Abstract
Background
The crucial factor in the production of bio-fuels is the choice of potent microorganisms used in fermentation processes. Despite the evolving trend of using bacteria, yeast is still the primary choice for fermentation. Molecular characterization of many genes from baker’s yeast (Saccharomyces cerevisiaea), and fission yeast (Schizosaccharomyces pombe), have improved our understanding in gene structure and the regulation of its expression. This in silico study was done with the aim of analyzing the promoter regions, transcription start site (TSS), and CpG islands of genes encoding for alcohol production in S. cerevisiaea S288C and S. pombe 972h-.
Results
The analysis revealed the highest promoter prediction scores (1.0) were obtained in five sequences (AAD4, SFA1, GRE3, YKL071W, and YPR127W) for S. cerevisiaea S288C TSS while the lowest (0.8) were found in three sequences (AAD6, ADH5, and BDH2). Similarly, in S. pombe 972h-, the highest (0.99) and lowest (0.88) prediction scores were obtained in five (Adh1, SPBC8E4.04, SPBC215.11c, SPAP32A8.02, and SPAC19G12.09) and one (erg27) sequences, respectively. Determination of common motifs revealed that S. cerevisiaea S288C had 100% coverage at MSc1 with an E value of 3.7e−007 while S. pombe 972h- had 95.23% at MSp1 with an E value of 2.6e+002. Furthermore, comparison of identified transcription factor proteins indicated that 88.88% of MSp1 were exactly similar to MSc1. It also revealed that only 21.73% in S. cerevisiaea S288C and 28% in S. pombe 972h- of the gene body regions had CpG islands. A combined phylogenetic analysis indicated that all sequences from both S. cerevisiaea S288C and S. pombe 972h- were divided into four subgroups (I, II, III, and IV). The four clades are respectively colored in blue, red, green, and violet.
Conclusion
This in silico analysis of gene promoter regions and transcription factors through the actions of regulatory structure such as motifs and CpG islands of genes encoding alcohol production could be used to predict gene expression profiles in yeast species.
Keywords: Alcohol production, CpG islands, Motifs, Saccharomyces cerevisiaea S288C, Schizosaccharomyces pombe 972h-
Background
The scarcity and rising prices of fossil fuels, geo-political instability in countries that hold most of the proven oil reserves together with apprehension about the environmental harm created by them, have resulted in increasing efforts to search for alternative energy sources [1]. Hence, production and use of bio-fuels for transport fuel has recently attracted significant attention worldwide. Likewise, Ethiopia’s sustainable development and the national fuel security can only be realized with increased production and utilization of renewable fuels. Substituting the demand for fossil fuel by locally produced fuels such as bio-ethanol and bio-diesel is paramount importance for the country’s economic use of scarce energy resources.
Bio-ethanol has been made since ancient times by fermenting sugars, and most bio-ethanol used for fuel and alcoholic drinks, and most industrial ethanol, is made by this process (Licht 2001; as cited in [2]). Great strides in research together with the development of new yeast strains have led to demands to model a new yeast strain which can withstand and produce at higher levels of alcohol, temperatures, and pH. This requires immense knowledge of the fermentation processes to improve its efficiency which is dependent on various factors, namely, process design, molasses quality, yeast strain, contamination, nutrient availability, and raw material purity [2]. Yeast alcohol is one of the most valuable products originating from the biotechnological industry with respect to both value and amount [3]. Yeast selection for fuel ethanol production over the past two decades and most bio-ethanol-related researches in developing tropical countries have focused primarily on the isolation of local Saccharomyces yeasts and their use for industrial ethanol production [4–6].
The development of DNA transformation in yeast has made possible the rapid molecular isolation of many genes from baker’s yeast (Saccharomyces cerevisiaea) and fission yeast (Schizosaccharomyces pombe). Concomitantly, our understanding of many aspects of gene structure and regulation of gene expression in these organisms has improved. Although the two organisms are similar in that they are both spore-forming yeasts capable of strong alcoholic fermentation, S. pombe actually has diverged significantly from S. cerevisiaea [7]. In recent years, genome mining and in silico analysis of gene sequences and their products have become a key methodology to identify gene expression patterns, sequences responsible for development of new molecules, leading to the discovery of dozens of novel compounds [8, 9]. A variety of computational tools have been developed to support scientists in this field. Most of the available tools are dedicated to the in silico analysis of specific gene and gene products [8].
Gene expression varies among tissues and even different cell types but also in response to specific signals (physiological, environmental, etc.). The main mechanism of transcriptional regulation is orchestrated by proteins called transcription factors (TFs), which promote (as activators) or block (as repressors) the recruitment of the RNA polymerase II (Pol II complex) [10]. The promoter is a DNA sequence that the transcription apparatus recognizes and binds. It indicates which of the two DNA strands is to be read as the template and the direction of transcription [11]. It is a functional region containing complex regulatory elements for determining the transcription initiation of genes [9, 10, 12, 13]. DNA-binding sites or motifs refer to short DNA sequences (typically 4 to 30 base pairs long, but up to 200 bp for recombination sites) that are explicitly bound by one or more DNA-binding proteins or protein complexes [14]. It is often associated with specialized proteins known as transcription factors and is thus linked to transcriptional regulation. A structural feature that has proven useful in the detection of promoters is the so called CpG islands, i.e., regions that are rich in CpGs, which are important because of their strong link with gene regulation[15]. CpG islands are playing an important role in gene regulation through epigenetic changes [16].
Therefore, the aim of this study is to predict promoter and regulatory elements of genes encoding alcohol production in yeast species (Saccharomyces cerevisiaea S288C and Schizosaccharomyces pombe 972h-) thereby providing basic information which could support the effort of improving them for a commercial-scale bio-ethanol production.
Methods
Determination of transcription start sites and promoter regions for genes encoding alcohol production
Gene sequences of yeast species (Saccharomyces cerevisiaea S288C and Schizosaccharomyces pombe 972h-) for genes encoding alcohol production were retrieved as FASTA file from NCBI Genome Browser (https://www.ncbi.nlm.nih.gov/gene). Gene sequences starting by ATG (starting codon) were identified, and coding sequences were used in this analysis. However, for S. pombe 972h-, all the sequences retrieved from direct NCBI web were not having the functional gene structure (no ATG in the beginning and many stop codons in the middle). Therefore, sequences used for the current study were retrieved via NCBI Reference Sequences (RefSeq). This section includes genomic Reference Sequences (RefSeqs) from all assemblies on which these genes were annotated, such as RefSeqs for chromosomes and scaffolds (contigs) from both reference and alternate assemblies. To this end, gene sequences were taken as identical protein annotated from PomBase annotation Provider for Eukaryotic Annotation Propagation Pipeline.
Generally, 23 and 21 sequences of alcohol dehydrogenase were retrieved for S. cerevisiaea S288C and S. pombe 972h-, respectively. To determine respective transcriptional start sites (TSSs) for all gene sequences, about 1-kb sequences upstream of the start codon were excised from all genes except for ADH1 of S. cerevisiaea S288C which was at 2-kb upstream of start codon. Similarly, all gene sequences of S. pombe 972h-, except three genes (erg27 at 2.5 kb and SPBC8E4.04 and SPBC337.11 at 2 kb), had TSSs at 1-kb sequences upstream of their start codons.
