Abstract
COVID-19 is now a pandemic and the knowledge gap on SARS-CoV-2, i.e., the COVID-19 disease agent, dispersion persists. The US Centers for Disease Control and Prevention suggests fomites may not be the main route through which the novel coronavirus spreads. Supporting the same view, the latest the World Health Organization report recommends wearing masks for every individual in public, highlighting the transmission through the air. In the current study AERMOD, one of the most validated and tested models suggested by the USEPA, is used to model SARS-CoV-2-laden PM10 in a hypothetical outdoor environment. Multiple scenarios including particle size, wind speed, source height variations as well as and combined scenarios were modeled to estimated how exposure risk changes with the above-mentioned variables. The results reveal that wind speed majorly narrows infectious plume rather than transferring the peak concentration. The particle size variation indicated that small particles, i.e.,0.01 − 2.5 μm, could reach more than 9 m away from the source in concentration range of 10 − 20 (μg/m3). On the other hand, source height contributes to peak plume shift rather than dispersing the infected particles. This idea was further studies by using combined scenarios which indicated height difference can impact peak plume displacement rather than wind speed. In the worst-case scenario, the results indicate that the virus-laden particles can travel outdoors more than 8 m away from an infected source. The video output of the model results clearly shows the dynamic of viral peak shifts in several scenarios. The results also indicate that in specific conditions the airborne SARS-CoV-2 can be transported to 9 m away from the source. These findings can be useful for individuals as well as decision-makers to mitigated exposure risk in real-world conditions.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40201-020-00602-9.
Keywords: COVID-19, SARS-CoV-2, AERMOD, Transmission through the air, Virus-laden particles, Outdoor exposure
Introduction
A pneumonia-like syndrome of an unidentified source was reported in Wuhan City, Hubei province in China on 3st December 2019 [12]. On 30st January [24] World Health Organization (WHO) expressed concern over the spread of COVID-19 for public health. COVID-19 has now become a pandemic and infected more than six million people and killed more than 1,335,263 people and infected 55,513,456 on a global scale [25]. Many studies have focused on the transmissibility of the SARS-CoV-2 virus in indoor environments. Following stay-at-home orders and lockdown interventions may cause issues including the sick building syndrome [8], despite reducing viral transmission risk; however, can the SARS-CoV-2 remain infectious in outdoor environments? Investigating the transmissibility of the virus in the outdoor environment is a continuing concern within understanding the transmissibility dynamics and virus-laden particles outdoors. The present study is an effort to address this gap of knowledge in literature.
While there are four known routes of transmission of SARS-CoV-2, i.e., air [13], fomites [15], wastewater [27], and possibly water based on its predecessor SARS-CoV-1 [9], the airborne transmission of SARS-CoV-2 is still overlooked in many countries and scientific communities [14]. Morawska and Cao [13] suggest that there is a significant chance of airborne transmission of the virus. The findings of Bourouiba [2] indicate micro-droplets after sneezing can travel 7–8 m in an indoor setting.
Lewis [11] argues that even if the RNA of SARS-CoV-2 is found in air samples it is not indicative of the virus being viable. More recent studies found viable SARS-CoV-2 in air samples suggesting the virus can infect via the airborne route in even longer distances in indoor environments [10]. These findings are in light of using an air sampler device that operates based on the water vapor condensation principle, which does not inactivate the virus during the sampling process. The virus can remain viable for hours on aerosols [21] and convert them into bio-aerosols. Wu et al. [26] found a significant increase in mortality rate with the increase of PM2.5 concentration.
The current literature studied the propagation of the virus by free-fall models [18] or multiphase turbulent gas cloud model [2] in indoor environments. Hence, there is a scientific gap to understand how the concentration of the virus-laden particles may vary based on wind speed, particle size, and source (infected sneeze) height in outdoor environments. These variables are important in the sense that any potentially vulnerable individual can reduce the risk of infection by repositioning him/herself based on envisioning how the concentration/risk may vary when some potentially-infected host sneezes. The current study employs the AERMOD to model infected aerosols concentrations in an outdoor environment based on varying meteorological conditions, source height variations, and combined scenarios. The findings of this research may help decision-makers as well as individuals to redefine social distancing rules regarding instantly-measurable meteorological variables as well as individuals.
Methodology
Aerosolized SARS-CoV-2 characteristics
To model how the bio-aerosols may disperse in the air there is a need to estimate the physical properties of a sneeze including emission rate, expiration velocity, and temperature, emission rate, and source area.
