Abstract
Despite its convenience and precision, CRISPR-based gene editing approaches still suffer from off-target effects and low efficiencies, which are partially rooted in Cas9, the nuclease component of the CRISPR/Cas9 system. In this study, we showed how mouse genome editing efficiency can be improved by constitutive and inheritable expression of Cas9 nuclease. For this goal, a transgenic mouse line expressing the Cas9 protein (Cas9-mouse) was generated. For in vitro assessment of gene editing efficiency, the Cas9-mice were crossed with the EGFP-mice to obtain mouse embryonic fibroblasts (MEF) expressing both EGFP and Cas9 (MEFCas9-EGFP). Transfection of these cells with in vitro transcribed (IVT) EGFP sgRNA or phU6-EGFPsgRNA plasmid led to robust decrease of Mean Fluorescent Intensity (MFI) to 8500 ± 1025 a.u. and 13,200 ± 1006 a.u. respectively. However, in the control group, in which the MEFEGFP cells were transfected with a pX330-EGFPsgRNA plasmid, the measured MFI was 16,800 ± 2254 a.u. For in vivo assessment, the Cas9-zygotes at two pronuclei stage (2PN) were microinjected with a phU6-HhexsgRNA vector and the gene mutation efficiency was compared with the wild-type (WT) zygotes microinjected with a pX330-HhexsgRNA plasmid. The analysis of born mice showed that while the injection of Cas9-zygotes resulted in 43.75% Hhex gene mutated mice, it was just 15.79% for the WT zygotes. In conclusion, the inheritable and constitutive expression of Cas9 in mice provides an efficient platform for gene editing, which can facilitate the production of genetically-modified cells and animals.
Keywords: CRISPR-Cas9, Gene knock-out, Genetically-modified cells and mice, Microinjection
Introduction
Animal models are routinely employed for better comprehension of diseases, gene functions, and molecular mechanisms underlying exclusive phenotypes. These provide a more precise understanding of the molecular basis of diseases and also render the development of new therapeutic approaches possible (Cong et al. 2013). Among various types of animal models, some characteristics of mice including genetic similarities to humans, short gestation period, availability of genetically inbred strains, and well-developed sets of embryo manipulation technologies make them an ideal choice for human disease simulation (Nagy et al. 2003; Sosa et al. 2010).
Nuclease-based gene-editing tools such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) have provided scientists with efficient gene-editing systems to produce animal models. However, the recent easy-to-handle, highly specific and efficient clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system has revolutionized the process of genetically manipulated mouse production and put an end to former conventional methods. There are three approaches to introduce CRISPR/Cas9 components into the living cells including (1) transfection of DNA plasmids encoding Cas9 and synthetic guide RNA (sgRNA); (2) delivery of both Cas9 and sgRNA transcripts and (3) transfection of CRISPR/Cas9 components as a ribonucleoprotein complex (RNP) into the cells of interest (Lino et al. 2018). Scientists have encountered some troubles during target site selection and sgRNA design, and also some challenges including the possibility of an off-target double-strand break, low efficiency of homology directed repair (HDR) in comparison to that of non-homologous end joining (NHEJ), and inconsistency in Cas9 activity and delivery methods’ efficiency, which altogether jeopardize this platform’s efficacy. CRISPR/Cas9 components delivery has been conducted through physical methods (microinjection, electroporation, and hydrodynamic delivery), and also through the employment of viral vectors (engineered adeno-associated virus and lentiviruses) and non-viral vectors (lipid nanoparticles, cell-penetrating peptides, DNA nano clew and gold nanoparticles) (Lino et al. 2018), all of which offer some benefits, while posing serious drawbacks.
The delivery method has remained as one of the major obstacles for CRISPR/Cas9 in vivo applications. These challenges include incompatibility in the incorporation of CRISPR/Cas9 components with the delivery systems. The large size of Cas9 protein is in discordance with the limited capacity of viral vectors (Schirmbeck et al. 2008; Wu et al, 2010). Moreover, viral delivery of components might create additional problems such as immune responses induction and random integration into the host genome (Kim and Ahituv, 2013; Xue et al. 2014; Yin et al; 2014).
