As a consequence, PBK promotes the enrichment of MSL complex on CD276 promoter, leading to the increased histone H4 K16 acetylation and the activation of CD276 transcription. In addition, we show that CD276 is highly upregulated and associated with immune infiltrating levels in NPC. Collectively, our findings describe a novel PBK/MSL1/CD276 signaling axis, which may play an important role in immune evasion of NPC and may be targeted for cancer immunotherapy.
Subject terms: Head and neck cancer, Tumour immunology
Introduction
Nasopharyngeal carcinoma (NPC) is one of the most common head and neck cancers in southeast Asia and north Africa1,2. The standard treatment for patients with NPC is concurrent chemoradiation preceded or followed by systemic chemotherapy, according to the National Comprehensive Cancer Network (NCCN) guidelines. Although the local control rate has been significantly improved, approximately 30–40% of patients with locoregionally advanced NPC eventually develop distant metastasis after receiving radical treatment3. Novel strategies are alarmingly needed for NPC patients with a high risk of distant metastasis.
Evading immune destruction is described as the hallmark of cancer4. NPC originated from the epithelium of the nasopharyngeal, which is regarded as a highly immunogenic tumor characterized by heavy tumor-infiltrating lymphocytes (TILs), therefore at one time called “lymphoepithelial neoplasia”5. Furthermore, Epstein–Barr virus (EBV) infection is a major risk factor for the development of NPC in the endemic regions6,7, upregulation of programmed cell death-ligand 1 (PDL1) was found in EBV-driven NPC cells, it is reported that PDL1 (CD274) expression on NPC is associated with a poor outcome8, this suggests that the development of NPC may be closely related to the immune escape of tumor cells. In pursuit of the novel effective treatment, several single-arm trials investigate that PD1 (CD279) inhibitors are effective in only 20–30% of recurrent or metastatic NPC patients9,10, it is urgent to develop more biomarkers to reconstruct the immune surveillance, which would be exceedingly beneficial for the clinical intervention.
CD276, also known as B7–H3, is a type I membrane protein with its sequence similarity to the extracellular domain of other B7 family members, which modulate T-cell function in a co-stimulatory or co-inhibitory manner11–13. The CD276 protein is rarely expressed and is only found at low levels in normal human tissues14,15. By contrast, recent studies found aberrant high CD276 expression on the many common malignancies, including stomach16, lung17, prostate18, kidney19, ovary20, and endometrium21, and the high expression of CD276 was also correlated with advanced disease and poor outcome22. Moreover, CD276 protein is also frequently overexpressed on tumor vessels of human lung, breast, colon, endometrial, renal, and ovarian cancer, but not in the angiogenic vessels of the normal ovary15. Notably, an increasing number of studies support a pro-oncogenic role for CD276 in various human cancer types that is independent of its immune function15,23,24. Overexpression of CD276 makes it an attractive target for the development of therapeutic agents, and in fact, CD276-targeting therapies are currently under clinical investigation in several children and adult tumors (NCT02982941, NCT01391143, NCT01099644, NCT02381314, and NCT04185038)15,25–31. However, the cellular mechanisms that promote CD276 expression are poorly understood.
A PDZ-binding kinase (PBK), also known as lymphokine-activated killer T-cell-originated protein kinase (TOPK), is a member of the novel MEK3/6-related mitogen-activated protein kinase–kinase (MAPKK) family32 and directly phosphorylates ERK, H2AX (Ser139), peroxiredoxin 1 (PRX1, Ser32), JNK1 (Thr183/Tyr185), and p53-related protein kinase (PRPK, Ser250)33–35. Thus, PBK activates downstream signaling cascades via its phosphorylated substrates and plays an important role in many kinds of cellular processes, including growth, development, apoptosis, and inflammation36. Previous studies including our group have highlighted that PBK was required for malignant phenotypes and was associated with poor prognosis of human common cancers, including NPC, oral cancer, breast cancer, colorectal cancer, leukemia and lymphoma, ovarian cancer, lung cancer, and glioma34,36–40. Due to the broad expression of PBK across multiple tumor types, specific inhibitors have been developed to target PBK, including HI-TOPK-03241, OTS514/OTS96442, and ADA-0743, and show high therapeutic potency in the preclinical study. PBK has been revealed to contribute to the regulation of proliferation and cell cycle progression, whereas whether PBK involves in the tumor immune evasion is not reported so far. In the current study, we revealed that PBK promotes the transcription of CD276 via an epigenetic way, providing new insight into immune evasion in NPC.
Results
PBK expression is associated with immune infiltration
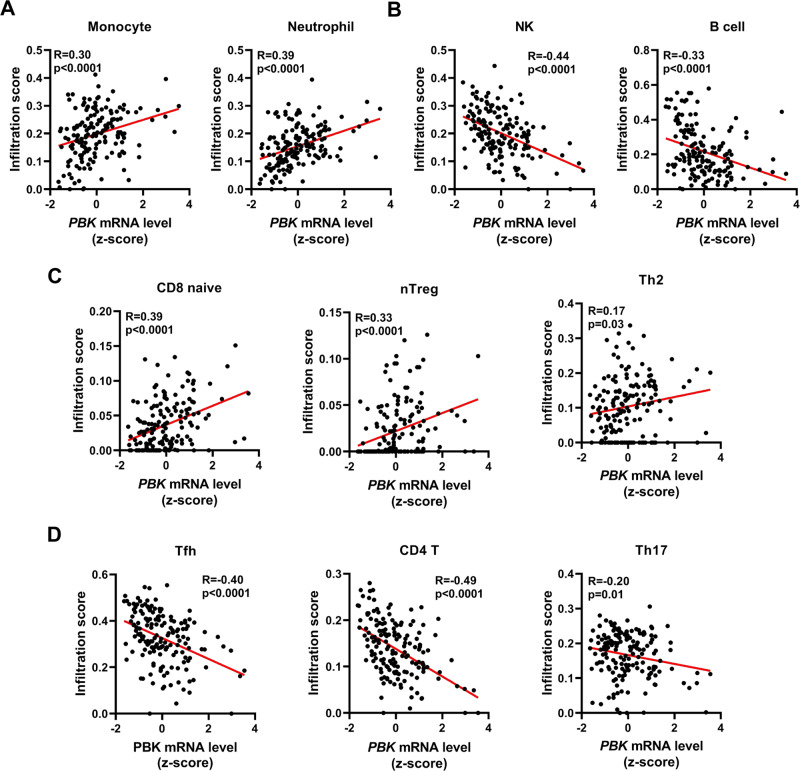
The level of TILs is an independent predictor of sentinel lymph node status and survival in many cancers including NPC44. To explore the role of PBK in immune regulation of NPC, we investigated whether PBK expression was correlated with immune infiltration levels in NPC. ImmuCellAI45, a highly accurate method, was used to estimate the infiltration abundance of immune cells from RNA-seq expression profiles of two NPC cohorts (GSE102349 and GSE68799). Interestingly, correlation analysis showed that PBK expression was significantly correlated to the infiltration of neutrophil and monocyte cells (Fig. 1a), which were reported to promote the immune evasion of tumors46–48, but negatively correlated to the infiltration of natural killer (NK) and B cells (Fig. 1b), which were reported to repress the immune evasion of tumors49–51. Moreover, ImmuCellAI was designed to estimate the abundance of 18 T-cell subsets that are major players in the tumor microenvironment. Notably, PBK expression has significant correlations with CD8-naive cell, nTreg cell, and Th2 cell (Fig. 1c), but was negatively correlated to the infiltration of Tfh cell, CD4+ T cell, and Th17 cell (Fig. 1d). Based on these results, it is supposed that high PBK expression is closely associated with compromised immune microenvironment in NPC.
Fig. 1. PBK mRNA level is associated with immune cell infiltration.
A The scatterplots show that the mRNA level of PBK (lymphokine-activated killer T-cell-originated protein kinase) is significantly correlated with the infiltration of neutrophil cells (left), monocyte cells in NPC (nasopharyngeal carcinoma). B The scatterplots show that the mRNA level of PBK is negatively correlated with the infiltration of NK (natural killer) cells (left) and B cells in NPC. C The scatterplots show that the mRNA level of PBK is significantly correlated with the infiltration of CD8-naive cell (left), nTreg cell (middle), and Th2 cell. D The scatterplots show that the mRNA level of PBK is negatively correlated with the infiltration of Tfh cell (left), CD4+ T cell (middle), and Th17 cell. The method called ImmuCellAI was used to estimate the abundance of 24 immune cell types from two integrated NPC RNA-seq data (GSE102349, GSE68799). A Pearson correlation test was used (two-tailed) (n = 159).