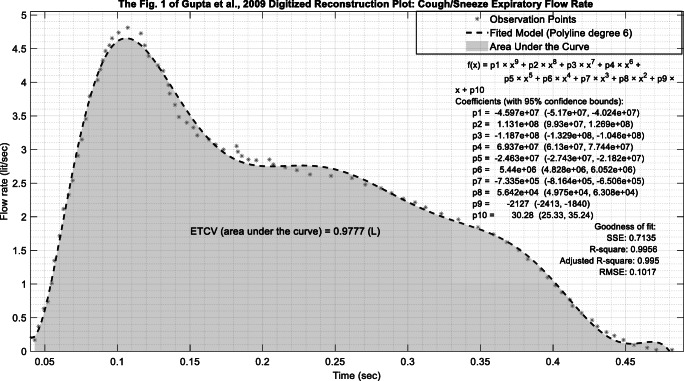
It is noteworthy to mention that each gram of sneeze droplet (≈1.007 cm3) can potentially contain millions of virus copies; however, due to the lack of research on the number of SARS-CoV-2 copies in a sneeze or cough, it was assumed each gram of the expired liquid contains at least one copy of SARS-CoV-2. The liquid emission rate was calculated based on the cough dynamic graph in “Fig. 1“ provided by Gupta et al. [7] in which they provide an estimate for a male cough volume (MCV) (see Eq. 1):
| 1 |
where, CPFR is cough peak flow rate. However, in the current study, the area under the curve of the digitized version of “Fig. 1“ in Gupta et al. [7] was used to estimate the mean cough volume and to approximate a typical cough flow rate based on time. Table 1 provides information on the main variables used in the current study to models an individual sneeze.
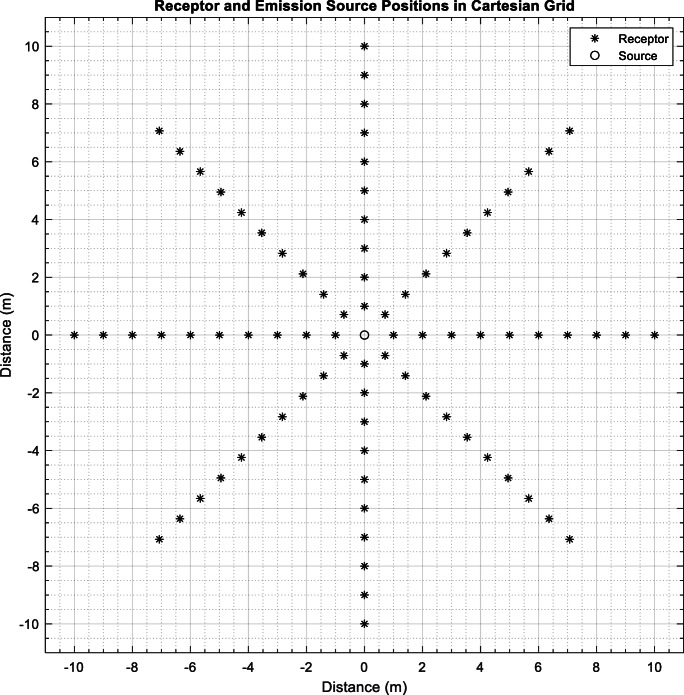
Fig. 1.
Emission source and receptors location
Table 1.
The variables used in the current study
| Variable name | Value | Reference |
|---|---|---|
| Mouth opening area | 3.4 cm2 | [3] |
| Fluid emission rate | 10.9 g/s | Calculated |
| Emission velocity | 11.2 m/s | [7] |
| Emission temperature | 34°C | [3] |
| Source height | 1.7 m | [18] |
| Receptor height | 1.7 m | [18] |
| Liquid volume fraction | 5.6 × 10−3 | Assumed |
| Droplet density | 993 kg/m3 | [3] |
| Mean wind speed and direction | 2 m/s and 0° | Assumed |
| Half-life in aerosols | 1.09 h | [21] |
Cartesian ARC receptors were located at a one-meter distance from each other in a circular pattern (see Fig. 1). Collectively, 80 receptors (eight directions each contain 10 receptors or potential non-infected one) were used to approximate the concentration of the virus-laden particles. It is important to note that AERMOD has been previously implemented for distances less than 10 m [23].