CRISPR/Cas9-mediated gene-editing platform is tedious. For example, even though plasmid-based gene editing approach is cost-effective, it suffers from insufficient efficiency. Scholars have reported that pronuclear injection of plasmid-harboring CRISPR elements resulted in birthing a high percentage of allelic complexity and mosaicism due to Cas9 transcriptional and translational delay and cell division (Horii et al. 2014; Mehravar et al. 2019). Moreover, in vitro transcription of long Cas9 mRNA molecules is expensive and laborious, because it needs excluding impurities such as incomplete transcripts from full-length RNAs (Karikó et al. 2011). Escherichia coli-expressed Cas9 protein requires several purification steps and might be contaminated with bacterial endotoxins. Therefore, the endotoxin level of Cas9 protein must be measured by commercial kits (Staahl et al. 2017). An interesting possibility to increase the efficiency of CRISPR/Cas9-mediated gene-editing is the generation of transgenic animals constitutively expressing the Cas9 protein. This could prevent the non-uniform distribution of Cas9 between embryonic blastomeres, which is responsible for the mosaicism and allelic complexity that normally occur in the early stages of embryonic development in CRISPR/Cas9-mediated transgenic mouse production (Zhang et al. 2016).
In this study, we developed a cloning-free CRISPR/Cas9 gene-editing platform by generating a transgenic mouse line, which constitutively expressed Cas9 nuclease. To overcome difficulties concerning the huge size of CRISPR/Cas9 macromolecules, a Cas9 encoding sequence was inserted into the mouse genome through pronuclear injection. Cas9-mice were generated and the functionality of the Cas9 nuclease was subsequently confirmed in vivo and in vitro. For in vitro confirmation, we documented a CRISPR/Cas9-mediated enhanced green fluorescent protein (EGFP) gene knockout through the transfection of in vitro transcribed (IVT) EGFPsgRNA into the mouse embryonic fibroblasts (MEFs) expressing both Cas9 and EGFP (MEFCas9-EGFP). For in vivo assessment, Cas9-embryos were microinjected with hematopoietically-expressed homeobox (Hhex)-targeting sgRNA (HhexsgRNA), which ultimately led to offspring-carrying mutations in this gene. Multiple sites within the growing embryo express Hex gene, which plays an indispensable role in the development of the gut endoderm into the liver. It mainly acts in response to signaling pathways that mediate the specification of the hepatic lineage (Kubo et al. 2010). Hence, the Cas9-mice/cells were demonstrated to be applicable as a straightforward, efficient, and standard genetic manipulation tool to accelerate the generation of any customized transgenic cell/animal in a short time.
Methods
Ethics statement
The study protocol was approved by the Research Ethics Committee of Iran University of Medical Sciences with ethical code number IR.IUMS.REC.1398.770. All of the experiments conducted on mice were in compliance with the ARRIVE guidelines and in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
Generation of Cas9-mouse
Construction of Cas9 vector
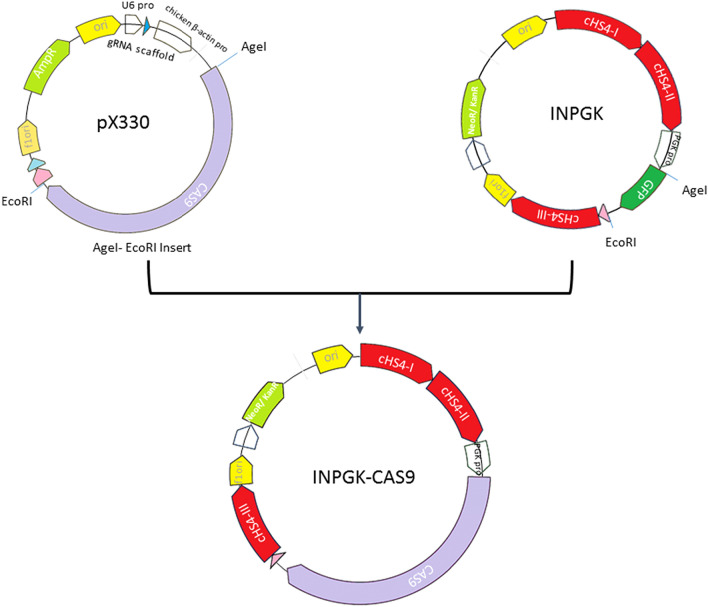
Cas9-expressing vector (INPGK-CAS9) was constructed using standard molecular biology techniques. Briefly, DNA fragment encoding Cas9 was extracted from pX330 plasmid (Addgene, ID #42230) using AgeI and EcoRI enzymes and was purified with QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). The fragment was then inserted into the pINPGK-EGFP vector, which had been linearized with the same enzymes to replace the EGFP gene, as previously reported by Farzaneh and colleagues (2019). The Cas9 cassette in the resulting pINPGK-CAS9 vector was flanked by chicken cHS4 insulators.