PBK regulates the transcription of CD276
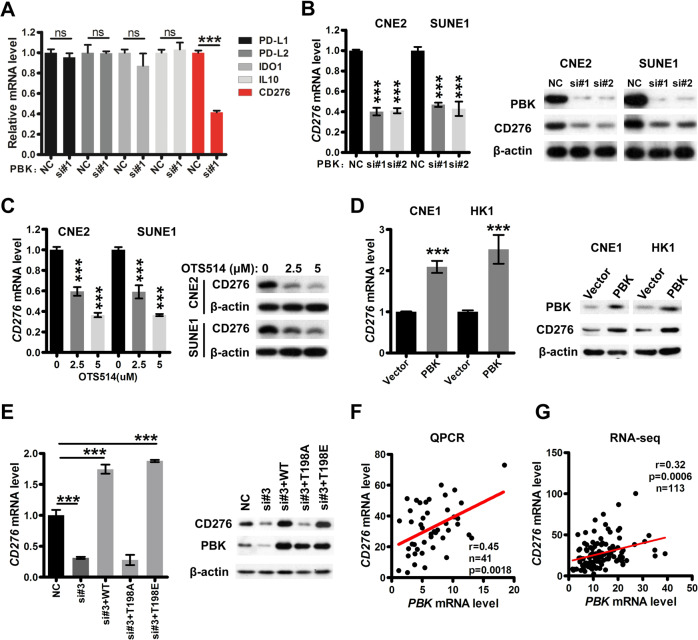
Given the pivotal roles of T-cell subsets in cancer initiation, progression, and immunotherapy52–54, we focus on how PBK modulates the infiltration of T-cell subsets. Accumulating evidence has indicated that cancer cells could upregulate a series of immune inhibitory factors to repress the proliferation, cytokine production, and effector activities of T cells, in turn leading to immune evasion55–57. Thus, we investigated whether previously reported immune inhibitory molecules of T cells, including PDL1 (CD274), PDL2 (CD273), CD276, IL10, and IDO1, could be downstream targets of PBK. The results of quantitative polymerase chain reaction (qPCR) showed that only CD276 was significantly downregulated in CNE2 cells after PBK knockdown by siRNA in our primary screening (Fig. 2a and Fig. S1). Further, qPCR and immunoblotting assays revealed that the mRNA and protein level of CD276 were downregulated in CNE2 and SUNE1 cells with PBK depletion by two siRNAs (Fig. 2b and Fig. S2a), but were upregulated in HK1 and CNE1 cells with PBK overexpression (Fig. 2c and Fig. S2b), suggesting that CD276 is a bona fide downstream target of PBK.
Fig. 2. PBK regulates the transcription of CD276 dependent on its kinase activity.
A CNE2 cells were transfected with one PBK siRNA or NC (nonspecific siRNA) for 48 h and qPCR (quantitative polymerase chain reaction) was performed to measure the mRNA expression of indicated genes. B CNE2 and SUNE1 cells were transfected with two PBK siRNAs or NC (nonspecific siRNA) for 48 h. qPCR and immunoblotting were conducted to measure the mRNA and protein expression of CD276. Left, qPCR. Right, immunoblotting with indicated antibodies. C CNE2 and SUNE1 cells were treated with DMSO (dimethylsulfoxide) or OTS514 (2.5 and 5 μM) for 24 h. qPCR and immunoblotting were conducted to measure the mRNA and protein expression of CD276. Left, qPCR. Right, immunoblotting with indicated antibodies. D CNE1 and HK1 cells were infected by lentiviruses carrying PBK coding region to establish the PBK-overexpression stable cell lines. qPCR and immunoblotting were conducted to measure the mRNA and protein expression of CD276. Left, qPCR. Right, immunoblotting with indicated antibodies. E CNE2 and SUNE1 cells were transfected with one siRNA targeting PBK 3′UTR (untranslated region) region or NC (nonspecific siRNA). After 24 h, the cells were transfected with PBK constructs, including WT (wild type), kinase-dead (T198A), or kinase consistently activated (T198E) for another 24 h. qPCR and immunoblotting were conducted to measure the mRNA and protein expression of CD276. Left, qPCR. Right, immunoblotting with indicated antibodies. A–E In qPCR assays, ACTB was used as the loading control, and the mRNA level of indicated genes is normalized to ACTB. The results are shown as the fold change over NC (nonspecific siRNA). Data present as the mean ± SEM (standard error of the mean) (n = 3). ***P < 0.001 (student’s t test for two groups, one-way ANOVA for three groups). All the experiments were performed independently three times with similar results, and the data are representative of three independent experiments. F–G Correlation analyses between the PBK and CD276 mRNA level were performed using our qPCR data (F) or public RNA-seq data (G). A Pearson correlation test was used (two-tailed).
To investigate whether the PBK kinase activity is indispensable for the regulation of CD276, we treated CNE2 and SUNE1 cells with OTS514, a PBK kinase-specific inhibitor58,59, followed by qPCR and immunoblotting assays, and found that compared to DMSO treatment, OTS514 treatment decreased the mRNA and protein level of CD276 in a dose-dependent manner (Fig. 2d and Fig. S2c). According to previous reports39, we construct two PBK mutants, including kinase-active mutants (PBK-T198E) and kinase-dead mutants (PBK-T198A). Both qPCR and immunoblotting assays revealed that PBK-WT and PBK-T198E but not PBK-T198A could rescue the repression of CD276 mediated by a PBK siRNA that targets the 3′UTR region (Fig. 2e and Fig. S2d). These data show that PBK regulates CD276 transcription dependent on its kinase activity.
Furthermore, correlation analysis showed that the mRNA expression of PBK and CD276 is positively correlated in our NPC cohort detected by qPCR (r = 0.45, n = 41, p = 0.0018, Fig. 2f) as well as in the NPC RNA-seq dataset (r = 0.32, n = 113, p = 0.0006, Fig. 2g). Notably, pan-cancer correlation analysis between PBK and CD276 was conducted using the mRNA expression data from TCGA. The result reveals that the mRNA level of PBK is positively correlated with CD276 in several common cancers (Fig. S3), including adrenocortical carcinoma, brain lower-grade glioma, kidney renal clear-cell carcinoma, kidney renal papillary cell carcinoma, liver hepatocellular carcinoma, lung adenocarcinoma, pancreatic adenocarcinoma, prostate adenocarcinoma, and thyroid carcinoma, indicating that this is a general mechanistic upregulation of CD276 by PBK in cancers.