AERMOD
AERMOD (version 14,134, https://www.epa.gov/scram/air-quality-dispersion-modeling-preferred-and-recommended-models) is a Gaussian plume model that is recommended to estimate short-range (less than 50 km) pollution concentration by the USEPA. Terrain features and meteorological parameters are pre-processed by AERMAP and AERMET, respectively [16]. More specifically, AERMET estimates planetary boundary layer height, Monin-Obukhov length, surface heat flux along with physical atmospheric properties, etc., and writes it in an SFC file format. Meteorological profile data including wind speed, wind direction, etc., recorded at different heights are written in a PFL file format. The SFC and PFL files together provide atmospheric characteristics to AERMOD. AERMAP processes terrain elevation data and assigns each receptor an elevation. In stable conditions, AERMOD estimates concentration C by Eq. 2 [16, 28]:
| 2 |
where, Ue, Hs, σz are the effective wind speed, the effective stack height, and the vertical dispersion parameter, respectively. Q and F(x, y) are the source strength and the weighted horizontal distribution function. AERMOD assumes 360° concentration dispersion in low-wind conditions; however, in the current study, “LOWWIND1” was used to disable horizontal meander (when the horizontal wind random component is less than σv = 0.5 m/s). The potentially infected person is considered to be the source and its properties including emission rate, height, etc., were considered in AERMOD based on Table 1. The receptors or vulnerable non-infected individual is considered in AERMOD as receptor where Fig. 1 shows their position in a hypothetical situation. Otherwise, in low-wind conditions, the concentration variations would be at 360°. polarplot3d function (Ken [6]) was used to visualize AERMOD output files in each video frame. As sneezing/coughing is usually released in a horizontal direction “POINTHOR” was used as a non-DEFAULT/BETA option. The POINTHOR type of source is introduced in version 06341 [19] modeling horizontal (HOR) point source (POINT) releases, altogether indicated as POINTHOR. As AERMOD does not calculate concentrations for shorter than 1-h intervals, USEPA [20] and Venkatram [22], suggest Eq. 3, that has been widely used in many studies [5, 17], to calculate model concentration in shorter intervals.
| 3 |
where C(t) is the concentration of the shorter averaging time (i.e., 1 s) and C(t0) is the concentration of the longer averaging time (i.e., 60 min). t0 and t is the shorter and the longer averaging time, respectively.
Sensitivity analysis
A MATLAB script was written to perform a sensitivity analysis of four variables, i.e., wind speed, particle size, and source and receptor height. The script generates 100 hourly SFC, PFL, and ADI (AERMOD input) files. The ADI files are a list of information presenting the location of each input file (e.g., AERMET outputs) and source and receptor characteristics. Besides, a post-processor script was written to fetch AERMOD POST files together. These four parameters (see Table 2) were selected based on the fact that they can readily be analyzed in real-world conditions. The rest of the variables in the AERMOD model, including albedo, Bowen ratio, receptor height, and roughness length were assumed to be constant, as the modeling area is assumed to be constant over time.
Table 2.
The variables range under sensitivity analysis
| Variable name | Range |
|---|---|
| Source height | 0.5 − 3.4 m |
| Wind speed | 0.1 − 11.5 m/s |
| Particle size | 0.01μm − 10μm |
The goodness of fit metrics
To estimate the accuracy of the fitted polyline compared to its original cough curve, i.e., (please see Fig. 2), coefficient of determination (R2), root mean squared error (RMSE), adjusted coefficient of determination (R2 _ Adj), and sum squares error (SSE) were calculated by Eqs. 4 to 7:
| 4 |
| 5 |
| 6 |
| 7 |
where, Oi and Pi are the ith observation and prediction, respectively, and n and p are the total number of observations and number of the predictor(s), respectively.
Fig. 2.
Total cough volume estimation from “Fig. 1“of Gupta et al. [7]
Results and discussion
Emission rate estimation
Emission rate estimation is carried out by calculating the area under the curve of “Fig. 1“in Gupta et al. [7]. Accordingly, it was estimated that the total (droplets and air) cough volume (ETCV) of a typical cough is ≈0.98 (l). As a normal cough takes about 0.49 s [7], it was estimated that in 1 s a normal cough volume would be about 1.98 (l). Ultimately, considering the volume fraction of 5.6 × 10−3 and liquid density of a cough 993 kg/m3 will yield 10.9 g/s of liquid expiration in 1 s.