Mouse preparation and pronuclei injection
NMRI and B6DF1 mice were purchased from Pasteur Institute of Iran (Tehran, Iran). Mice were kept under the conditions of 50% humidity and a 14:10 h light:dark cycle. They were fed a standard pellet diet and tap water supplied throughout the day. A ketamine/xylazine injection method was employed for mice euthanasia. The vasectomized and pseudopregnant mice were prepared as described previously (Ghassemi et al. 2018). For superovulation, the female B6D2 mice (4-weeks-old) were intraperitoneally injected with 7 IU Pregnant Mare's Serum Gonadotropin (PMSG) (ProSpec, East Brunswick, New Jersey, USA). Human chorionic gonadotropin (hCG) (ProSpec, East Brunswick, New Jersey, USA) was applied the same as the PMSG 46 h later and the mice were immediately caged with B6D2 males. The following morning, two pronuclei (2PN) embryos were extracted from the vaginal plug mice, washed in M2 medium (Sigma, St. Louis, Missouri, USA) and kept in M6 medium (Sigma, St. Louis, Missouri, USA) in a 5% CO2 incubator. The Cas9-cassette with insulators was extracted from the INPGK-CAS9 plasmid using AseI and NotI enzymes and were injected into the male pronucleus at the concentration of 5 ng/µL using an inverted microscope (Olympus, Tokyo, Japan) equipped with a micromanipulator (Eppendorf, Hamburg, Germany). The embryos were then incubated overnight at 37 °C in M16 drops and the following day 35–40 two-cell embryos were transferred into the oviduct of surrogate NMRI mice. Surrogates were kept under standard conditions until the delivery of pups.
Cas9 transgenic mouse genotyping
Pups were screened by polymerase chain reaction (PCR) and also by sequencing to confirm the insertion of the Cas9 encoding gene into their genome. DNA was prepared from the tail biopsy of 3-week-old pups using proteinase K treatment (Thermofisher, Waltham, Massachusetts, USA) and subsequent standard phenol extraction. 50 ng of the DNA was amplified using Taq DNA Polymerase Master Mix RED (Ampliqon, Odense, Denmark) and then electrophorized on 1% agarose gel. PCR products were further cloned using TA Cloning® Kit (Thermofisher, Waltham, Massachusetts, USA) and the resulting vector was sequenced by the Sanger method. The mice carrying the Cas9 sequence were nominated as transgenic founders (F0). To investigate the germline transmission of the Cas9 gene, the F0 mice were crossed with wild-types (WT) and the inheritance of Cas9 gene was traced in the F1 animals. Moreover, the probable cytotoxic effect of the Cas9 expression throughout the mouse body was evaluated by comparing the weight of the whole body with organs of the same aged Cas9 and WT mice.
Western blotting
Cas9-transgenic mice were analyzed for the expression of Cas9 protein by western blotting. Tissue samples from Cas9 and WT mice were obtained and treated in a RIPA buffer (5 mM Tris, pH 8.0, 15 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM EDTA). About 20–30 µg of whole lysate per lane were loaded on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) for 3 h. For immunoblotting, proteins were transferred to a polyvinylidene fluoride (PVDF) membrane using an electrophoretic transfer apparatus (Bio-Rad, Hercules, California, USA). The membrane was blocked in 5% non-fat milk and incubated with anti- Cas9 antibody (Abcam, Cambridge, UK) at the dilution of 1:5000 at 4 °C overnight. Signals were visualized by ECL detection kit (Pierce. Rockford, Illinois, United States) and chemiluminescence was recorded on the Kodak BioMax MS film. Beta-actin (Abcam, Cambridge, UK) was used as loading control for each sample.