The Neural Network Promoter Prediction (NNPP version 2.2) tool set was used with the minimum standard predictive score (between 0 and 1) [17]. For those regions containing more than one TSS, the one with the highest value of prediction score was considered to have trustable and truthful prediction. Promoter regions were defined as 1-kb region upstream of each TSS. For those regions containing more than one TSS, the highest value of prediction score will be considered so as to have a more accurate prediction.
Determination of common motifs and TFs for genes encoding alcohol production in the promoter region
Analysis of conserved motifs for genes encoding for alcohol productions for both yeast species was performed by MEME (Multiple Em for Motif Elicitation) software version 3.5.4 (http://meme.sdsc.edu). This online web-based analysis was performed with minimum and maximum motif width of 6 and 50 residues, respectively, for both yeast species whereas a maximum number of motifs for S. cerevisiaea S288C and S. pombe 972h- were 23 and 21, respectively, which were used to identify probable promoter regulatory elements (motifs), keeping the rest of the parameters at default. The MEME output in HTML showed the motifs as local multiple alignments of the input sequences, as well as in several other formats. Buttons on the MEME HTML output were allowed one or all of the motifs to be forwarded for additional investigation. Descriptions the identification of motifs by TOMTOM [18] web server were designated where numerous sequence databases can be searched for sequences matching the identified motif, in which the output of TOMTOM will include LOGOS on behalf of the alignment of two motifs, the p value and q value (a measure of false discovery rate) of the match [18]. TOMTOM showed that the query motif closely resembles the binding motif in the set of genes encoding for alcohol production promoter regions.
Search for CpG islands for genes encoding alcohol production promoter regions
To search CpG islands, first, the stringent search criteria were used in the Takai and Jones algorithm: GC content ≥ 50%, Obs CpG/ExpCpG ≥ 0.60, and length ≥ 200 bp [20]. For this purpose, the CpG island searcher program (CpGi130) available at web link (http://www.bioinformatics.org/sms/cpg_island.html) was used. The CpG island graphs were plotted using EMBOSS Cpgplot (https://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/) which identify and plot CpG islands in nucleotide sequence(s). Secondly, the CLC Genomics Workbench ver. 3.6.5 (http://clcbio.com, CLC bio, Aarhus, Denmark) was used for restriction enzyme MSpI cutting sites (fragment sizes between 40 and 220 bp).
Phylogenetic analysis
Phylogenetic analysis of gene sequences of both yeast species (S. cerevisiaea S288C and S. pombe 972h-) was conducted using the Molecular Evolution Genetic Analysis 6 (MEGA6) tool by the neighbor-joining tree-making method. Similarly, Tajima’s neutrality test of selection was conducted using the same software to find nucleotide diversity. The p-distance model was applied with transition and trans-version nucleotide substitution. Bootstrap values of the super tree were computed with 2000 repetitions with uniform rate among sites and complete deletion of gaps/missing data were used to analyze the sequences.
Results
Determination of transcription start sites and promoter regions for genes encoding alcohol production
Promoter region analysis of genes encoding for alcohol productions of both yeast species (S. cerevisiaea S288C and S. pombe 972h-) showed a great variation in the number of TSS. The highest promoter prediction scores (1.0) for TSS of S. cerevisiaea S288C alcohol dehydrogenase were obtained for five gene sequences (AAD4, SFA1, GRE3, YKL071W, andYPR127W) while the lowest promoter prediction scores (0.8) were obtained for three gene sequences (AAD6, ADH5, and BDH2) (Table 1). In addition, the result of promoter predictions for S. cerevisiaea S288C sequences with score cutoff 0.80 showed that out of twenty-three gene sequences used in this analysis only ADH1 and ADH7 (8.70%) had showed a single TSS while the remaining (91.30%) showed multiple TSS. In these scenarios, TSSs with the highest prediction scores were considered for further uses. TSSs of genes encoding for alcohol production in S. cerevisiaea S288C were mostly located in the upstream region of − 31 to − 1545 bp, with the relatively highest incidence of occurrence in the upstream region of − 1 to − 200 bp (10 sequences, 43%) followed by − 201 to − 400 bp and − 601 to − 800 bp (4 sequences each, 17.4%) from the transcription start site, while the lowest occurrence was observed at − 801 to − 1000 bp and above − 1000 bp (only 1 sequence each).
Table 1.
TSS number, its promoter predictive score values, and distance from the start codon of S. cerevisiaea S288C
| Gene name | Gene ID | Location | Number of TSS | Predictive score at cutoff value of 0.8 | Distance from start codon (ATG) |
|---|---|---|---|---|---|
| Alcohol dehydrogenase ADH1 | 854068 | Chromosome XV; NC_001147.6 (c160594-159548) | 1 | 0.99 | − 1545 |
| Alcohol dehydrogenase ADH2 | 855349 | Chromosome XIII; NC_001145.3 (c874337-873291) | 4 | 0.98, 0.93, 0.92, 0.91 | − 130, − 467, − 197, − 177 |
| S-(Hydroxymethyl) glutathione dehydrogenase SFA1 | 851386 | Chromosome IV; NC_001136.10 (159604-160764) | 3 | 1.00, 0.94, 0.89 | − 789, − 159, − 531 |
| Alcohol dehydrogenase ADH3 | 855107 | Chromosome XIII; NC_001145.3 (434788-435915) | 3 | 0.91, 0.89, 0.88 | − 31, − 639, − 657 |
| Alcohol dehydrogenase ADH4 | 852636 | Chromosome VII; NC_001139.9 (15159-16307) | 2 | 0.89, 0.86 | − 95, − 120 |
| Alcohol dehydrogenase ADH5 | 852442 | Chromosome II, NC_001134.8 (533762-534817) | 6 | 0.96, 0.91, 0.86, 0.84, 0.8, 0.8 |
− 90, − 737, − 224, − 703, − 688, − 715 |
| Alcohol dehydrogenase ADH6 | 855368 | Chromosome XIII, NC_001145.3 (c912143-911061) | 3 | 0.94, 0.93, 0.89 | − 215, − 822, − 71 |
| NADP-dependent alcohol dehydrogenase (ADH7) | 850469 | Chromosome III, NC_001135.5 (309070-310155) | 1 | 0.93 | − 63 |
| Putative aryl-alcohol dehydrogenase (AAD3) | 850471 | Chromosome III, NC_001135.5 (313890-314981) | 2 | 0.98, 0.98 | − 133, − 199 |
| Putative aryl-alcohol dehydrogenase AAD4 | 851354 | Chromosome IV, NC_001136.10 (c17577-18566) | 4 | 1.00, 0.96, 0.92, 0.90 | − 759, − 356, − 245, − 42 |
| Aryl-alcohol dehydrogenase (NADP+) activity (AAD6) | 850488 | Chromosome VI, NC_001138.5 (c14305-15431) | 5 | 0.97, 0.89, 0.86, 0.85, 0.80 | − 101, − 518, − 734, − 457, − 28 |