However, this value was not directly inputted in AERMOD as in a real-world scenario one does not cough/sneeze continuously during an hour. In this vein, 10.9 g/s was divided by 3600 to yield a hypothetical hourly emission of 10.9 g/h. As AERMOD does not calculate concentration for shorter than 1-h intervals, we followed the approach suggested by USEPA to interpolate calculated concentration for a shorter interval of 1-s [20, 22]. Following Eq. 3, the model output concentration was multiplied by C(1/60) = C(t0) × (60/(1/60))0.2 which yields ~5.14 to interpolate 1-h average concentration to 1-s average concentration. As Fig. 2 shows, the fitted polyline, based on the goodness of fit metrics, is in good agreement with observations (Fig 2).
COVID-19 exposure variations
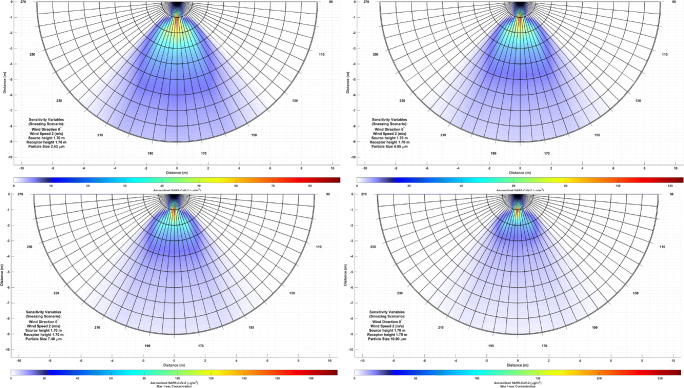
In four scenarios wind speed, particle size, receptor height, and source height were varied to observe aerosolized SARS-CoV-2 variations. Figures 3, 4, 5, 6 and 7 presents the AERMOD response to the above-mentioned variations (interested readers can refer to the supplementary materials for the colored video version of the outputs). The results of particle size variations indicate that smaller particles (less than 2.5 μm) can reach the concentration range of 10 − 20 (μg/m3) in more than 9 m away from the source. Blocken et al. [1] who found the droplets in the size of 40 μm, which is far larger than the studied particles, can travel more than 1.5 away from a runner at speed of 4 m/s using computational fluid dynamics approach.
Fig. 3.
COVID-19 exposure variation due to particle size variation
Fig. 4.
COVID-19 exposure variation due to source height variation
Fig. 5.
COVID-19 exposure variation due to wind speed variation
Fig. 6.
COVID-19 exposure variation due to wind speed increase and source height increase
Fig. 7.
COVID-19 exposure variation due to wind speed decrease and source height increase
The results of wind speed variation suggest an increase in wind speed concentrates bio-aerosol rather than moving the peak concentration away from the source. In this case, peak concentration may move to 2 to 5 m away from the source at a wind speed higher than 10 m/s. However, as the plume contracts with wind speed, higher concentrations could be found at longer distances, i.e., more than 7 m away from the source. These findings are in agreement with Bourouiba [2] who virus-laden particles can travel up to 7 − 8 m away from the source. Dbouk and Drikakis [4] found at the wind speed of ~4 m/s the droplets can travel up to 6 m a using computational fluid dynamics approach.
In low-wind conditions, the result is different. In this case, the peak concentration shape does not follow the Gaussian distribution (LOWWIND1 option is used to disable horizontal meander in low-wind conditions); however, the peak concentration does not disperse more than 2 m away from the source. On the other hand, the width of peak concentration spreads over 1 m. These findings are in agreement with Stariolo [18] who found in indoor conditions virus-laden particles may not travel more than 3 m away from the source.
The source height variations at low levels (less than 1.7 m) are majorly affected by source (or hypothetical horizontal point) aerodynamic downwash and near ground-level turbulence. At higher levels (>1.7 m) as the bio-aerosolized virus plume spreads, the concentration reduces. It can be seen that source height has more impact on peak plume concentration displacement than wind velocity (<10 m/s). Based on this, the higher the infected source position, the higher the distance peak concentration can travel.
The combined wind speed and source position variation were modeled in two scenarios, i.e., wind speed increase and wind speed decrease while the source height increases. These two scenarios provide insight into the dynamics of the infected plume and how much an infected plume can travel in the worst-case scenario. In the first scenario, as wind speed and source height increase the peak concentration moves away from the infected source; however, under low-wind conditions and low-level source height, the peak concentration does not spread further 1.5 m away. In this case, when wind speed and source height reach their maximum allowable values, the plume shifts to more than 9 m away from the source. Not to mention the maximum concentration drastically decreases as both variables increase.