EGFP and Hhex knockout model development
gRNA design and vector construction
Suitable gRNAs targeting EGFP and Hhex genes were selected using CHOPCHOP online tool (www.chopchop.cbu.uib.no) (Table 1). The oligodeoxyribonucleotides (oligos) encoding gRNAs were designed to have 5′-CACC and 5′-AAAC overhangs at the 5′ end of sense and antisense strands, respectively for insertion into the BbsI site of the pX330 plasmid (Addgene, ID # 42230). An additional G nucleotide was placed at the 5′ end of sense oligonucleotides to ensure optimal expression by the U6 promoter. The complementary DNA oligos were hybridized each other in a thermal cycler as follow: 95 °C for 5 min and then ramp down to 25 °C slowly at 5 °C/min (Ghassemi et al. 2018). Using the BbsI enzyme, the annealed sgRNA oligos were cloned into the both pX330 and phU6-gRNA (Addgene, ID # 53188) plasmids according to the procedure provided by Ran et al. (2013). The gRNAs sequences are shown in Table 1.
Table 1.
Oligonucleotides sequences used in this study
| Oligonucleotides | Application | Sequence (5′–3′) |
|---|---|---|
| GFP sense strand | sgRNA cloning | CACCGGAGCGCACCATCTTCTTCA |
| GFP anti sense strand | sgRNA cloning and cloning confirmation | AAACTGAAGAAGATGGTGCGCTCC |
| Hhex sense strand | sgRNA cloning | CACCGTAAATGGAGAAGACTGAAAC |
| Hhex antisense strand | sgRNA cloning and cloning confirmation | AAACGTTTCAGTCTTCTCCATTTAC |
| U6-Fwd | GFP and Hhex sgRNA cloning confirmation | ACTATCATATGCTTACCGTAAC |
In vitro transcription of EGFPsgRNA
EGFPsgRNA sequence cloned into the pX330 vector was used as a template for PCR amplification alongside T7 promoter-attached primers. PCR with 5′-GAAATTAATACGACTCACTATAGGAGCGCACCATCTTC-3′ and 5′- AAAAGCACCGACTCGGTGCC -3′ primers introduced the T7 promoters into the sgRNA-DNA template. The T7-sgRNA PCR products were used as a template for in vitro transcription using a HiScribe™ T7 High Yield RNA Synthesis Kit (New England Biolabs, Ipswich, Massachusetts, United States). The transcribed sgRNAs were purified with Qiagen RNA purification kit (Qiagen, Hilden, Germany) and quantified using a Nano Drop (Thermo Scientific, Waltham, Massachusetts, United States). Finally, the integrity of sgRNAs was confirmed by running the purified RNAs on a 2% agarose gel.
MEF isolation and lipid-mediated transfection of genetic material
Cas9-mice were mated with EGFP-mice to produce MEFCas9-EGFP cells. Briefly, embryos were harvested from 12.5 days post coitum (d.p.c.) pregnant mice and washed in PBS and well minced in a dish. The samples went under treatment with 0.05% trypsin–EDTA (Invitrogen, Carlsbad, California, United States) to yield single cells. After trypsin inactivation, single cells were transferred to the DMEM high glucose with 2 mM l-glutamine, 10% FBS, non-essential amino acids and 1% antibiotics. The MEFs were seeded in a 6-well plate and were separately transfected at 70% confluency with the IVT EGFPsgRNA (group 1) phU6-EGFPsgRNA plasmid (group 2). As the control, MEFEGFP cells-derived from 12.5 d.p.c. EGFP-fetus were transfected with the pX330-EGFPsgRNA (group 3). Transfection was performed using the Lipofectamine 3000 (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s protocol. The decline in mean fluorescent intensity (MFI) was quantified by a FACScan Flow cytometry (ABI Biosystem, Foster City, California, USA).
Generation of Hhex knockout mouse
The Cas9-embryos were harvested from B6D2 mice and were injected into the male pronuclei with the phU6-HhexsgRNA plasmid. As control, the WT fertilized embryos were microinjected with the pX330-HhexsgRNA vector. The manipulated embryos were transferred to the NMRI surrogate mice at the two-cell stage (Fig. 1).
Fig. 1.
Construction of INPGK-CAS9 vector from pX330 and INPGK vectors. The Cas9 encoding sequence was obtained from a pX330 plasmid through EcoRI and AgeI restriction enzymes digestion and subsequently replaced the EGFP gene in a INPGK plasmid, which served as the backbone vector. The resulting vector, INPGK-CAS9, was incorporated into mice genome to generate the Cas9 transgenic mice
Statistical analysis
All quantitative data were analyzed using the t test and the p ≤ 0.05 was considered significant statistically.