| Putative aryl-alcohol dehydrogenase AAD10 | 853620 | Chromosome X, NC_001142.9 (727405-728271) | 3 | 0.97, 0.86, 0.83 | − 654, − 359, − 171 |
| Putative aryl-alcohol dehydrogenase AAD14 | 855385 | Chromosome XIV, NC_001146.8 (c16118-17248) | 2 | 0.94, 0.81 | − 663, − 452 |
| Putative aryl-alcohol dehydrogenase AAD15 | 853999 | Chromosome XV, NC_001147.6 (c1647-2078) | 2 | 0.94, 0.86 | − 492, − 955 |
| D-Arabinose 1-dehydrogenase (NAD(P)(+)) ARA1 | 852446 | Chromosome II, NC_001134.8 (539987..541021) | 2 | 0.98, 0.95 | − 147, − 92 |
| Glucose-6-phosphate dehydrogenase (ZWF1) | 855480 | Chromosome XIV, NC_001146.8 (c196426..197943) | 4 | 0.99, 0.94, 0.86, 0.85 | − 318, − 145, − 636, − 120 |
| Glycerol 2-dehydrogenase (NADP(+)) GCY1 | 854287 | Chromosome XV, NC_001147.6 (551114-552052) | 4 | 0.99, 0.97, 0.92, 0.91 | − 357, − 85,− 899, − 829 |
| Trifunctional aldehyde reductase/xylose reductase/glucose 1-dehydrogenase (NADP(+))GRE3 | 856504 | Chromosome VIII, NC_001140.6 (323409-324392) | 3 | 1.00, 0.96, 0.90 | − 355, − 778, − 679 |
| Putative dehydrogenase BDH2 | 851238 | Chromosome I, NC_001133.9 (33448-34701) | 2 | 0.99, 0.8 | − 146, − 154 |
| Glucose 1-dehydrogenase (NADP(+)) YPR1 | 851974 | Chromosome IV, NC_001136.10 (1213904-1214842) | 3 | 0.87, 0.83, 0.83 | − 97, − 10, − 252 |
| Hypothetical protein YKL071W | 853792 | Chromosome XI, NC_001143.9 (305114-305884) | 3 | 1.00, 0.95, 0.93 | − 73, − 513, − 881 |
| Hypothetical protein YFL041W-A | 1466401 | Chromosome VI, NC_001138.5 (48734..48925) | 2 | 0.97, 0.84 | − 866, − 115 |
| Pyridoxine 4-dehydrogenase (YPR127W) | 856245 | Chromosome XVI, NC_001148.4 (790083..791120) | 2 | 1.00, 0.83 | − 437, − 776 |
Note: Figures in italics were taken as cutoff point and respective distances for more than one TSS
Likewise, twenty-one sequences of genes encoding for alcohol production for S. pombe 972h- (fission yeast) were retrieved from NCBI Genome Browser. Accordingly, the result of TSS and promoter analysis had showed a significant variation in the number of TSS and the distance of TSS from the start codons (Table 2). The numbers of TSSs were varied from 1 to 3 with majority of sequences (71.43%) having more than one TSS. In particular to this fact, out of twenty-one sequences six, seven, and eight sequences had one, two, and three TSSs respectively. The relative locations of all TSS with respect to start codon were given in Table 2. The nearest TSS were recorded for SPCC13B11.04c (− 39) followed by Adh1 (− 77) while the far-flanged TSS were observed for egr27 (− 2308) followed by SPBC337.11 (− 1636) upstream of the start codons of their respective genes. The current analysis also revealed that the relatively highest frequency of occurrence in the upstream region of − 1 to − 200 bp and − 201 to − 400 bp (6 sequences each, 57.1% share) followed by − 401 to − 600 bp (4 sequences, 19.04%) from the transcription start site, while the lowest occurrences were observed at − 601 to − 800 and − 801 to − 1000 bp (only 1 sequence each) and whereas three sequences had their TSS at above − 1000 bp (Table 1). The predictive score at a cutoff value of 0.8 ranged from 0.99 to 0.88.
Table 2.
TSS number, its promoter predictive score values, and distance from start codon for S. pombe 972h-
| Gene name | Gene ID | Location | Number of TSS | Predictive score at cutoff value of 0.8 | Distance from start codon (ATG) |
|---|---|---|---|---|---|
| Alcohol dehydrogenase Adh1 | 2538902 |
Chromosome III, NC_003421.2 (158942-1592889) |
1 | 0.99 | − 77 |
| Alcohol dehydrogenase Adh4 | 2542714 |
Chromosome I, NC_003424.3 (c156430-158279) |
2 | 0.98, 0.89 | − 565, − 85 |
| Putative alcohol dehydrogenase (SPBC1773.06c) | 2540139 | Chromosome II, NC_003423.3 (c293225-294725 | 2 | 0.97, 0.87 | − 120, − 39 |
| Pyridoxal reductase Plr1 | 2542917 | Chromosome I, NC_003424.3 (4461697-4462901) | 2 | 0.95, 0.90 | − 89, 246 |
| Putative aldo/keto reductase (SPBC8E4.04) | 2541256 | Chromosome II, NC_003423.3 (c4432247-4433501) | 1 | 0.99 | − 1632 |
| Putative glutathione-dependent formaldehyde dehydrogenase SPCC13B11.04c | 2538802 | Chromosome III, NC_003421.2 (c1596549-1599028) | 3 | 0.92, 0.80, 0.80 | − 39, − 59, − 295 |
| NADH/NADPH-dependent indole-3-acetaldehyde reductase AKR3C2 (SPAC19G12.09) | 2542483 | Chromosome I, NC_003424.3 (4059980-4061192) | 3 | 0.99, 0.96, 0.92 | − 211, − 42, − 560 |
| Putative dehydrogenase SPAC2E1P3.01: | 2541784 | Chromosome: I; NC_003424.3 (2922761..2924720) | 2 | 0.98, 0.95 | − 126, − 848 |
| Putative 3-oxoacyl-[acyl-carrier-protein] reductase Oar2 | 2543512 | Chromosome: I; NC_003424.3 (c3180426-3181164) | 3 | 0.97, 0.89, 0.83 | − 402, − 746, 795 |
| Aldose reductase ARK13 family YakC (yak3) | 2541648 | Chromosome I, NC_003424.3 (4248777-4250231) | 1 | 0.92 | − 260 |
| Putative xylose and arabinose reductase (SPAP32A8.02) | 2541584 | Chromosome I, NC_003424.3 (3405955-3409780) | 2 | 0.99, 0.97 | − 273, − 353 |
| NADP-dependent glucose 1-dehydrogenase (SPAC26F1.07) | 2542088 | Chromosome I; NC_003424.3 (c5170789..5172112) | 3 | 0.98, 0.96, 0.84 | − 227, − 47, 290 |
| Hypothetical protein SPBC16A3.02c | 2540039 | Chromosome II, NC_003423.3 (4296979-4298874) | 3 | 0.97, 0.93, 0.86 | − 493, − 602, − 731 |
| Aldo-keto reductase family protein (SPAC977.14c) | 2543325 | Chromosome I, NC_003424.3 (c59614-60907) | 1 | 0.89 | − 783 |
| Putative pyridoxal reductase (SPCC1281.04) | 2539165 | Chromosome III, NC_003421.2 (1386524-1387961) | 3 | 0.95, 0.90, 0.87 | − 291, − 345, − 433 |
| Aldo/keto reductase family protein (SPAC750.01) | 3361570 | Chromosome I, NC_003424.3 (5555716-5556768) | 1 | 0.98 | − 927 |
| Putative xylose and arabinose reductase (SPAC2F3.05c) | 2541958 | Chromosome I, NC_003424.3 (c3928625-3929950) | 2 | 0.92, 0.81 | − 520, − 494 |
| Putative pyridoxal reductase (SPAC3A11.11c) | 2543167 | Chromosome I, NC_003424.3 (3446262-3448081) | 2 | 0.97, 0.95 | − 146, − 878 |
| Aldo/keto reductase family protein (SPBC215.11c) | 2540698 | Chromosome II, NC_003423.3 (c4050360-4051532) | 3 | 0.99, 0.97, 0.94 | − 380, − 688, − 753 |
Note: Figures in italics were taken as the cutoff point and respective distances for more than one TSS
Determination of common motifs and TFs for genes encoding alcohol production in the promoter region
The promoter regions shared by majority of the gene sequences used in the current study revealed that S. cerevisiaea S288C had 100% coverage among the gene sequences at MSc1 with an E value of 3.7e−007 and 15 motif widths (Table 3).
