Abstract
The newly emerged severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) coronavirus initiated a pneumonia outbreak (COVID-19) that rapidly spread worldwide and quickly became a public health emergency of international concern; However to date, except Remdesivir, there are no clinically approved specific or effective medicines to prevent or treat COVID-19. Therefore, the development of novel treatments against coronavirus infections caused by the current SARS-CoV-2 virus, as well as other highly pathogenic human coronaviruses, represents an urgent unmet need. Stimulator of interferon genes (STING) plays a central role in host defense mechanisms against microbial infections. STING activation leads to the induction of both type I interferon and autophagy responses, which elicit strong inhibitory effect against the infections caused by a broad range of microbial pathogens. However, whether STING activation can impact infections from SARS-CoV-2 or other coronaviruses remains largely unknown. In this study, we investigated the anti-coronavirus activity triggered by STING activation. We discovered that dimeric amidobenzimidazole (diABZI), a synthetic small molecule STING receptor agonist, showed potent anti-coronavirus activity against both the common cold human coronavirus 229E (HCoV-229E) and SARS-CoV-2 in cell culture systems. In addition, we demonstrated that the antiviral activity of diABZI was dependent on the interferon pathway in HCoV-229E infected normal human fibroblast lung cells (MRC-5) and reconstituted primary human airway air-liquid interface (ALI) cultures. Furthermore, low-dose of diABZI treatment at 0.1 μM effectively reduced the SARS-CoV-2 viral load at the epithelial apical surface and prevented epithelial damage in the reconstituted primary human bronchial airway epithelial ALI system. Our findings have thus revealed the therapeutic potential of STING agonists, such as diABZI, as treatments for SARS-CoV-2 and other human coronavirus infections.
Keywords: STING agonist, SARS-CoV-2, HCoV-229E, Coronavirus, diABZI
1. Introduction
In December 2019, a new coronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), caused an outbreak of the novel coronavirus disease COVID-19 in the city of Wuhan, China. This disease has since spread to more than 200 countries with approximately 50 million confirmed cases and over 1.2 million confirmed deaths worldwide as of Dec 8, 2020 (WHO COVID-19 weekly epidemiological update). The WHO declared the coronavirus outbreak a public health emergency of international concern. Currently, no clinically effective vaccines or specific antiviral drugs are available for the prevention or treatment of SARS-CoV-2 infections (Shang et al., 2020).
Coronaviruses (CoVs) are a highly diverse family of enveloped, positive-sense, single-stranded RNA viruses. Seven human coronaviruses (HCoVs) have been identified thus far, namely, HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV) and the novel coronavirus (SARS-CoV-2). While SARS-CoV, MERS-CoV, and SARS-CoV-2 are highly pathogenic, the others generally cause mild to moderate upper-respiratory tract illness and contribute to 15%–30% of the cases of common colds in human adults (Paules et al., 2020). The prototypic stain HCoV-229E, which has long been known to circulate in the population and cause mild symptoms, was first isolated in the 1960s and has been used as an essential tool for the coronavirus research field (V'Kovski et al., 2020).
A number of therapeutic agents are currently being evaluated for the treatment of COVID-19, but their reported clinical efficacies are limited. Remdesivir (RDV), an inhibitor of the viral RNA-dependent RNA polymerase, demonstrated certain clinical efficacy in shortening the time to recovery in adults hospitalized with COVID-19 with evidence of lower respiratory tract infection (Beigel et al., 2020). Dexamethasone has been shown to decrease mortality in only patients receiving invasive mechanical ventilation (Group et al., 2020). Type I interferon (IFN) alpha/beta are broad spectrum antivirals that can both elicit direct inhibitory effect against viral replication and also induce immune responses to further restrict virus infection (Wang and Fish, 2019). Several studies recently reported that the pre-/post-treatment of type I interferon inhibited the cytopathic effect (CPE) effect of SARS-CoV-2 in cell culture models and improved recovery and decreased 28-day mortality in COVID-19 patients (Clementi et al., 2020; Davoudi-Monfared et al., 2020; Lokugamage et al., 2020; Monk et al.). Overall, the clinical efficacies of these drugs are far from satisfactory. Thus, the discovery and development of broad spectrum antivirals with improved efficacy in the control and prevention of current and emerging coronavirus diseases are urgent unmet medical needs.