Results
The Cas9-mouse generation by microinjection
The endotoxin-free pINPGK-CAS9 plasmid was extracted from E. coli. The Cas9 cassette accompanied by the flanking insulators was exited from the plasmid backbone using AseI and NotI enzymes and microinjected into the mouse 2PN stage embryos (Fig. 2a). The transgenic mice were screened by PCR on the genome and the presence of Cas9 gene was confirmed in three out of 30 pups by amplification and sequencing of a 500 bp fragment of the transgene. For germinal transfer, the Cas9 founders (F0) were crossed with WT mice to obtain F1 pups and the PCR confirmed that all the three founders transferred the Cas9 transgene to the next generation. Western blotting was employed to detect the expression of Cas9 protein in the F1 mice tissues. The results revealed that all samples taken from various organs of transgenic mouse expressed the Cas9 protein (Fig. 2b). The toxic effect of Cas9 expression on mice was investigated by comparing the weight of the whole body and key organs between the WT and Cas9 mice. As data in Table 2 illustrate, there were no meaningful changes between the two groups.
Fig. 2.
a Schematic representation of Cas9 transgenic mouse generation. The Cas9-expressing cassette along with the flanked insulators (which appeared as a 7230 bp fragment on the gel electrophoresis) was injected into 2PN stage zygotes, and then the zygotes implanted into a surrogate mouse until the breeding of offspring. b Western blot confirmed the Cas9 expression. Each well represents samples of various body organs derived from either Cas9 or WT mice. Lanes 1–6 belonging to Cas9 mouse’s kidney, liver, testis, bone marrow, hearts, and brain, respectively, and lanes 7–9 being derived from WT mouse’s kidney, liver, and testis. Beta actin was used as loading control
Table 2.
Comparing the weight of whole body and internal organs between the WT and transgenic mice
| Genotype | Whole body | Heart | Lung | Spleen | Liver | Kidney |
|---|---|---|---|---|---|---|
| WT-mice | 34.733 ± 2.173 | 0.176 ± 0.017 | 0.215 ± 0.011 | 0.122 ± 0.01 | 1.735 ± 0.145 | 0.449 ± 0.043 |
| Cas9-mice | 33.706 ± 1.847 | 0.171 ± 0.01 | 0.213 ± 0.007 | 0.124 ± 0.01 | 1.603 ± 0.094 | 0.426 ± 0.022 |
Cas9 nuclease functional assay in transgenic mice
The functionality of endogenous Cas9 nuclease in somatic cells was investigated through EGFP gene knock-out in the MEFCas9-EGFP cells. By crossing the Cas9 and EGFP mice with each other and extracting MEF cells, the coexistence of Cas9 and EGFP genes was respectively confirmed by PCR and visualization of the green fluorescent emission (Fig. 3). The MEFCas9-EGFP cells were divided into two groups and were individually transfected by the IVT EGFPsgRNA in group 1 and the phU6-EGFPsgRNA plasmid in group 2. In group 3, the MEFEGFP cells were transfected with the pX330-EGFPsgRNA. The data revealed a reduction of the MFI in group 1 cells to about 8500 ± 1025, which was the most measured decrement among the groups. The MFI in group 2 cells was calculated to be around 13,200 ± 1006. MFI value in the control group 3 cells was almost 16,800 ± 2254 (Fig. 4).
Fig. 3.
CRISPR/Cas9-mediated EGFP knocking out in MEF cells. a The figure represents schematic strategy for in vitro EGFP gene knockout. As displayed, the Cas9-mice were crossed with the EGFP mice and the MEFCas9-EGFP cells were isolated from 12.5 days old embryos and cultured. The IVT EGFPsgRNA and phU6-EGFPsgRNA plasmid were transfected separately into the MEFCas9-EGFP cells. The pX330-EGFPsgRNA vector was also transfected into the MEFEGFP cells which served as the control of this study. b The diagram represents the measured MFI in three MEF populations using flow cytometry. As shown, the level of MFI in the different groups of MEFs was measured as IVT EGFPsgRNA < phU6-EGFPsgRNA < pX330-EGFPsgRNA
Fig. 4.