Table 3.
List of discovered motifs and the number and proportion of promoter-containing motifs for S. cerevisiaea S288C
| Discovered motif | Number (%) of promoter-containing motifs | E value | Motif width | Total number of binding sites |
|---|---|---|---|---|
| MSc1 | 23 (100%) | 3.7e−007 | 15 | 23 |
| MSc2 | 18 (78.26%) | 1.8e+004 | 29 | 18 |
| MSc3 | 8 (34.78%) | 2.5e+004 | 10 | 8 |
| MSc4 | 3 (13.04%) | 4.8e+004 | 29 | 3 |
| MSc5 | 7 (30.43%) | 1.6e+004 | 29 | 7 |
MSc motif for S. cerevisiaea S288C
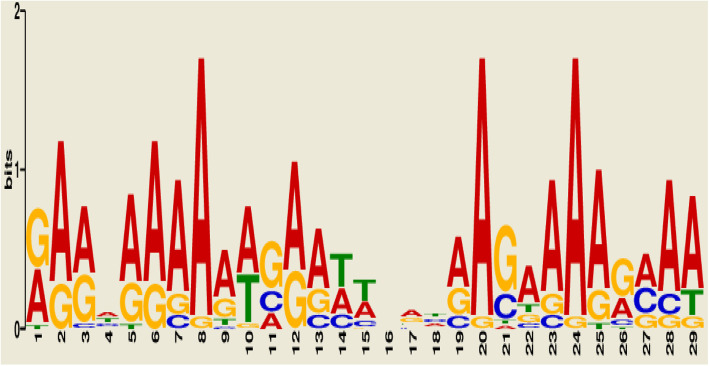
To determine motifs which are functionally important, motifs which were shared by majority of promoter regions of S. cerevisiaea S288C genes encoding for alcohol production were chosen. Accordingly, MSc1 was revealed as the common promoter motif for all (100%) genes that serves as binding sites for transcription factors involved in the expression regulation of these genes. Sequence logo for MSc1 generated by MEME is presented in Fig. 1
Fig. 1.
Sequence logo for the identified common promoter motif MSc1 gene for gene encoding alcohol production gene in S. cerevisiaea S288C
Likewise, S. pombe 972h- promoter sequences had 95.23% conserved motif at MSp1 with E value of 2.6e+002 and 29 motif widths (Table 4). The common motifs of MSp1 shared by majority of the genes encoding for alcohol production sequences of S. pombe 972h- as generated by MEME revealed in Fig. 2.
Table 4.
List of discovered motifs and the number and proportion of promoter-containing motifs for S. pombe 972h-
| Discovered motif | Number (%) of promoter-containing motifs | E value | Motif width | Total number of binding sites |
|---|---|---|---|---|
| MSp1 | 20 (95.23%) | 2.6e+002 | 29 | 20 |
| MSp2 | 4 (19.04%) | 1.5e+004 | 48 | 4 |
| MSp3 | 13 (61.90%) | 1.8e+004 | 15 | 13 |
| MSp4 | 6 (28.57%) | 2.2e+004 | 21 | 6 |
| MSp5 | 5 (23.81%) | 1.8e+005 | 19 | 5 |
MSp motif for S. pombe
Fig. 2.
Sequence logo for the identified common promoter motif MSp1 for gene encoding alcohol production gene in S. pombe
Furthermore, candidate transcription factor proteins were identified for motifs (MSc1 and MSp1) of both yeast species. They were then compared to the registered motifs in publicly available database of Jaspar2018_Core_Fungi_Non-Redundant DNA so as to see if they are similar to known regulatory motifs using TOMTOM web application [19]. The output from TOMTOM also links back to the parent motif database for more detailed information on biological functions of the matched motif. As a result, 13 motifs out of 176 common promoter motif/transcription factors were identified for MSc1 while only 9 motifs out of 176 in MSp1 were being found matched with known motifs found in JASPAR 2018 CORE fungi motif databases (Table 2). In addition, the comparison of the identified transcription factor proteins indicated that almost 88.88% (8 out of 9) of the MSp1 were exactly similar to that of MSc1. This could be due to the fact that these two yeast species (S. cerevisiaea S288C and S. pombe 972h-) shared common promoters and their subsequent transcription factor protein were highly conserved regions of the genes encoding for alcohol production. TOMTOM is a motif comparison algorithm that ranks the target motifs in a given database according to the estimated statistical significance of the match between the query and the target. In similar manner, TOMTOM provides LOGOS that represents the alignment of two motifs and a numeric score for the match between two motifs together with a statistical significance [21].