PBK regulates CD276 transcription dependent on MSL1
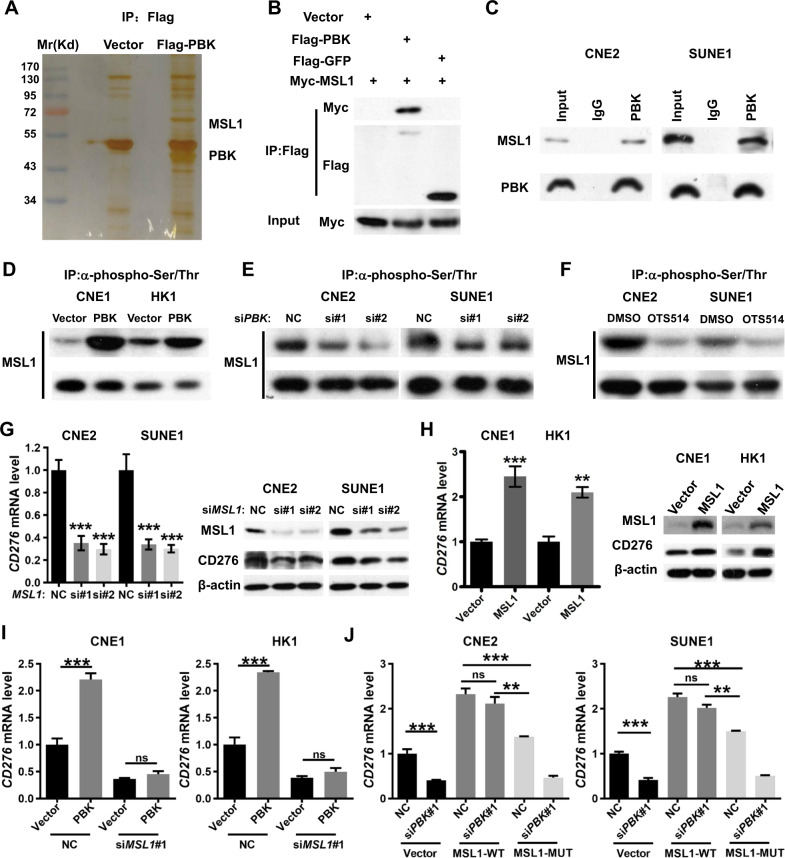
Considering that PBK is a kinase, not a transcription regulator, there is a mediator involved in CD276 transcription regulation by PBK. In order to identify the mediator, we carried out immunoprecipitation and mass spectrometry analysis and identified MSL1 as an interacting partner of PBK (Fig. 3a). Co-immunoprecipitation assays validated the exogenous interaction between PBK and MSL1 in 293T cells (Fig. 3b). To further confirm the in vivo interaction, total proteins from CNE2 and SUNE1 cells were extracted, and immunoprecipitation was performed with an antibody against PBK followed by immunoblotting with an antibody against MSL1, demonstrating that MSL1 was efficiently co-immunoprecipitated with PBK (Fig. 3c). Since PBK has serine/threonine kinase activity, we investigated whether MSL1 is a phosphorylation target of PBK. Immunoprecipitation assays using an antibody against phosphorylated serine/threonine followed by immunoblotting analysis using MSL1 antibody demonstrated that the phosphorylation level of MSL1 was enhanced in PBK-overexpression cells (Fig. 3d and Fig. S4a), but was decreased in PBK-knockdown cells (Fig. 3e and Fig. S4b) as well as in cells treated with PBK inhibitor (Fig. 3f and Fig. S4c). These data show that PBK indeed interacts with and phosphorylates MSL1.
Fig. 3. PBK regulates the expression of CD276 dependent on MSL1.
A SDS/PAGE and silver staining analysis of proteins pulled down by Flag-PBK or vector, via anti-Flag affinity purification. Mass spectrometry analysis recognized MSL1 (male-specific lethal 1 homolog) as an interacting partner of PBK. B 293T cells were co-transfected Myc-MSL1 with vector, Flag-GFP (green fluorescent protein), or Flag-PBK for 36 h, and immunoprecipitation assays and immunoblotting analyses with the indicated antibodies were performed. C Endogenous immunoprecipitation assay in CNE2 and SUNE1 cells with an anti-PBK antibody followed by immunoblotting with antibodies against the indicated proteins. D Immunoprecipitation assays and immunoblotting analyses with the indicated antibodies were performed in PBK-overexpression stable cell lines or control cell lines. E CNE2 and SUNE1 cells were transfected with PBK siRNAs or NC (nonspecific siRNA) for 48 h. Immunoprecipitation assays and immunoblotting analyses with the indicated antibodies were performed. F CNE2 and SUNE1 cells were treated with DMSO or OTS514 (5 μM) for 24 h. Immunoprecipitation assays and immunoblotting analyses with the indicated antibodies were performed. G CNE2 and SUNE1 cells were transfected with two MSL1 siRNAs or NC (nonspecific siRNA) for 48 h. qPCR and Immunoblotting were conducted to measure the mRNA and protein expression of CD276. Left, qPCR. Right, immunoblotting with indicated antibodies. H CNE1 and HK1 cells were infected by lentiviruses carrying MSL1 coding region to establish the MSL1 overexpression stable cell lines. qPCR and immunoblotting were conducted to measure the mRNA and protein expression of CD276. Left, qPCR. Right, immunoblotting with indicated antibodies. I PBK-overexpression cell lines or control cell lines were transfected with one MSL1 siRNA or NC (nonspecific siRNA) for 48 h, respectively. qPCR assays were conducted to measure the mRNA level of CD276. Left, CNE1 cell lines, right, HK1 cell lines. J MSL1 wild type or serine/threonine residues' mutant overexpression cell lines or control cell lines were transfected with a PBK siRNA or NC (nonspecific siRNA) for 48 h. qPCR assays were conducted to measure the mRNA level of CD276. Left, CNE2 cell lines, right, SUNE1 cell lines. J In qPCR assays, ACTB was used as the loading control, and the mRNA level of CD276 is normalized to ACTB. The results are shown as the fold change over NC (nonspecific siRNA). Data present as the mean ± SEM (n = 3). **P < 0.01, ***P < 0.001, ns not significant (Student’s t test for two groups, one-way ANOVA for three groups). All the experiments were performed independently three times with similar results and the data are representative of three independent experiments.
Furthermore, qPCR and immunoblotting results showed that MSL1 depletion decreased the expression of CD276 (Fig. 3g and Fig. S4d) and MSL1 overexpression increased the expression of CD276 (Fig. 3h and Fig. S4e), suggesting that MSL1 is involved in the transcriptional regulation of CD276. More importantly, the knockdown of MSL1 repressed the upregulation of CD276 mediated by PBK overexpression (Fig. 3i and Fig. S4f). MSL1 contains multiple serine/threonine residues, six of which (Ser66, Ser126, Ser205, Ser393, Thr396, and Ser442) were reported as phosphorylation sites60,61. Given that the target sites of MSL1 phosphorylated by PBK are unknown, we replaced all of those MSL1 serine/threonine residues noted above individually with an alanine residue (S/T > A-MSL1). The result of qPCR showed that MSL1 wild-type but not MSL1-S/T > A mutant overexpression could rescue the downregulation of CD276 after PBK depletion in CNE2 and SUNE1 cells (Fig. 3j and Fig. S4g), indicating that PBK controls the transcription of CD276 dependent on the phosphorylation status of MSL1.
PBK promotes the enrichment of MSL complex on CD276 promoter
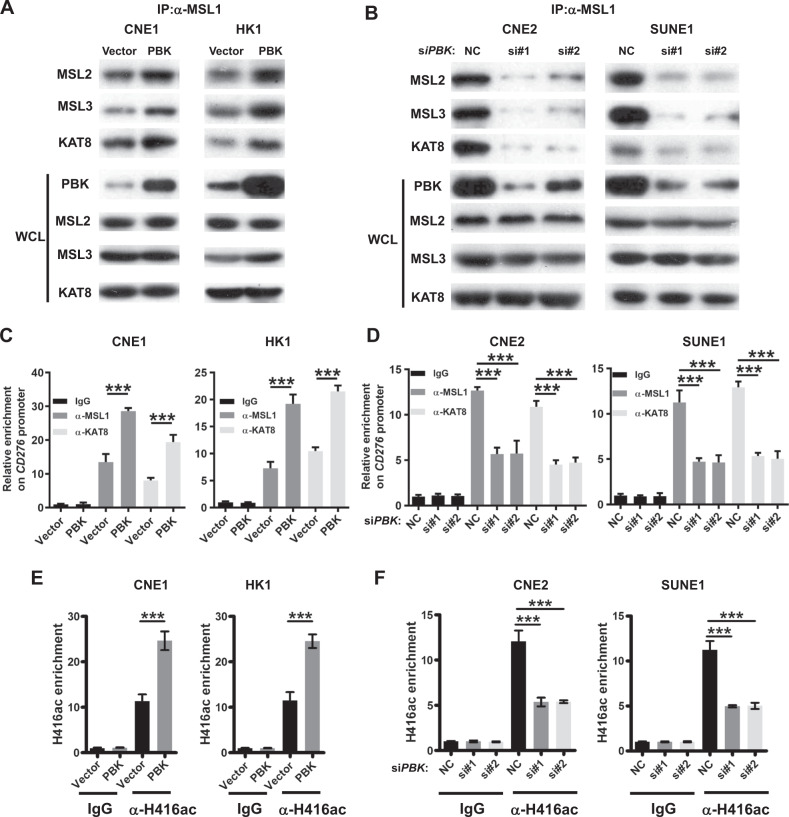
MSL1 acts as a scaffold to tether MSL2, MSL3, and KAT8 (MOF) together for the formation of the MSL histone acetyltransferase complex, in turn leading to histone H4 Lys16 acetylation (forming H4K16ac), thereby efficiently increasing the gene expression62,63. Apart from the dosage-compensation role of the MSL complex, accumulating evidence suggests that MSL proteins may be involved in additional functions, such as transcription regulation64–67. To address the role of PBK in the MSL complex, we conducted immunoprecipitation experiments using an antibody against MSL1 followed by immunoblotting with antibodies against the components of the MSL complex in PBK-overexpression or PBK-knockdown cells, respectively. The results demonstrated that PBK overexpression could enhance the physical association between MSL1 and MSL2, MSL3, and KAT8 (Fig. 4a and Fig. S5a), whereas PBK depletion obviously inhibited these interactions (Fig. 4b and Fig. S5b). Moreover, quantitative chromatin immunoprecipitation (qChIP) analysis using specific antibodies against MSL1 or KAT8 showed that the occupancy of MSL1 and KAT8 on the CD276 promoter region is increased in PBK-overexpression cells (Fig. 4c) but decreased in PBK- deficient cells (Fig. 4d). Consistently, the acetylation level of histone H4 Lys16 on the promoter region is increased in PBK-overexpression cells but decreased in PBK-deficient cells, as indicated by qChIP results (Fig. 4e, f).