In the second scenario, as wind speed decreases, esp., <10 m/s and source height increases, the aerosolized SARS-CoV-2 plume contracts drastically; however, as soon as source height increases plume expands and concentration reduces. This behavior indicates the source downwash effect is more capable of transporting infected aerosols than wind speed. On the other hand, as wind speed reduces to 2 m/s the infected plume propagates in the left and right directions.
Conclusion
As the COVID-19 pandemic prolongs, the need for understanding the dynamics of aerosolized SARS-CoV-2 dispersion in the outdoor environment increases. Since its introduction, AERMOD is one of the most tested and validated models recommended by the USEPA for atmospheric pollution dispersion modeling. The correct understanding of the novel coronavirus dispersion can help to take preventative measures to control and contain it in outdoor environments. In this regard, five scenarios of SARS-CoV-2 infection spatial variation were demonstrated. However, in some cases, the relationship of bio-aerosol intake with wind velocity and source height may not fit our expectations.
The results suggest that source height can impact the displacement of the peak plume concentration more than wind speed due to the source downwash effect. Wind speed contracted the plume rather than transferring the peak concentration. Although the results may need comprehensive time-intensive validation, at this point, it is believed that the results can change the current understanding of SARS-CoV-2 exposure risk. Hence, in the modeled scenarios, a vulnerable individual may consider height difference rather than wind velocity and direction to reduce exposure risk. The particle size variation indicates less exposure to large particles at long distances and relatively higher exposure to small particles at long distances.
In equal height conditions, wind speed can be a major infection factor; however, not in large distances, i.e., more than 8 m away from the potential source. In these conditions, the peak concentration position may not shift drastically (the peak concentration was found to be located at 1 − 2 m away from the source), but the plume shape contracts in a manner that relatively-high concentrations could be found at even 9 m away from the potential source. Hence, social distancing rules may not apply in these conditions, and wearing facial masks for both non-infected and infected individuals is recommended.
Limitations of the study
The present study, similar to many environmental modeling studies, is subjected various to limitations, which will give rise to further investigation. As the current study was carried out in AERMOD, the accuracy of the model output requires an experimental air sampling approach to be accessed. This fact due to the gap of knowledge on the virus behavior and special sampling instruments (to capture viable airborne SARS-CoV-2) requires more time to be explored. There are many sources of uncertainty in the modeling process; however, the overall shape of the plume is expected to be similar to the model.
The results of the present study are only indicative of a relative aerosolized SARS-CoV-2 concentration and exposure risk. Therefore, the results do not indicate the received dose of virus-laden particles, at any distance from the source, is enough to make an individual infected. A comprehensive risk assessment may unveil if these concentrations are enough to make an individual infected or not. Also, the viability of the infected particles need to be addressed at long distances, typically, more than 8 m away from the source.
Supplementary Information
(MP4 5547 kb)
(MP4 5679 kb)
(MP4 4324 kb)
(MP4 5898 kb)
(MP4 4628 kb)
Acknowledgments
We would like to extend our appreciation to all healthcare workers dealing with the pandemic around the world.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Blocken, B., Malizia, F., Van Druenen, T. & Marchal, T. 2020. Towards aerodynamically equivalent COVID19 1.5 m social distancing for walking and running. Questions and Answers. Website Bert Blocken, Eindhoven University of Technology (The Netherlands) and KU Leuven (Belgium). Disponibile su: http://www.urbanphysics.net/COVID19.html (ultimo accesso 21 aprile 2020).
- 2.Bourouiba L. Turbulent gas clouds and respiratory pathogen emissions: potential implications for reducing transmission of COVID-19. JAMA. 2020;323:1837–1838. doi: 10.1001/jama.2020.4756. [DOI] [PubMed] [Google Scholar]
- 3.Bourouiba L, Dehandschoewercker E, Bush JWJJOFM. Violent expiratory events: on coughing and sneezing. 2014;745:537–63.
- 4.Dbouk T, Drikakis DJPOF. On coughing and airborne droplet transmission to humans. 2020;32:053310. doi: 10.1063/5.0011960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Melo AMV, Santos JM, Mavroidis I, Junior NCR. Modelling of odour dispersion around a pig farm building complex using AERMOD and CALPUFF. Comparison with wind tunnel results. Build Environ. 2012;56:8–20. doi: 10.1016/j.buildenv.2012.02.017. [DOI] [Google Scholar]
- 6.Garrard K. 3D Polar Plot. MATLAB central file exchange. 2020
- 7.Gupta J, Lin CH, Chen QJIA. Flow dynamics and characterization of a cough. 2009;19:517–25. [DOI] [PubMed]
- 8.Hosseini MR, Fouladi-Fard R and Aali R. COVID-19 pandemic and sick building syndrome. Indoor and Built Environment. 2020;29(8):1181–1183.