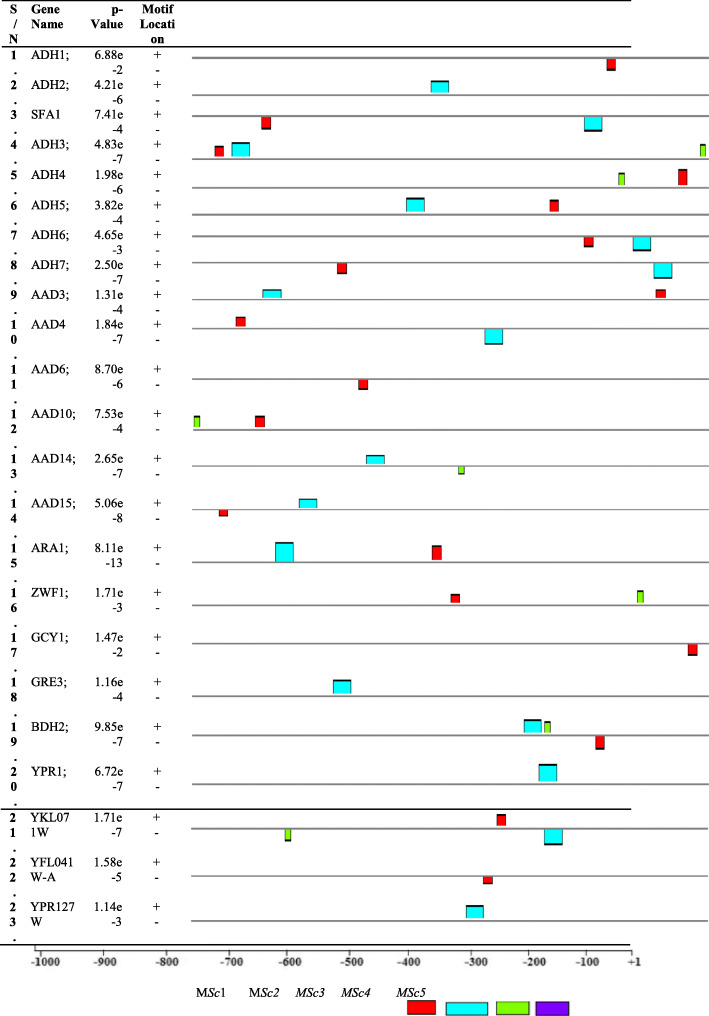
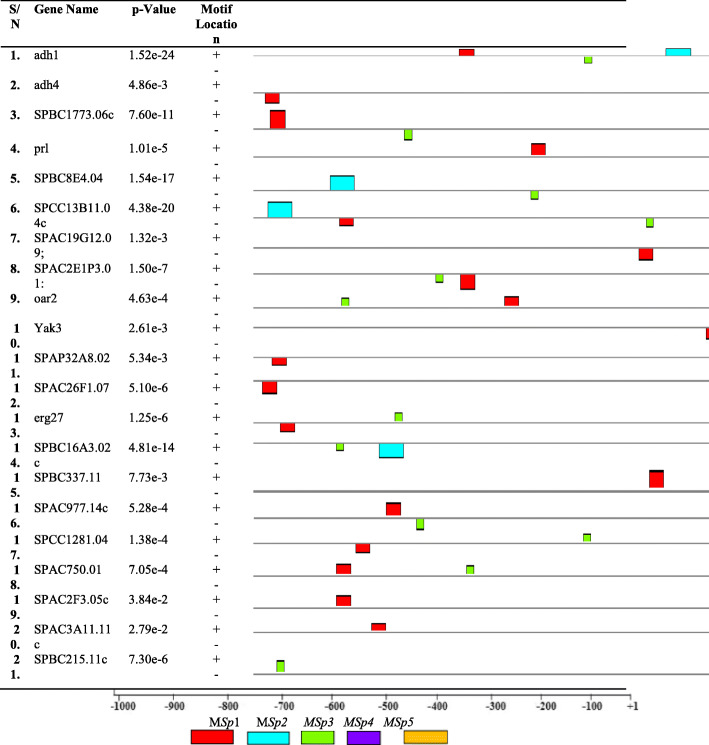
The total numbers of motifs discovered in S. cerevisiaea S288C for genes encoding alcohol production promoter regions were about 60 out of which relatively, higher distributions of motifs were found also in positive (39) than in negative (21) strands (Fig. 3). The location and distribution of these motifs were ranged from − 998 to − 1 while higher concentration of motifs was found between − 850 and − 50 bp of the transcription start sites (TSSs). In the same view, only 48 motifs were discovered in S. pombe 972h- out of which relatively, higher distributions of motifs were found also in negative (25) than in positive (23) strands (Fig. 4). The location and distribution of these motifs were ranged from − 996 to − 2 while higher concentration of motifs was found between − 800 and − 100 bp of the transcription start sites (TSSs).
Fig. 3.
The relative positions of motifs in alcohol dehydrogenase for S. cerevisiaea S288C sequences relative to TSSs
Fig. 4.
The positions and strand orientation of motifs of S. pombe alcohol production gene relative to TSSs
Search for CpG islands for genes encoding alcohol production in the promoter regions
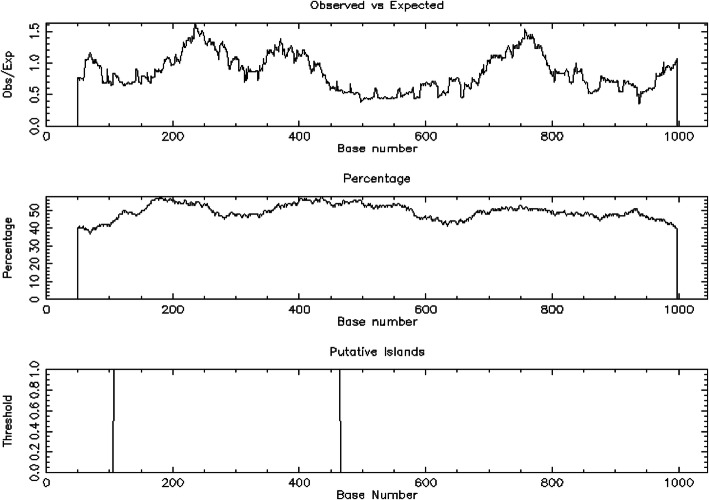
In the current study, revealed CpG islands were examined in the promoter and gene body regions of both yeast species using both Takai and Jones algorithm using parameters as indicated in this section of this study and CLC Genomics Workbench ver. 3.6.5. Accordingly, as per the stringent criteria of Takai and Jones [17] as indicated in this section, there were only five (ADH1 (Fig. 5), ADH2, ADH5, ZWF1, and BDH2) (21.73%) CpG islands observed in the gene body regions in analogous to only six (ADH1, SFA1, ADH3, ZWF1, BDH2, and YPR127W) out of twenty-three (26.08%) gene sequences used for the analysis in promoter regions of S. Cerevisiaea S288C yeast species. Likewise, only one (adh1) had CpG island in the promoter region and six (adh1, SPBC1773, SPCC13B11.04c, SPAC2E1P3.01, Yak3, and SPBC16A3.02c) CpG islands were observed in the gene body of genes encoding for alcohol production of S. pombe 972h-.