Fig. 4. PBK promotes the formation of MSL complex on the promoter of CD276.
A Immunoprecipitation assays with MSL1 antibody and immunoblotting analyses with the indicated antibodies were performed in PBK-overexpression stable cell lines. Left, CNE1, right, HK1. B CNE2 and SUNE1 cells were transfected with PBK siRNAs or NC (nonspecific siRNA) for 48 h. Immunoprecipitation assays with MSL1 antibody and immunoblotting analyses with the indicated antibodies were performed. Left, CNE2, right, SUNE1. C ChIP-qPCR (chromatin immunoprecipitation-quantitative PCR) analysis of CD276 promoter using antibodies against MSL1 and KAT8 in PBK-overexpression or control cell lines. Left, CNE1, right, HK1. D ChIP-qPCR analysis of CD276 promoter using antibodies against MSL1 and MOF in PBK-knockdown or control cells. Left, CNE2, right, SUNE1. E, F ChIP-qPCR analysis of CD276 promoter using antibodies against H4K16ac (acetylation of histone H4 on lysine 16) in PBK-overexpression (E) or -knockdown cells (F). In C–F, ChIP-qPCR results were presented as the fold change over the vector/IgG or NC/IgG group. Error bars represent the mean ± SD (standard deviation) for three biological replicates. ***P < 0.001 (student’s t test for two groups, one-way ANOVA for three groups). All the experiments were performed independently three times with similar results and the data are representative of three independent experiments.
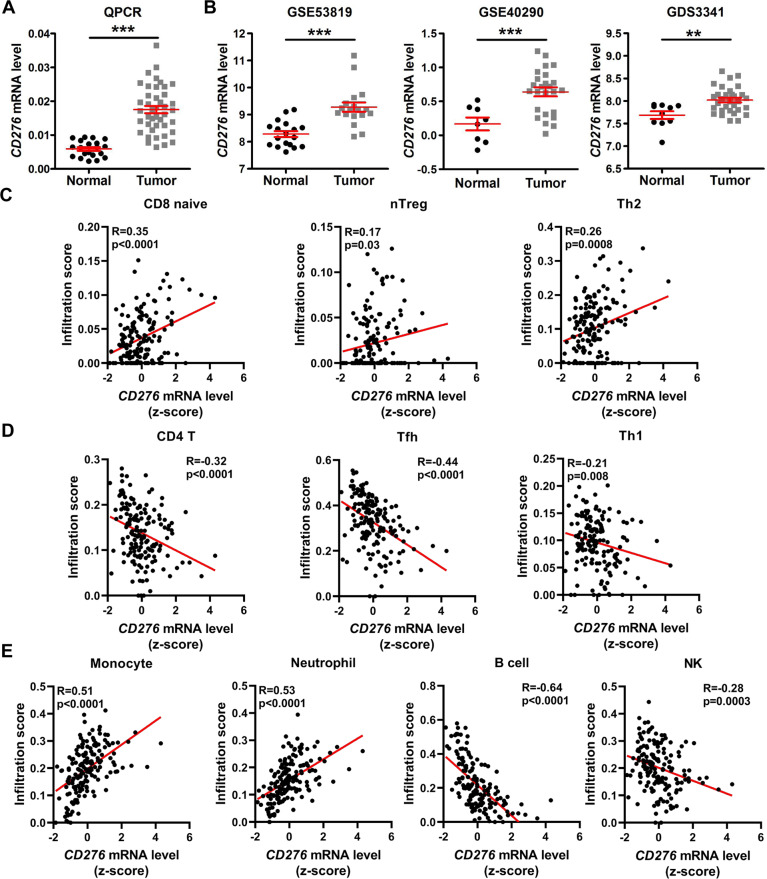
High expression of CD276 is associated with the immune infiltration in NPC
Since the role of CD276 in NPC remains unknown, we performed a qPCR assay and found that CD276 mRNA expression in NPC tissues was higher than in the noncancerous NP tissues (Fig. 5a). In addition, in agreement with our qPCR result, gene expression profiles derived from the GEO (GDS3341, GSE40290, and GSE5381968) showed that compared to noncancerous NP tissues, the CD276 mRNA level was obviously elevated in the NPC tissues (Fig. 5b). Previous reports showed that CD276 has been associated with co-stimulatory as well as co-inhibitory functions in regulating T-cell responses12,14,69,70. In NPC, correlation analysis showed that CD276 mRNA level was positively associated with the infiltration level of CD8-naive cell, nTreg cell, and Th2 cell (Fig. 5c), but was negatively associated with the infiltration level of CD4 T cell, Tfh cell, and Th1 cell (Fig. 5d). In addition, CD276 expression has a significant correlation with monocyte and neutrophil cells and a negative correlation with NK and B cells (Fig. 5e). Taken together, our findings demonstrated that CD276 is upregulated in NPC and associated with immune cell infiltration.
Fig. 5. CD276 is upregulated and associated with immune infiltration in NPC.
A The graphs show that CD276 is significantly upregulated in NPC samples (n = 41) compared to normal samples (n = 20) via qPCR analysis. ***P < 0.001 was determined by Student’s t test. B The graphs show that CD276 is significantly upregulated in NPC samples compared to normal samples, as indicated by four public datasets from GEO (Gene Expression Omnibus database). **P < 0.01, ***P < 0.001 was determined by Student’s t test. C Correlation analysis between the CD276 mRNA level and the tumor-promotion T-cell subset infiltration, including CD8-naive cell (left), nTreg cell (middle), and Th2 cell using publicly available NPC RNA-seq data. A Pearson correlation test was used (two-tailed). D Correlation analysis between the CD276 mRNA level and the tumor-repression T-cell subset infiltration, including CD4 T cell (left), Tfh cell (middle), and Th1 cell using publicly available NPC RNA-seq data. A Pearson correlation test was used (two-tailed). E Correlation analysis between the CD276 mRNA level and the infiltration level of monocyte, neutrophil, NK, and B cells using publicly available NPC RNA-seq data. A Pearson correlation test was used (two-tailed).