a. Microinjection-based strategy for in vivo gene knockout. The phU6-HhexsgRNA vector was microinjected into the Cas9 2PN zygotes. In parallel, the pX330-HhexsgRNA was injected into the WT embryos. The offspring were then assessed for the extent of genetic alterations in the Hhex locus. b Mutation positions throughout Hhex alleles in WT and Cas9 embryos’ offspring. Three out of 19 pups, which had been injected with the pX330-HhexsgRNA into the WT zygotes, carried mutant Hhex allele; while seven out of 16 pups, which had been injected by the phU6-HhexsgRNA into the Cas9-zygotes displayed indels at sgRNA targeting site
In vivo assessment of Cas9 activity
WT and Cas9 embryos were microinjected with HhexsgRNA-harboring vectors (Fig. 4a). Injection of the Cas9-embryos with a phU6-HhexsgRNA vector efficiently resulted in the birth of the mice with indel mutations in the Hhex locus. 7 out of 16 offspring (43.75%) showed indel mutation at the CRISPR/Cas9 cut site. For comparison, WT embryos were injected with the pX330-HhexsgRNA and the mutant Hhexs alleles were found in three out of 19 pups after sequencing (Fig. 4b).
Discussion
In this study, we developed a CRISPR/Cas9-mediated genome editing system for a convenient one-step generation of genetically-modified mice and cells. The Cas9 expression cassette was prepared and injected into the pronuclei of mouse zygotes. The Cas9 protein expression was detected in tissues of the resulting Cas9-mice by western blot. However, the western blot result showed variation in the expression of Cas9 protein among tissues. This heterogeneity in the expression of Cas9 protein might be in relation to the transgene integration site. Following pronuclear injection, foreign DNA is randomly integrated into the genome. The integration site during embryonic development might be converted to heterochromatin or euchromatin regions in the differentiated cells and tissues. This chromatin remodeling causes transgene expression heterogeneity or even gene silencing among body tissues. Hence, the transgenic cassettes are mostly equipped with special elements such as chromatin insulators and matrix-attachment regions (MARs) to neutralize or reduce the integration position effect (Rincón-Arano et al. 2007; Farzaneh et al. 2019; Pérez-González and Caro 2019). To achieve this goal in our study, the Cas9-expressing cassette was surrounded with three copies of cHS4 insulators.
It was demonstrated that the somatic cells derived from the Cas9-mice can be directly subjected to sgRNA transfection to produce genetically-modified cells. According to our results, higher suppression of the EGFP signal was measured in the MEFCas9-EGFP cells compared to the MEFEGFP cells. Similar results were obtained in our in vivo study. This indicates that the endogenous expression of Cas9 nuclease could be sufficient for high percentage induction of indel mutations using small genetic materials such as IVT sgRNAs. This Cas9 transgenic platform liberates the system from transcription and translation of vector-delivered Cas9, because Cas9 is already expressed to a sufficient extent within the host cells.
Generally speaking, our results indicate that the selected sgRNAs worked effectively alongside the endogenously-expressed Cas9 nuclease to induce NHEJ-mediated mutations. This genome editing approach introduces several benefits in comparison with conventional CRISPR/Cas9. Cas9-mouse system can yield genome site-specific cleavage with a higher efficiency relative to its counterpart system. This platform enables us to develop several knockout lines of mice and cells in a short time schedule. It is also more cost-effective compared to the commercially available methods which employ Cas9 nuclease in forms of protein or mRNA. Moreover, endogenous expression of Cas9 protein permits a cloning-free CRISPR-mediated genetic engineering method since the sgRNA can be synthesized either chemically or in vitro. The Cas9 expressing organisms/cells can be conveniently used in multiplex gene-editing format through co-transfection of multiple sgRNAs targeting several genes simultaneously. Endogenous Cas9 nuclease decreases the chance of mosaicism among F0 offspring as well as allelic complexity, which appears during the routine Cas9 mRNA/sgRNAs oocyte injection strategy. This will guarantee the germ cell transmission of the edited alleles to the next generations (Zhang et al. 2016).
Availability of data and material
The data that support the findings of this study are available in the manuscript and also from the corresponding authors upon reasonable request.
Author contributions
JK and MSH designed the study and wrote and edited the draft of the manuscript. BG and MJ conducted the experiments and wrote the manuscript. MS helped JK and MSH supervise the project. GS and MK helped carry out the experiments. JK is the corresponding author of this manuscript.