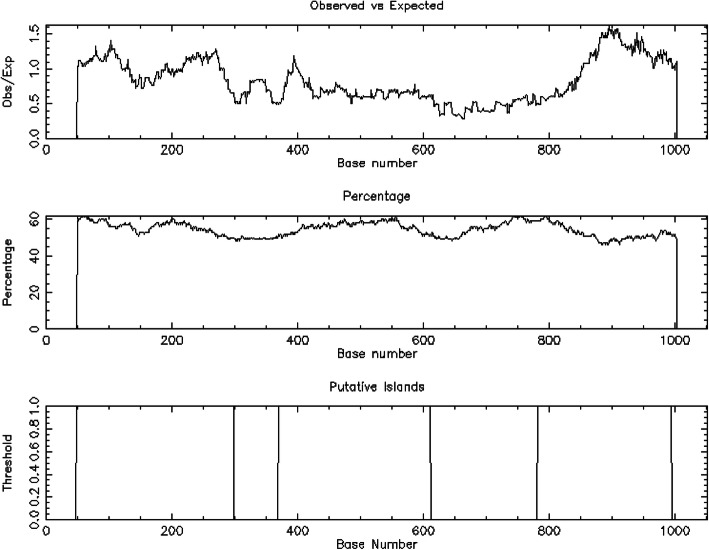
Fig. 5.
CPGPLOT islands of ADH1, with length 358 (108–465)
On the other hand, CLC genomics workbench ver 3.6.1 using restriction enzyme MSpI (C/CGG sequence) cutting sites with standard fragment sizes between 40 and 220 bp revealed that in S. cerevisiaea S288C five (ARA1, BDH2, GCY1, SFA1, and GRE3) and only one (GRE3) CpG islands were found in gene body and promoter regions, respectively. In contrarily, the number of CpG islands in the gene body was only one (SPBC16A3.02c) while four (adh1 (Fig. 6), adh4, SPAP32A8.02, and SPBC337.11) in the promoter regions of S. pombe 972h- in alcohol dehydrogenase (Table 5). This indicates the poor occurrence of CpG islands in both gene body and promoter regions which may affect the access of promoter region of genes to their transcription factors, hence preventing their expression.
Fig. 6.
CPGPLOT islands of three regions of adh1 gene; from 1 to 1053 with length 251 (49–299), length 243
Table 5.
Identification of MSpI cutting sites and fragment sizes for genes encoding for alcohol production body and promoter regions of S. cerevisiaea S288C and S. pombe 972h-
| S. Cerevisiaea S288C | S. pombe 972h- | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene body | Promoter regions | Sequence name | Gene body | Promoter regions | |||||
| Sequence name | No. of cut sites (cut position/s) | Fragment size | No. of cut sites (cut position/s) | Fragment size | No. of cut sites (cut position/s) | Fragment size | No. of cut sites (cut position/s) | Fragment size | |
| ADH1 | 1 (632) | – | adh1 | 2 (644, 743) | 99 | ||||
| ADH3 | 0 | – | 1 (827) | – | adh4 | 0 | – | 3 (71, 501, 570) | 69 |
| ADH4 | 1 (146) | – | 2 (276, 650) | – | SPBC1773.06c | 1 (940) | – | 0 | – |
| ADH5 | 2 (88, 539) | – | 1 (827) | – | prl | 1 (289) | – | 0 | – |
| ADH6 | 1 (992) | – | 2 (271, 979) | – | SPBC8E4.04 | 0 | – | 1 (736) | – |
| ADH7 | 0 | – | 1 (507) | – | SPCC13B11.04c | 1 (553) | – | 3 (95, 316, 506) | – |
| AAD10 | 0 | – | 1 (489) | – | SPAC19G12.09 | 1(401) | – | 0 | – |
| AAD14 | 3 (74, 314, 991) | – | 1 (440) | – | SPAC2E1P3.01 | 1 (825) | – | 0 | – |
| AAD15 | 1 (322) | – | 0 | – | Yak3 | 1 (511) | – | 0 | – |
| AAD3 | 0 | – | 0 | – | SPAP32A8.02 | 1 (353) | – | 2 (886, 933) | 47 |
| AAD4 | 2 (176, 928) | – | 2 (236, 902) | – | SPAC26F1.07 | 2 (74, 941) | – | 0 | – |
| AAD6 | 1 (1065) | – | 1 (428) | – | erg27 | 2 (29, 787) | – | 0 | – |
| ARA1 | 2 (307, 454) | 147 | 0 | – | SPBC16A3.02c | 2 (238, 431) | 193 | 0 | – |
| BDH2 | 3 (315, 481, 806) | 166 | 2 (411, 918) | – | SPBC337.11 | 2 (331, 577) | – | 2 (855, 948) | 93 |
| GCY1 | 2 (697, 737) | 40 | 2 (198, 977) | – | SPAC977.14c | 0 | – | 1 (739) | – |
| GRE3 | 1 (456) | – | 2 (331, 496) | 185 | SPCC1281.04 | 0 | – | 0 | – |
| SFA1 | 2 (221, 326) | 105 | 1 (273) | – | SPAC750.01 | 0 | – | 0 | – |
| YFL041W–A | 0 | – | 1 (768) | – | SPAC2F3.05c | 0 | – | 0 | – |
| YKL071W | 1 (550) | – | 1 (142) | – | SPAC3A11.11c | 1 (809) | – | 1 (942) | – |
| YPR127W | 1 (868) | – | 0 | – | SPBC215.11c | – | 0 | – | |
| YPR1 | 2 (206, 268) | 62 | 1 (81) | – | – | ||||
Phylogenetic analysis
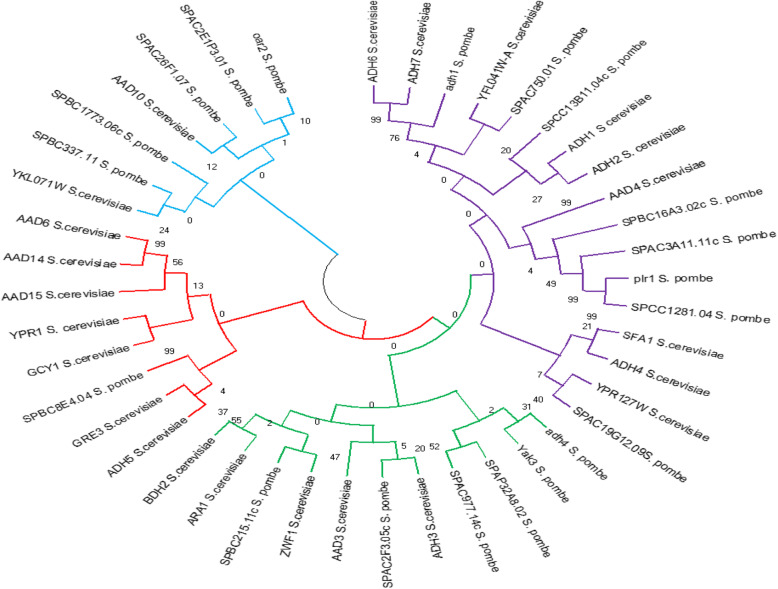
A combined analysis of all the data weighted equally resulted in a single most-parsimonious cladogram (Fig. 7). Statistics for this tree revealed that in general, relationships on the tree are very strongly supported. A phylogenetic tree was generated using the neighbor-joining (NJ) as well as minimum-evolution method of MEGA 6.0. As illustrated in Fig. 7, all sequences from both S. cerevisiaea S288C and S. pombe 972h- were divided into four subgroups (I, II, III, and IV). The four clades are respectively colored in blue, red, green, and violet. The phylogenetic tree indicated that some gene sequences irrespective of source organism clustered together within each sub-group suggest a close evolutionary relationship among the genes rather than the whole species.