PBK reduces T-cell-mediated cytotoxicity dependent on CD276
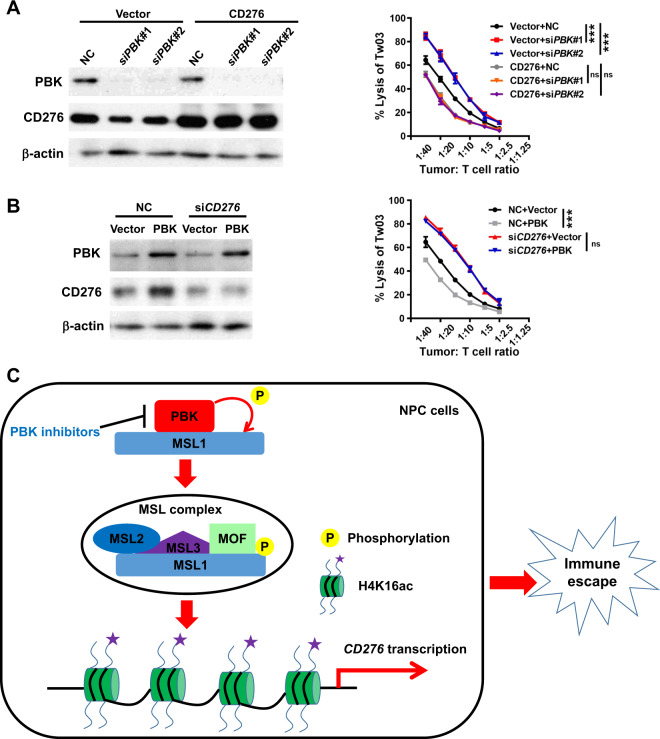
In order to investigate whether the PBK/CD276 axis plays an important role in tumor immune evasion, we conducted the cytotoxic T-lymphocyte assay. Since the NPC cell lines used above did not express HLA-A2 that is important for tumor cell recognition by HLA-A2+ T cells, we selected another NPC cell line TW03 with HLA-A2 positive for the cytotoxic T-lymphocyte assay. The results showed that PBK knockdown significantly increased the percentage of lysis TW03 cells when the tumor cells were cocultured with CD3/CD28-activated human HLA-A2+ T lymphocytes, and this increase was markedly reversed by the recovery of CD276 (Fig. 6A). On the other hand, PBK overexpression significantly reduced the percentage of dead TW03 cells, and this decrease was markedly rescued by CD276 knockdown (Fig. 6B). Collectively, these data confirm that PBK contributes to the limitation of T-lymphocyte attack on NPC cells dependent on CD276.
Fig. 6. PBK modulates the T-cell-mediated killing in NPC cells mediated by CD276 in vitro.
A TW03 cells were infected by lentiviruses carrying CD276 coding region to establish the CD276-overexpression (OE) stable cell lines. Control and CD276-OE cells were transfected with two PBK siRNAs or NC (nonspecific siRNA) for 48 h. Then, the prepared tumor cells and activated T cells were cocultured at a ratio of 1:40–1:1.25 (tumor cells:T cells) in triplicates and the cytolysis of tumor cells was evaluated using the xCELLingence real-time cell analysis (RCTA) system. Left, immunoblotting analysis using indicated antibodies. Right, the cure displayed the percentage of lysis tumor cells at a ratio of 1:40–1:1.25 (tumor cells:T cells). Data shown are mean ± SD and the P values were calculated by one-way ANOVA. ***P < 0.01, ns (not significant) means P > 0.05. B TW03 cells were infected by lentiviruses carrying PBK coding region to establish the PBK-overexpression (OE) stable cell lines. Control and PBK-OE cells were transfected with one CD276 siRNA or NC (nonspecific siRNA) for 48 h. Then, the cytotoxic T-lymphocyte assay was performed as mentioned above. Left, immunoblotting analysis using indicated antibodies. Right, the data shown are mean ± SD and the P values were calculated by Student’s t test (two-tailed). ***P < 0.01, ns (not significant) means P > 0.05. All the experiments were performed independently three times with similar results and the data are representative of three independent experiments. C Proposed model of the PBK/MSL1/CD276 signaling pathway that regulates immune invasion in NPC cells, as well as the intervention strategy.
Discussion
Herein, our findings provide new insight into understanding the potential role of PBK in tumor immunology. We demonstrate that PBK expression is correlated with infiltration levels of diverse immune cells in NPC. Specifically, the infiltration levels of tumor-promoting leukocytes, including neutrophil, monocyte, CD8-naive, nTreg, and Th2 cells, were positively correlated with PBK mRNA expression. Simultaneously, the infiltration levels of tumor-repressing leukocytes, including NK, B, Tfh, CD4+ T cells, and Th17, were negatively correlated with PBK mRNA expression. These correlation patterns are at least partly attributed to the transcription regulation of the CD276 gene by PBK. Although the role of CD276 in antitumor immunity has been controversial with conflicting co-stimulatory and co-inhibitory functions, CD276 may exert a protumor effect on tumor progression in NPC based on the results that the expression of CD276 is much higher in tumor samples than normal samples and is associated with the compromised immune microenvironment. Since several immunomodulatory agents, especially anti-PD1 and anti-PDL1 antibodies, have shown great promise in treating advanced NPC, the role of the PBK/CD276 axis in immunotherapy response is further to be elucidated.
PBK, a dual-specificity serine/threonine kinase, plays an important role in the activation of the Raf/MEK/ERK pathway to promote cell proliferation, colony formation, and cancer development. For example, PBK facilitates transformation by upregulating and activating ERK2 through an increase in AP-1 (c-Jun) transcriptional activity and increased tumorigenesis of HCT116 colorectal cancer33,71. PBK can also directly bind to and phosphorylate AP-1 (c-Jun) after solar ultraviolet exposure72,73. Besides a potential central player of the MAPK cascade mechanism, PBK binds with other proteins and promotes various cancer-related processes, such as PBK promotes tumor dissemination by direct phosphorylation of p53-related protein kinase (PRPK)74. PBK regulates autophagy by phosphorylating ULK1 and promotes glioma resistance to TMZ (temozolomide)75. Thus, more research is necessary to identify potential downstream targets of PBK. Herein, we identify MSL1 as a previous unknown substrate of PBK. Indeed, PBK phosphorylates MSL1 and promotes the formation of the MSL complex. However, the specific phosphorylation site(s) of MSL1 and target genes of the PBK/MSL1 axis are needed to be explored.
CD276 mRNA is widely expressed in many tissues and notably aberrantly expressed in various types of cancer. However, compared to PDL1, the cellular mechanisms that regulate CD276 expression in cancer cells are poorly understood. So far, only a few findings identified revealed that overexpression of CD276 in tumor tissues was highly correlated with decreased expression of several miRNAs as compared to normal tissues, suggesting that a microRNA regulatory mechanism is involved in its differential expression76,77. In this report, we first present an epigenetic modulation mechanism of CD276 expression. Specifically, PBK phosphorylates MSL1 and promotes the enrichment of MSL complex on the promoter region of CD276, in turn leading to an increase in histone H4 Lys16 acetylation, thereby activating the transcription of CD276 and suppressing the cytotoxic T-cell function (Fig. 6C). Further investigations need to be conducted to clarify which transcription factors are essential for CD276 transcription.
In summary, what is noteworthy is that this is the first demonstration of PBK modulation of a co-inhibitory signal CD276 induction to escape from immunosurveillance in NPC. Our results highlight the potential clinical benefits of targeting both PBK and CD276 in NPC patients with a high risk of progression.
Materials and methods
Cell culture
The 293T cells and the human NPC cell lines, including CNE2, SUNE1, HK1, and CNE1, were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS at 37 °C and 5% CO2. All the cell lines were tested for negative mycoplasma contamination.
Sample collection
Fresh frozen tissues for qPCR analysis were resected from histopathologically and clinically diagnosed NPC patients. All samples were obtained from the tumor resource bank of Guangzhou Medical University Affiliated Cancer Hospital and Institute. All patients supplied informed consent.
Plasmids and mutagenesis
Full-length human PBK cDNA was amplified and cloned into a pcDNA6B-his-myc vector (ECORI and XholI) or plenti-puro vector (ECORI and XholI). Full-length human MSL1 cDNA was amplified and cloned into pCMV-N-flag (BamHI and ECORI). ClonExpress II One Step Cloning Kit (Vazyme) was used according to the manufacturer’s instructions.
pcDNA6B-his-myc-PBK(T198A), pcDNA6B-his-myc-PBK(T198E), and pCMV-N-flag-MSL1(S/T > A) mutants were constructed using a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Sequences of the constructs were all verified by Sanger sequencing.