Funding
This project was supported financially by research grant number 97-4-20-13610 from Cellular and Molecular Research Centre, Iran University of Medical Sciences and the National Institute for Medical Research Development (NIMAD) grant number 962825. The funders provided only financial support for the conduct of the research.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Ethical approval
The study protocol was approved by the Research Ethics Committee of Iran University of Medical Sciences with ethical code number IR.IUMS.REC.1398.770. All of the experiments conducted on mice were in compliance with the ARRIVE guidelines and in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
Contributor Information
Bita Ghassemi, Email: bghassem@uci.edu.
Monire Jamalkhah, Email: jamalkhah94m@gmail.com.
Gelareh Shokri, Email: gelareh.shokri@gmail.com.
Mousa Kehtari, Email: mousakehtari@gmail.com.
Masoud Soleimani, Email: soleim_m@modares.ac.ir.
Mehdi Shamsara, Email: shamsa@nigeb.ac.ir.
Jafar Kiani, Email: ja.kiani@gmail.com.
References
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzaneh F, Mirzapoor Z, Jahangirian E, Heidari F, Hashemi E, Rahim-Tayefeh A, Fatemi N, Jamshidizad A, Dashtizad M, Shamsara M. The chicken hypersensitive site-4 insulator cannot fully shield the murine phosphoglycerate kinase-1 promoter from integration site effects in transgenic mice. 3 Biotech. 2019;9:255. doi: 10.1007/s13205-019-1786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemi B, Shamsara M, Soleimani M, Kiani J, Rassoulzadegan M. Pipeline for the generation of gene knockout mice using dual sgRNA CRISPR/Cas9-mediated gene editing. Anal Biochem. 2018;568:31–40. doi: 10.1016/j.ab.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Horii T, Arai Y, Yamazaki M, Morita S, Kimura M, Itoh M, Abe Y, Hatada I. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci Rep. 2014;4:4513. doi: 10.1038/srep04513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Ahituv N. The hydrodynamic tail vein assay as a tool for the study of liver promoters and enhancers. Methods Mol Biol. 2013;1015:279–289. doi: 10.1007/978-1-62703-435-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Kim YH, Irion S, Kasuda S, Takeuchi M, Ohashi K, Iwano M, Dohi Y, Saito Y, Snodgrass R, Keller G. The homeobox gene Hex regulates hepatocyte differentiation from embryonic stem cell-derived endoderm. Hepatology. 2010;51:633–641. doi: 10.1002/hep.23293. [DOI] [PubMed] [Google Scholar]
- Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25:1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehravar M, Shirazi A, Nazari M, Banan M. Mosaicism in CRISPR/Cas9-mediated genome editing. Dev Biol. 2019;445:156–162. doi: 10.1016/j.ydbio.2018.10.008. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Pérez-González A, Caro E. Benefits of using genomic insulators flanking transgenes to increase expression and avoid positional effects. Sci Rep. 2019;9:8474. doi: 10.1038/s41598-019-44836-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón-Arano H, Furlan-Magaril M, Recillas-Targa F. Protection against telomeric position effects by the chicken cHS4 beta-globin insulator. Proc Natl Acad Sci USA. 2007;104:14044–14049. doi: 10.1073/pnas.0704999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmbeck R, Reimann J, Kochanek S, Kreppel F. The immunogenicity of adenovirus vectors limits the multispecificity of CD8 T-cell responses to vector-encoded transgenic antigens. Mol Ther. 2008;16:1609–1616. doi: 10.1038/mt.2008.141. [DOI] [PubMed] [Google Scholar]
- Sosa MAG, Gasperi RD, Elder GA. Animal transgenesis: an overview. Brain Struct Funct. 2010;214:91–109. doi: 10.1007/s00429-009-0230-8. [DOI] [PubMed] [Google Scholar]
- Staahl BT, Benekareddy M, Coulon-Bainier C, Banfal AA, Floor SN, Sabo JK, Urnes C, Munares GA, Ghosh A, Doudna JA. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol. 2017;35:431–434. doi: 10.1038/nbt.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, Zhang F, Anderson DG, Sharp PA, Jacks T. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou J, Han J, Hu B, Hou N, Shi Y, Huang X, Lou X. Generation of an oocyte-specific Cas9 transgenic mouse for genome editing. PLoS ONE. 2016;11:e0154364. doi: 10.1371/journal.pone.0154364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available in the manuscript and also from the corresponding authors upon reasonable request.