Fig. 7.
Phylogenetic tree of genes encoding alcohol production sequence from S. cerevisiaea S288C and S. pombe 972h-
Discussion
Sequence-specific DNA-binding transcription factors (TFs) are often termed as “master regulators” which bind to DNA and either activate or repress gene transcription. Determination of a gene’s transcriptional start site underlies the identification of the proximal promoter region and thus facilitates the subsequent analysis of gene expression. In the current in silico analysis, majority of the sequences in both species had multiple transcription start sites. This could give the alternative transcription potential for the gene sequence under consideration. However, for better prediction, TSS with a higher cutoff value was considered in the current study. Many authors [22–24] have reported the presence of multiple transcription sites in genes encoding for alcohol production. The comparison of the two yeast species with this regard showed that S. cerevisiaea S288C had a nearby site than S. pombe 972h-. This may be due to the fact that S. cerevisiaea S288C heavily relies on genes encoding for alcohol production to convert aldehydes and ketones into alcohols and NADH to NAD+ that the yeasts can use for energy [25]. This process of yeasts turning aldehydes and ketones into alcohols is called fermentation. In general, promoter regions are located at the immediately upstream of a transcription start site (TSS) and have a variety of sequence motifs that participate in gene regulation [26].
The establishment and maintenance of temporal and spatial patterns of gene expression are achieved primarily by transcription regulation. Functionally important motifs are usually short conserved sequence pattern associated with distinct functions of DNA often serve as transcription factor biding sites. In the current in silico analysis using MEME, the conserved short sequences of S. cerevisiaea S288C had confirmed to have higher occurrencesthan that of S. pombe 972h-. Common motifs, short DNA segments, are binding sites for transcription factors [27]. In both cases, the probability of finding a well-conserved pattern in random sequences as evidenced by an E value is significantly higher than expected value. It is generally believed that genes having similar expression patterns contain common motifs in their promoter regions [28, 29]. Common promoter motifs are the key signatures for a family of co-regulated genes and are usually present in the regions where complex protein interactions occur [30]. However, in some cases, single motifs can bind various transcription factors thereby bringing the genes under multiple regulatory controls [31, 32]. Extensive studies on 500 bp upstream regions of yeast promoters suggest that regulatory elements are commonly present in those regions [33].
It is well reported that CGIs are highly involved in gene regulatory processes [34]. They are present at or near the gene’s transcription start site and are often associated with the promoters of most house-keeping genes and many tissue-specific genes, and thus have important regulatory functions and can be used as gene markers [35].
A phylogenetic tree generated in the current study showed that all sequences from both S. cerevisiaea S288C and S. pombe 972h- were divided into four subgroups. This could be due to the fact that some vital genes for alcohol fermentation of yeast strains. Similar studies by other authors [34, 35] have revealed that one organism which is equally amenable to genetic manipulation as is S. cerevisiaea is the fission yeast S. pombe. Although the two organisms are similar in that they are ascospore-forming yeasts capable of strong alcoholic fermentation, S. pombe actually has diverged significantly from S. cerevisiaea [36, 37]. In fact, the available information suggests that S. pombe may actually be more closely related to filamentous fungi such as Neurospora and Asperglus than it is to budding yeast [9, 38].
Conclusion
The enhancement of DNA transformation in yeast has resulted in revolutionized molecular tools and facilitated ease of thought of aspects of gene structure and regulation of gene expression of many genes from baker’s yeast (S. cerevisiaea) and fission yeast (S. pombe). Generally, the regulation of alcohol dehydrogenase enzyme which is critically important for the survival and enhanced efficiency of yeast species can better be examined and explained with the use of increasing technological advancement, genome mining, and in silico analysis of gene sequences. Majority of the genes encoding for alcohol production sequences have multiple TSS in both yeast species suggesting alternative gene regulation.
The result of this analysis could be critically important to understand the nature of promoter regions, the motif discovered in line with the transcription factor binding proteins, and the strength of the genome to different transcriptions via following the frequency of CpG islands. The phylogenetic analysis revealed that majority of genes is clustered together irrespective of the source organism suggesting a close evolutionary relationship among the gene rather than the whole species.
In general, this in silico analysis of genes encoding for alcohol production of S. cerevisiaea S288C and S. pombe could be helpful to add knowledge about the species molecular data and supportive to identify gene regulatory elements in the promoter regions. It could also help to predict gene expression profiles in various yeast species which in turn could be helpful to improve efficiencies of these organisms for domestic and commercial production of bio-fuel with higher rate of recovery.
Acknowledgements
The authors acknowledge Adama Science and Technology University, School of Applied Natural Science, for funding the research.
Abbreviations
- TSS
-
Transcription start site
MSc1
Motif of Saccharomyces cerevisiaea 1
MSp1
Motif of Schizosaccharomyces pombe
S. cerevisiaea
Saccharomyces cerevisiaea
S. pombe
Schizosaccharomyces pombe
TFs
Transcription factors
MEME
Multiple Em for Motif Elicitation
NCBI
National Center for Biotechnology Information
bp
Base pair
NNPP
Neural Network Promoter Prediction
Authors’ contributions
JA designed and performed the experiment, analyzed the data, and prepared the draft manuscript. MK designed the experiment, supervised the research, revised the manuscript, and is the correspondence of the paper. The authors read and approved the final manuscript.
Funding
This work was financially supported by the graduate program of Adama Science and Technology University.
Availability of data and materials
The qualitative and quantitative data of this manuscript are available through the first author.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declared that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jemal Aman Beshir, Email: jemalbeshir@gmail.com.
Mulugeta Kebede, Email: kmulugetak@yahoo.com, Email: mulugeta.kebede@astu.edu.et.
References
- 1.P.Hazell, “B ioenergy and agriculture: promises and challenges. The International Food Policy Research Institute (IFPRI),” Focus 14 • Brief 2 Of 12 • Report uploaded by Peter Hazell on February 2015..
- 2.Ngwenya TT (2012) An industrial perspective of factors affecting molasses fermentation by Saccharomyces cerevisiae. J Brew Distill 3(2). 10.5897/jbd12.002
- 3.Nissen TL, Kielland-Brandt MC, Nielsen J, Villadsen J. Optimization of ethanol production in Saccharomyces cerevisiae by metabolic engineering of the ammonium assimilation. Metab Eng. 2000;2(1):69–77. doi: 10.1006/mben.1999.0140. [DOI] [PubMed] [Google Scholar]
- 4.V. Ansanay-galeote, B. Blondin, S. Dequin, and J. Sablayrolles, “Stress effect of ethanol on fermentation kinetics by stationary-phase cells of Saccharomyces cerevisiae Stress effect of ethanol on fermentation kinetics by stationary-phase cells of Saccharomyces cerevisiae,” 2001, 10.1023/A.