The primers used are listed as follows:
1. PBK with pcDNA6B-his-myc vector:
forward: 5′-TAGTCCAGTGTGGTGGAATTCATGGAAGGGATCAGTAATTTCAAGA-3′
reverse: 5′-CGCGGGCCCTCTAGACTCGGACATCTGTTTCCAGAGCTT-3′
2. PBK with plenti-puro vector:
forward: 5′-CTACCGGACTCAGATCTCGAGATGGAAGGGATCAGTAATTTCAAGA-3′
reverse: 5′-GTCATCCTTGTAATCGAATTCGACATCTGTTTCCAGAGCTTCAACA-3′
3. MSL1 with pCMV-N-flag:
forward: 5′-GATAAGAGCCCGGGCGGATCCATGGAAAGGCGGATGCAGC-3′
reverse: 5′-TCTGTCGACGATATCGAATTCCTATTTCCTACACGTCCGGTGAG-3′
PBK inhibitor
The PBK inhibitor OTS514 was purchased from Selleck (S7652) and dissolved in DMSO with a stocked concentration (50 mM). For the functional assays, NPC cell lines CNE2 and SUNE1 were treated with the OTS514 at two different concentrations, including 2.5 and 5 μM.
siRNA transfection
The specific siRNAs and control nonspecific siRNA (NC) were obtained from Guangzhou RiboBio Co., Ltd. Cancer cells were counted and seeded into 6-well plates with 2 × 105 cells/well. After 24 h, the cells were 30–40% confluent and transfected with siRNAs using RNAiMAX transfection reagents (Invitrogen) according to the manufacturer’s instructions. The cells were harvested for further experiments after 24 or 48 h. The siRNA targeting sequences were as follows:
siPBK#1: 5′-GAATATGGCAAGAGGGTTAAA-3′
siPBK#2: 5′-GGGAACTAGGCCACCTATTAA-3′
siPBK#3: 5′-GAAGTGTGGCTTGCGTAAATA-3′
siMSL1#1: 5′-CACCGGACGTGTAGGAAATAG-3′
siMSL1#1: 5′-ATGTTATCACTCGCTGATAAT-3′
siCD276: 5′-AAAGAAGATGATGGACAAG-3′.
RNA extraction and qPCR
Total RNA was extracted using the TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. Reverse transcription was performed using a cDNA Synthesis Kit (Thermo, K1622). The quantitative real-time polymerase chain reaction was conducted using ChamQ SYBR Color qPCR Master Mix (Vazyme, Q411-02). The sequences of the PCR primers used for amplification were as follows:
ACTB forward, 5′-AAGGTCATCC CTGAGCT GAA-3′
ACTB reverse, 5′-TGACAAAGTG GTCGTTG AGG-3′
PBK forward, 5′-GCGGTGAGACTCTGGACTGA-3′
PBK reverse, 5′-CTGCATAAACGGAGAGGCCG-3′
CD276 forward, 5′-GGAGAATGCAGGAGCTGAGG-3′
CD276 reverse, 5′-GCCAGAGGGTAGGAGCTGTA-3′
PDL1 forward, 5′-ACATGTCAGGCTGAGGGCTA-3′
PDL1 reverse, 5′-TTGGTGGTGGTGGTCTTACC-3′
PDL2 forward, 5′-CAGTGTTCTGCGCCTAAAGC-3′
PDL2 reverse, 5′-GGTCCTGGGTTCCATCTGAC-3′
IDO1 forward, 5′-CACTTTGCTAAAGGCGCTGT-3′
IDO1 reverse, 5′-CCCTTCATACACCAGACCGT-3′
IL10 forward, 5′-ATCAAGGCGCATGTGAACTC-3′
IL10 reverse, 5′-CATTCTTCACCTGCTCCACGG-3′
Immunoblotting
Protein lysates were electrophoresed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% gel) followed by iBlot transfer to polyvinylidene fluoride (Thermo Fisher Scientific). Membranes were blocked in 5% skim milk powder in TBS with 0.1% Tween-20 and then incubated overnight with primary antibody at 4 °C. The following primary antibodies were used: anti-PBK (PTG, 13739-1-AP), anti-CD276 (PTG, 14453-1-AP), anti-MSL1 (PTG, 24373-1-AP), anti-β-actin (PTG, 20536-1-AP), and anti-mouse and anti-rabbit peroxidase-conjugated secondary antibodies (CST, HAF007, and HAF008).
Co-immunoprecipitation and mass spectrometry analysis
For co-immunoprecipitation (co-IP), treated cells were lysed in IP lysis buffer (50 mM Tri-Cl (pH 7.4), 150 mM NaCl, 0.5% NP-40, and 5 mM EDTA) supplemented with PMSF (Sigma, USA) and protease inhibitor cocktail (Roche). The cell lysates were incubated with the indicated antibodies or Flag-beads (Sigma) overnight at 4 °C and washed with lysis buffer 4 times. Proteins were eluted and detected using immunoblotting assays with mouse antibody to Flag (Sigma, M2), an antibody to MSL1, and other appropriate antibodies.
For mass spectrometry analysis, 293T cells were transfected with Flag-PBK or vector control for 48 h. Then treated cells were lysed in IP lysis buffer (50 mM Tri-cl (pH 7.4), 150 mM NaCl, 0.5% NP-40, and 5 mM EDTA) supplemented with PMSF (Sigma, USA) and protease inhibitor cocktail (Roche) and incubated with Flag-tagged affinity agarose beads (Sigma, M2) overnight at 4 °C. The beads were then washed 4 times with IP lysis buffer. The immunoprecipitates were eluted and separated by SDS-PAGE and then stained via silver staining, and the indicated bands were subjected to mass spectrometry analysis.
qChIP assay
The ChIP assays were carried out with a ChIP assay kit (Upstate Biotechnology, Lake Placid, NY, USA) according to the manufacturer’s instructions. The cultured NPC cells were fixed with 1% formaldehyde for 10 min at room temperature. Then, the ultrasonic breaker was set to 10 s per ultrasonic cycle with 10-s intervals with 15 cycles. Subsequently, the fragments underwent centrifugation (30,237×g) at 4 °C (part of the DNA fragments were used as input). The supernatant was collected and added with IgG control (1 ug/ml) or indicated antibodies, including anti-MSL1 (1 ug/ml, Merckmillipore, ABE469), anti-MOF (1 µg/ml, PTG, 13842-1-AP), and anti-H4K16ac (1 µg/ml, CST, #13534), followed by incubation at 4 °C overnight. Protein Agarose/Sepharose was used to precipitate the endogenous DNA–protein complex. After a short period of centrifugation (1000×g), the supernatant was removed, and the nonspecific complex was washed. Following de-cross-linking at 65 °C overnight, DNA fragments were extracted, purified, and retrieved with phenol/chloroform. The enrichment of MSL1 or MOF or H4K16ac to CD276 promoter region was detected via qPCR assay, and the specific primers of CD276 promoter region were as follows:
5′-GGTGAATGCGCTTTTGCAGG-3′
5′-CAGAGTGGACACGCCTAACC-3′
T-lymphocyte preparations
HLA-A2 expression of the T lymphocytes from healthy donors was screened via flow cytometry and only positive individuals subjected to leukapheresis collections. T lymphocytes were then isolated from peripheral blood lymphocytes by depletion of non-T lymphocytes using a Pan T Cell Isolation Kit (Cat# 130-096-535, Miltenyi Biotec). Isolated human T-lymphocyte cells were activated in a T-cell culture medium supplemented with Interleukin-2 (Cat# 200-02, Peprotech) in the pre-coated plate with anti-CD3 (Cat#300313, BioLegend) and anti-CD28 (Cat#302913, BioLegend).
T-cell killing assay
The prepared tumor cells and activated T cells were cocultured at a ratio of 1:40–1:1.25 (tumor:T cells) in triplicate and the cytolysis of tumor cells was evaluated using the xCELLingence real-time cell analysis system.
Data acquisition
Comprehensive immune cell abundance prediction is estimated via ImmuCellAI. The following RNA expression data of NPC samples were retrieved from GEO: GSE102349, GSE68799, GDS3341, GSE40290, and GSE53819.
Statistical analysis
All the experiments were conducted by three biological replicates with similar results. Student’s t test was used to compare two independent groups of data. One-way analysis of variance was used to analyze the significance among multi groups. Pearson correlation analysis was performed as indicated. P value < 0.05 was considered statistically significant in all cases. Asterisk (*) means P < 0.05, asterisks (**) mean P < 0.01, and asterisks (***) mean P < 0.001. SEM means standard error of mean and SD means standard deviation. All statistical analyses were performed using the SPSS 16.0 or Graphpad 8.01.