- 9.Hung LSJJJJOTRSOM. 2003. The SARS epidemic in Hong Kong: what lessons have we learned? 96, 374–378. [DOI] [PMC free article] [PubMed]
- 10.Lednicky JA, Lauzardo M, Fan ZH, Jutla A, Tilly TB, Gangwar M, Usmani M, Shankar SN, Mohamed K, Eiguren-Fernandez A, Stephenson CJ, Alam MM, Elbadry MA, Loeb JC, Subramaniam K, Waltzek TB, Cherabuddi K, Morris JG, Wu, C.-Y. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int J Infect Dis. 2020;100:476–482. doi: 10.1016/j.ijid.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis D. Is the coronavirus airborne? Experts can’t agree. Nature. 2020;580:175. doi: 10.1038/d41586-020-00974-w. [DOI] [PubMed] [Google Scholar]
- 12.Lu H, Stratton CW, Tang YWJJOMV. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. 2019. [DOI] [PMC free article] [PubMed]
- 13.Morawska L, Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ Int. 2020;139:105730. doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nayereh Rezaie Rahimi, Reza Fouladi-Fard, Rahim Aali, Ali Shahryari, Mostafa Rezaali, Yadollah Ghafouri, Mohammad Rezvani Ghalhari, Mahdi Asadi Ghalhari, Babak Farzinnia, Maria Fiore, Oliveri Conti Gea, (2020) Bidirectional Association Between COVID-19 and the Environment: a Systematic Review. Environmental Research:110692 [DOI] [PMC free article] [PubMed]
- 15.Ong SWX, Tan YK, Chia PY, Lee TH, Ng OT, Wong MSY, Marimuthu KJJ. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. 2020 [DOI] [PMC free article] [PubMed]
- 16.Pandey G, Sharan MJAE. Accountability of wind variability in AERMOD for computing concentrations in low wind conditions. 2019;202:105–16.
- 17.Santos JM, Griffiths RF, Reis NC, Mavroidis I. Experimental investigation of averaging time effects on building influenced atmospheric dispersion under different meteorological stability conditions. Build Environ. 2009;44:1295–1305. doi: 10.1016/j.buildenv.2008.09.020. [DOI] [Google Scholar]
- 18.Stariolo DA. COVID-19 in air suspensions. arXiv preprint arXiv: 2020;2004:05699
- 19.USEPA 2004. User's guide for the AMS/EPA regulatory model–AERMOD. Office of air Quality Planning and Standards Washington, DC,, USA.
- 20.USEPA . Guidance notes for using the regulatory air pollution model AERMOD in Victoria. 2013. [Google Scholar]
- 21.Van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed]
- 22.Venkatram A. Accounting for averaging time in air pollution modeling. Atmos Environ. 2002;36:2165–2170. doi: 10.1016/S1352-2310(02)00200-5. [DOI] [Google Scholar]
- 23.Venkatram A, Isakov V, Yuan J, Pankratz DJAE. Modeling dispersion at distances of meters from urban sources. 2004;38:4633–41.
- 24.WHO. Considerations for quarantine of individuals in the context of containment for coronavirus disease (COVID-19): interim guidance, 19 March 2020: World Health Organization; 2020a.
- 25.WHO. 2020b. WHO COVID-19 live update [Online]. Available: https://www.worldometers.info/coronavirus/? [Accessed November 17, 2020].
- 26.Wu X, Nethery RC, Sabath BM, Braun D, Dominici FJM. Exposure to air pollution and COVID-19 mortality in the United States. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeo C, Kaushal S, Yeo DJTLG, Hepatology. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? 2020;5:335–7. [DOI] [PMC free article] [PubMed]
- 28.Zou B, Benjamin Zhan F, Gaines Wilson J, Zeng Y. Performance of AERMOD at different time scales. Simul Model Pract Theory. 2010;18:612–623. doi: 10.1016/j.simpat.2010.01.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(MP4 5547 kb)
(MP4 5679 kb)
(MP4 4324 kb)
(MP4 5898 kb)
(MP4 4628 kb)