- 5.Fadel M, Keera AA, Mouafi FE, Kahil T. High Level Ethanol from Sugar Cane Molasses by a New Thermotolerant Saccharomyces cerevisiae Strain in Industrial Scale. Biotechnol Res Int. 2013;2013:1–6. doi: 10.1155/2013/253286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart GG, Panchal C, Russell I, Sills AM. Biology of Ethanol-Producing Microorganisms. Crit Rev Biotechnol. 2008;1(3):161–188. doi: 10.3109/07388558309077977. [DOI] [Google Scholar]
- 7.Russell BDHPR. The primary structure of the alcohol dehydrogenase gene from the fission yeast Schizosaccharomyces pombe. J Biol Chem. 1983;10(1):143–149. doi: 10.1016/S0021-9258(18)33232-0. [DOI] [PubMed] [Google Scholar]
- 8.Weber T. In silico tools for the analysis of antibiotic biosynthetic pathways. Int J Med Microbiol. 2014;304(3–4):230–235. doi: 10.1016/j.ijmm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Ziemert N, Alanjary M, Weber T. The evolution of genome mining in microbes-a review. Nat Prod Rep. 2016;33(8):988–1005. doi: 10.1039/c6np00025h. [DOI] [PubMed] [Google Scholar]
- 10.González C, Salces-Ortiz J, Calvo JH, Serrano MM. In silico analysis of regulatory and structural motifs of the ovine HSP90AA1 gene. Cell Stress Chaperones. 2016;21(3):415–427. doi: 10.1007/s12192-016-0668-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peirce BA (2010) Genetics: A Conceptual Approach, 4th edn. W. H. Freeman ISBN-10 : 1429232501
- 12.Abeel T, Saeys Y, Rouzé P, Van De Peer Y (2008) ProSOM : core promoter prediction based on unsupervised clustering of DNA physical profiles. 24:24–31. 10.1093/bioinformatics/btn172 [DOI] [PMC free article] [PubMed]
- 13.Korkuć P, Schippers JHM, Walther D. Characterization and identification of cis-regulatory elements in arabidopsis based on single-nucleotide polymorphism information. Plant Physiol. 2014;164(1):181–200. doi: 10.1104/pp.113.229716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borneman AR, Gianoulis TA, Zhang ZD. Divergence of Transcription Factor Binding Sites Across Related Yeast Species. Science. 2007;317:815–820. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 15.Robertson KD. DNA methylation and chromatin - Unraveling the tangled web. Oncogene. 2002;21(35 REV. ISS. 3):5361–5379. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 16.Du X, Han L, Guo AY, Zhao Z (2012) Features of methylation and gene expression in the promoter-associated CpG islands using human methylome data. Comp Funct Genomics 2012. 10.1155/2012/598987 [DOI] [PMC free article] [PubMed]
- 17.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS (2007) Quantifying similarity between motifs. 8(2). 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed]
- 18.Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001;26:51–56. doi: 10.1016/S0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 19.Bailey TL et al (2009) MEME S UITE : tools for motif discovery and searching:1–7. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed]
- 20.Takai D, Jones PA (2002) Comprehensive analysis of CpG islands in human chromosomes 21 and 22. 99(6):3740–3745 [DOI] [PMC free article] [PubMed]
- 21.Sandelin A, Alkema W, Lenhard B (2004) JASPAR : an open-access database for eukaryotic transcription factor binding pro ® les. 32(iii). 10.1093/nar/gkh012 [DOI] [PMC free article] [PubMed]
- 22.Irani M, Taylor WE, Young ET. Transcription of the ADH2 gene in Saccharomyces cerevisiae is limited by positive factors that bind competitively to its intact promoter region on multicopy plasmids. Mol Cell Biol. 1987;7(3):1233–1241. doi: 10.1128/mcb.7.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishida Y, Nguyen TTM, Izawa S. The yeast ADH7 promoter enables gene expression under pronounced translation repression caused by the combined stress of vanillin, furfural, and 5-hydroxymethylfurfural. J Biotechnol. 2017;252:65–72. doi: 10.1016/j.jbiotec.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 24.Mohanty B, Takahashi H, de los Reyes BG, Wijaya E, Nakazono M, Lee DY. Transcriptional regulatory mechanism of alcohol dehydrogenase 1-deficient mutant of rice for cell survival under complete submergence. Rice. 2016;9(1):1–8. doi: 10.1186/s12284-016-0124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Smidt O, Du Preez JC, Albertyn J. The alcohol dehydrogenases of Saccharomyces cerevisiae: A comprehensive review. FEMS Yeast Res. 2008;8(7):967–978. doi: 10.1111/j.1567-1364.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- 26.Halees AS, Leyfer D, Weng Z. PromoSer: A large-scale mammalian promoter and transcription start site identification service. Nucleic Acids Res. 2003;31(13):3554–3559. doi: 10.1093/nar/gkg549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das MK, Dai H (2007) A survey of DNA motif finding algorithms. 13:1–13. 10.1186/1471-2105-8-S7-S21 [DOI] [PMC free article] [PubMed]
- 28.Brazma A, Bergen N, Robinson A (1998) Using Gene Expression Data. 17:279–287
- 29.Klok EJ, et al. Expression profile analysis of the low-oxygen response in arabidopsis root cultures. Plant Cell. 2002;14(10):2481–2494. doi: 10.1105/tpc.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Dalkilic M, Kim S. Guiding motif discovery by iterative pattern refinement. Proc ACM Symp Appl Comput. 2004;1:162–166. doi: 10.1145/967900.967934. [DOI] [Google Scholar]
- 31.Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41(5):577–585. doi: 10.1023/A:1006319732410. [DOI] [PubMed] [Google Scholar]
- 32.Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7(12):555–560. doi: 10.1016/S1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- 33.Caselle M, Di Cunto F, Provero P. Correlating overrepresented upstream motifs to gene expression : a computational approach to regulatory element discovery in in eukaryotes. BMC Bioinformatics. 2002;3:1–10. doi: 10.1186/1471-2105-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye S, Asaithambi A, Liu Y. CpGIF: an algorithm for the identification of CpG islands. Bioinformation. 2008;2(8):335–338. doi: 10.6026/97320630002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riveros-Rosas H, Julián-Sánchez A, Villalobos-Molina R, Pardo JP, Piña E. Diversity, taxonomy and evolution of medium-chain dehydrogenase/reductase superfamily. Eur J Biochem. 2003;270(16):3309–3334. doi: 10.1046/j.1432-1033.2003.03704.x. [DOI] [PubMed] [Google Scholar]
- 37.Kuramae EE, Robert V, Snel B, Boekhout T (2006) Conflicting phylogenetic position of Schizosaccharomyces pombe. 88:387–393. 10.1016/j.ygeno.2006.07.001 [DOI] [PubMed]
- 38.Liu Q, Wang X. Characterization and phylogenetic analysis of the complete mitochondrial genome of a basidiomycetous yeast Cystobasidium sp. (Cystobasidiales: Cystobasidiaceae) Mitochondrial DNA Part B Resour. 2020;5(3):2449–2450. doi: 10.1080/23802359.2020.1777910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The qualitative and quantitative data of this manuscript are available through the first author.