Supplementary information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81702690), from the Guangzhou Key Medical Discipline Construction Project fund, and from the Research Fund for the State key laboratory of oncology in south China (No. HN2017-09). We apologize to all colleagues whose work could not be cited due to space constraints. We thank Prof. Chao-Nan Qian for kindly providing NPC cancer cell lines.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Meng-Yao Wang, Bin Qi, Fang Wang
Change history
4/30/2021
A Correction to this paper has been published: 10.1038/s41389-021-00325-y
Change history
11/8/2023
A Correction to this paper has been published: 10.1038/s41389-022-00399-2
Contributor Information
Jin-Quan Liu, Email: liujinquan2018@163.com.
Dong-Ping Chen, Email: dongpingchen_20@163.com.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41389-020-00293-9).
References
- 1.Carioli G, et al. Global trends in nasopharyngeal cancer mortality since 1970 and predictions for 2020: focus on low-risk areas. Int. J. Cancer. 2017;140:2256–2264. doi: 10.1002/ijc.30660. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Hui EP, et al. Lung metastasis alone in nasopharyngeal carcinoma: a relatively favorable prognostic group. A study by the Hong Kong Nasopharyngeal Carcinoma Study Group. Cancer. 2004;101:300–306. doi: 10.1002/cncr.20358. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Wenig BM. Lymphoepithelial-like carcinomas of the head and neck. Semin. Diagn. Pathol. 2015;32:74–86. doi: 10.1053/j.semdp.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–1024. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 7.Vaughan TL, et al. Nasopharyngeal cancer in a low-risk population: defining risk factors by histological type. Cancer Epidemiol. Biomark. Prev. 1996;5:587–593. [PubMed] [Google Scholar]
- 8.Minichsdorfer C, et al. PD-L1 expression on tumor cells is associated with a poor outcome in a cohort of caucasian nasopharyngeal carcinoma patients. Front. Oncol. 2019;9:1334. doi: 10.3389/fonc.2019.01334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu C, et al. Safety and antitumor activity of pembrolizumab in patients with programmed death-ligand 1-positive nasopharyngeal carcinoma: results of the KEYNOTE-028 study. J. Clin. Oncol. 2017;35:4050–4056. doi: 10.1200/JCO.2017.73.3675. [DOI] [PubMed] [Google Scholar]
- 10.Fang W, et al. Camrelizumab (SHR-1210) alone or in combination with gemcitabine plus cisplatin for nasopharyngeal carcinoma: results from two single-arm, phase 1 trials. Lancet Oncol. 2018;19:1338–1350. doi: 10.1016/S1470-2045(18)30495-9. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Kang FB, Shan BE. B7-H3-mediated tumor immunology: friend or foe? Int. J. Cancer. 2014;134:2764–2771. doi: 10.1002/ijc.28474. [DOI] [PubMed] [Google Scholar]
- 12.Chapoval AI, et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat. Immunol. 2001;2:269–274. doi: 10.1038/85339. [DOI] [PubMed] [Google Scholar]
- 13.Sun M, et al. Characterization of mouse and human B7-H3 genes. J. Immunol. 2002;168:6294–6297. doi: 10.4049/jimmunol.168.12.6294. [DOI] [PubMed] [Google Scholar]
- 14.Hofmeyer KA, Ray A, Zang X. The contrasting role of B7-H3. Proc. Natl Acad. Sci. USA. 2008;105:10277–10278. doi: 10.1073/pnas.0805458105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seaman S, et al. Eradication of tumors through simultaneous ablation of CD276/B7-H3-positive tumor cells and tumor vasculature. Cancer Cell. 2017;31:501–515 e508. doi: 10.1016/j.ccell.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CP, et al. Relationship between co-stimulatory molecule B7-H3 expression and gastric carcinoma histology and prognosis. World J. Gastroenterol. 2006;12:457–459. doi: 10.3748/wjg.v12.i3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altan M, et al. B7-H3 expression in NSCLC and its association with B7-H4, PD-L1 and tumor-infiltrating lymphocytes. Clin. Cancer Res. 2017;23:5202–5209. doi: 10.1158/1078-0432.CCR-16-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zang X, et al. B7-H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc. Natl Acad. Sci. USA. 2007;104:19458–19463. doi: 10.1073/pnas.0709802104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, et al. Overexpression of B7-H3 in CD14+ monocytes is associated with renal cell carcinoma progression. Med. Oncol. 2014;31:349. doi: 10.1007/s12032-014-0349-1. [DOI] [PubMed] [Google Scholar]
- 20.Fauci JM, Straughn JM, Jr, Ferrone S, Buchsbaum DJ. A review of B7-H3 and B7-H4 immune molecules and their role in ovarian cancer. Gynecol. Oncol. 2012;127:420–425. doi: 10.1016/j.ygyno.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Brunner A, Hinterholzer S, Riss P, Heinze G, Brustmann H. Immunoexpression of B7-H3 in endometrial cancer: relation to tumor T-cell infiltration and prognosis. Gynecol. Oncol. 2012;124:105–111. doi: 10.1016/j.ygyno.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 22.Flem-Karlsen K, Fodstad O, Nunes-Xavier CE. B7-H3 immune checkpoint protein in human cancer. Curr. Med. Chem. 2020;27:4062–4086. doi: 10.2174/0929867326666190517115515. [DOI] [PubMed] [Google Scholar]
- 23.Flem-Karlsen K, Fodstad O, Tan M, Nunes-Xavier CE. B7-H3 in cancer—beyond immune regulation. Trends Cancer. 2018;4:401–404. doi: 10.1016/j.trecan.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Tekle C, et al. B7-H3 contributes to the metastatic capacity of melanoma cells by modulation of known metastasis-associated genes. Int. J. Cancer. 2012;130:2282–2290. doi: 10.1002/ijc.26238. [DOI] [PubMed] [Google Scholar]
- 25.Fauci JM, et al. Monoclonal antibody-based immunotherapy of ovarian cancer: targeting ovarian cancer cells with the B7-H3-specific mAb 376.96. Gynecol. Oncol. 2014;132:203–210. doi: 10.1016/j.ygyno.2013.10.038. [DOI] [PubMed] [Google Scholar]
- 26.Kasten BB, et al. B7-H3-targeted (212)Pb radioimmunotherapy of ovarian cancer in preclinical models. Nucl. Med. Biol. 2017;47:23–30. doi: 10.1016/j.nucmedbio.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer K, et al. Compartmental intrathecal radioimmunotherapy: results for treatment for metastatic CNS neuroblastoma. J. Neurooncol. 2010;97:409–418. doi: 10.1007/s11060-009-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loos M, Hedderich DM, Friess H, Kleeff J. B7-h3 and its role in antitumor immunity. Clin. Dev. Immunol. 2010;2010:683875. doi: 10.1155/2010/683875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Souweidane MM, et al. Convection-enhanced delivery for diffuse intrinsic pontine glioma: a single-centre, dose-escalation, phase 1 trial. Lancet Oncol. 2018;19:1040–1050. doi: 10.1016/S1470-2045(18)30322-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed M, et al. Humanized affinity-matured monoclonal antibody 8H9 has potent antitumor activity and binds to FG loop of tumor antigen B7-H3. J. Biol. Chem. 2015;290:30018–30029. doi: 10.1074/jbc.M115.679852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loo D, et al. Development of an Fc-enhanced anti-B7-H3 monoclonal antibody with potent antitumor activity. Clin. Cancer Res. 2012;18:3834–3845. doi: 10.1158/1078-0432.CCR-12-0715. [DOI] [PubMed] [Google Scholar]
- 32.Aksamitiene E, Kholodenko BN, Kolch W, Hoek JB, Kiyatkin A. PI3K/Akt-sensitive MEK-independent compensatory circuit of ERK activation in ER-positive PI3K-mutant T47D breast cancer cells. Cell Signal. 2010;22:1369–1378. doi: 10.1016/j.cellsig.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh SM, et al. T-lymphokine-activated killer cell-originated protein kinase functions as a positive regulator of c-Jun-NH2-kinase 1 signaling and H-Ras-induced cell transformation. Cancer Res. 2007;67:5186–5194. doi: 10.1158/0008-5472.CAN-06-4506. [DOI] [PubMed] [Google Scholar]
- 34.Roh E, et al. Targeting PRPK and TOPK for skin cancer prevention and therapy. Oncogene. 2018;37:5633–5647. doi: 10.1038/s41388-018-0350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zykova TA, et al. T-LAK cell-originated protein kinase (TOPK) phosphorylation of Prx1 at Ser-32 prevents UVB-induced apoptosis in RPMI7951 melanoma cells through the regulation of Prx1 peroxidase activity. J. Biol. Chem. 2010;285:29138–29146. doi: 10.1074/jbc.M110.135905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herbert KJ, Ashton TM, Prevo R, Pirovano G, Higgins GS. T-LAK cell-originated protein kinase (TOPK): an emerging target for cancer-specific therapeutics. Cell Death Dis. 2018;9:1089. doi: 10.1038/s41419-018-1131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishikawa C, Senba M, Mori N. Mitotic kinase PBK/TOPK as a therapeutic target for adult Tcell leukemia/lymphoma. Int J. Oncol. 2018;53:801–814. doi: 10.3892/ijo.2018.4427. [DOI] [PubMed] [Google Scholar]
- 38.Pirovano G, et al. TOPK modulates tumour-specific radiosensitivity and correlates with recurrence after prostate radiotherapy. Br. J. Cancer. 2017;117:503–512. doi: 10.1038/bjc.2017.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang MY, et al. PDZ binding kinase (PBK) is a theranostic target for nasopharyngeal carcinoma: driving tumor growth via ROS signaling and correlating with patient survival. Oncotarget. 2016;7:26604–26616. doi: 10.18632/oncotarget.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang CF, et al. PBK/TOPK expression predicts prognosis in oral cancer. Int. J. Mol. Sci. 2016;17:1007. doi: 10.3390/ijms17071007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DJ, et al. Novel TOPK inhibitor HI-TOPK-032 effectively suppresses colon cancer growth. Cancer Res. 2012;72:3060–3068. doi: 10.1158/0008-5472.CAN-11-3851. [DOI] [PubMed] [Google Scholar]
- 42.Matsuo Y, et al. TOPK inhibitor induces complete tumor regression in xenograft models of human cancer through inhibition of cytokinesis. Sci. Transl. Med. 2014;6:259ra145. doi: 10.1126/scitranslmed.3010277. [DOI] [PubMed] [Google Scholar]
- 43.Gao G, et al. ADA-07 suppresses solar ultraviolet-induced skin carcinogenesis by directly inhibiting TOPK. Mol. Cancer Ther. 2017;16:1843–1854. doi: 10.1158/1535-7163.MCT-17-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Azimi F, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J. Clin. Oncol. 2012;30:2678–2683. doi: 10.1200/JCO.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- 45.Miao YR, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv. Sci. 2020;7:1902880. doi: 10.1002/advs.201902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szczerba BM, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566:553–557. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- 47.Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer. 2016;16:431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- 48.Olingy CE, Dinh HQ, Hedrick CC. Monocyte heterogeneity and functions in cancer. J. Leukoc. Biol. 2019;106:309–322. doi: 10.1002/JLB.4RI0818-311R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodgins JJ, Khan ST, Park MM, Auer RC, Ardolino M. Killers 2.0: NK cell therapies at the forefront of cancer control. J. Clin. Investig. 2019;129:3499–3510. doi: 10.1172/JCI129338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 2016;17:1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 51.Ding T, Yan F, Cao S, Ren X. Regulatory B cell: new member of immunosuppressive cell club. Hum. Immunol. 2015;76:615–621. doi: 10.1016/j.humimm.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Kochenderfer JN, et al. Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J. Clin. Oncol. 2015;33:540–549. doi: 10.1200/JCO.2014.56.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luckheeram RV, Zhou R, Verma AD, Xia B. CD4(+)T cells: differentiation and functions. Clin. Dev. Immunol. 2012;2012:925135. doi: 10.1155/2012/925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamphorst AO, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc. Natl Acad. Sci. USA. 2017;114:4993–4998. doi: 10.1073/pnas.1705327114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vinay DS, et al. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 2015;35:S185–S198. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin. Cancer Res. 2015;21:687–692. doi: 10.1158/1078-0432.CCR-14-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilky BA. Immune checkpoint inhibitors: the linchpins of modern immunotherapy. Immunol. Rev. 2019;290:6–23. doi: 10.1111/imr.12766. [DOI] [PubMed] [Google Scholar]
- 58.Alachkar H, et al. T-LAK cell-originated protein kinase presents a novel therapeutic target in FLT3-ITD mutated acute myeloid leukemia. Oncotarget. 2015;6:33410–33425. doi: 10.18632/oncotarget.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ikeda Y, et al. T-LAK cell-originated protein kinase (TOPK) as a prognostic factor and a potential therapeutic target in ovarian cancer. Clin. Cancer Res. 2016;22:6110–6117. doi: 10.1158/1078-0432.CCR-16-0207. [DOI] [PubMed] [Google Scholar]
- 60.Zhou H, et al. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 2013;12:260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 61.Bian Y, et al. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J. Proteom. 2014;96:253–262. doi: 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 62.Smith ER, et al. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat. Rev. Genet. 2012;13:123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- 64.Chelmicki T, et al. MOF-associated complexes ensure stem cell identity and Xist repression. Elife. 2014;3:e02024. doi: 10.7554/eLife.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, et al. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ravens, S. et al. Mof-associated complexes have overlapping and unique roles in regulating pluripotency in embryonic stem cells and during differentiation. Elife3, e02104 (2014). [DOI] [PMC free article] [PubMed]
- 67.Deng X, et al. Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life, and MOF-mediated H4K16 acetylation. Dev. Cell. 2013;25:55–68. doi: 10.1016/j.devcel.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bao YN, et al. Urokinase-type plasminogen activator receptor signaling is critical in nasopharyngeal carcinoma cell growth and metastasis. Cell Cycle. 2014;13:1958–1969. doi: 10.4161/cc.28921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suh WK, et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat. Immunol. 2003;4:899–906. doi: 10.1038/ni967. [DOI] [PubMed] [Google Scholar]
- 70.Prasad DV, et al. Murine B7-H3 is a negative regulator of T cells. J. Immunol. 2004;173:2500–2506. doi: 10.4049/jimmunol.173.4.2500. [DOI] [PubMed] [Google Scholar]
- 71.Zhu F, et al. Bidirectional signals transduced by TOPK-ERK interaction increase tumorigenesis of HCT116 colorectal cancer cells. Gastroenterology. 2007;133:219–231. doi: 10.1053/j.gastro.2007.04.048. [DOI] [PubMed] [Google Scholar]
- 72.Roh E, et al. Suppression of the solar ultraviolet-induced skin carcinogenesis by TOPK inhibitor HI-TOPK-032. Oncogene. 2020;39:4170–4182. doi: 10.1038/s41388-020-1286-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu K, et al. Sunlight UV-induced skin cancer relies upon activation of the p38alpha signaling pathway. Cancer Res. 2013;73:2181–2188. doi: 10.1158/0008-5472.CAN-12-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zykova TA, et al. The T-LAK cell-originated protein kinase signal pathway promotes colorectal cancer metastasis. EBioMedicine. 2017;18:73–82. doi: 10.1016/j.ebiom.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu H, et al. TOPK inhibits autophagy by phosphorylating ULK1 and promotes glioma resistance to TMZ. Cell Death Dis. 2019;10:583. doi: 10.1038/s41419-019-1805-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao J, et al. MicroRNA-187, down-regulated in clear cell renal cell carcinoma and associated with lower survival, inhibits cell growth and migration though targeting B7-H3. Biochem. Biophys. Res. Commun. 2013;438:439–444. doi: 10.1016/j.bbrc.2013.07.095. [DOI] [PubMed] [Google Scholar]
- 77.Nygren MK, et al. Identifying microRNAs regulating B7-H3 in breast cancer: the clinical impact of microRNA-29c. Br. J. Cancer. 2014;110:2072–2080. doi: 10.1038/bjc.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.