Physiologically relevant in vitro cell-based models are essential for basic coronavirus research and antiviral drug discovery. SARS-CoV-2 has been extensively studied in cell line infection models, such as the Vero E6 cells, which can easily replicate the virus and viral particles at high titers (Matsuyama et al., 2020). These cells were isolated from the kidney epithelial cells of an African green monkey and are deficient in innate immune pathways, such as Stimulator of interferon genes (STING), making them unsuitable for evaluating the antiviral function of innate immune modulators (Langereis et al., 2015; Osada et al., 2014). In addition, traditional submerged cell culture systems have major limitations because they cannot fully mimic the complicated structures and functions of the respiratory tract epithelium. To overcome these limitations, more advanced systems in which SARS-CoV-2 infection and CPEs were demonstrated have been developed (Pizzorno et al., 2020; Zhu et al., 2020a). The in vitro reconstituted primary human airway epithelium (HAE) with air-liquid interface (ALI) culture system exhibits a pseudo layered morphology and is composed of heterogeneous cell populations, including cilia and mucus secreting cells (Shirato et al., 2018). In addition, the expression of tight junction proteins and a high transepithelial electrical resistance (TEER) have been demonstrated in ALI cultures, acting as a protective barrier against inhalation injury, which mimics the human physiological environment (Prytherch et al., 2011). Thus, the primary human airway cell ALI based coronavirus infection system is highly complementary of traditional cell line derived models with high reproducibility for most of the structural, functional, and innate immune features as observed under in vivo conditions.
STING, a critical innate immunity component located in the endoplasmic reticulum (ER), is essential for the activation of host innate immune responses against microbial infections (Barber, 2015). Upon binding of its cyclic dinucleotides (CDNs) ligand, STING is activated, which leads to the recruitment and activation of TANK-binding kinase 1 (TBK1) and other downstream factors for the induction of antiviral genes, such as IFNs and interferon stimulated genes (ISGs) (Barber, 2015). In addition, STING activation can induce autophagy-related gene 5 (ATG5)-dependent autophagy, which can restrict the replication of certain RNA viruses (Zhu et al., 2020b). Furthermore, STING activation was found to effectively repress the replication of a broad range of DNA and RNA viruses as reviewed in (Ahn and Barber, 2019). Recently, a synthetic small molecule STING receptor agonist, dimeric amidobenzimidazole (diABZI), with much higher cellular potency than CDNs was reported (Ramanjulu et al., 2018). In our previous study, we showed that diABZI induced STING activation elicited potent antiviral effects against parainfluenza virus type 3 (PIV3) and human rhinovirus 16 (HRV16), two representative respiratory viral pathogens, through distinct mechanisms (Zhu et al., 2020b).
In this paper, we evaluated the potential anti-coronavirus ability of diABZI using cellular and reconstituted HAE ALI models. The STING agonist diABZI showed significant inhibitory effects against both HCoV-229E and SARS-CoV-2, indicating diABZI as a potential broad-spectrum anti-coronavirus agent.
2. Materials and methods
2.1. Small molecules
diABZI (Ramanjulu et al., 2018) was synthesized >99% purity according to patents WO2017175156 and WO2017175147. The TBK1 inhibitor BX795, 2′,3′-cGAMP and digitonin were purchased from Sigma (SML0694, SML1229 and 300410). Autophagy inhibitor chloroquine and Remdesivir (RDV) were purchased from Selleck Chemical (S4157 and S8932).
2.2. Cell lines and antibodies
MRC-5 cells (ATCC, CCL-171) were cultured in MEM supplemented with 10% FBS (Gibco) and 1% Penicillin/Streptomycin (Sigma). The IFNR antibody (21385-1) was purchased from PBL.
2.3. Virus
HCoV-229E virus was purchased from ATCC (VR-740) and propagated on MRC-5 cells according to the manufacturer's protocol. The Coronavirus SARS-COV-2 (Beta CoV/France/IDF0571/2020 strain) was provided and operated by Epithelix Sàrl (Geneva, Switzerland). All viruses were stored in −80 °C until use.
2.4. HCoV-229E CPE inhibition assay
MRC-5 cells were seeded at 10,000 cells per well in a 96-well plate. After incubation overnight, the cells were infected with HCoV-229E virus at a multiplicity infection (MOI) of 0.01. Immediately following the addition of virus, the compound was added to the culture medium at the indicated concentration, and the cells were returned to 35 °C incubator for 3 days. Following incubation, cell viability was determined using a Cell Counting Kit-8 (CCK8) kit (Dojindo, Cat.CK04) and an Envision reader (PE) according to the manufacturer's protocol.
2.5. Real-time PCR assay
Total RNA was isolated from cells with an RNeasy Mini Kit (Qiagen), and first-strand cDNA was generated from total RNA using random primers and a reverse transcriptase system (Invitrogen). Real-time PCR was performed on a Roche Light Cycler 480 system with TaqMan qPCR Master Mix Universal (ABI). The primer and probe sequences used to detect HCoV-229E (N gene CDS for genomic and subgenomic RNA level) and SARS-CoV-2 (NSP14 CDS for genomic RNA level) are shown below.
HCoV-229E-Forward primer: 5′- TGGCACAGGACCCCATAAAG- 3’.
HCoV-229E-Reverse primer: 5′- CAACCCAGACGACACCTTCA- 3’.
HCoV-229E-Probe: FAM 5′- TGCAAAATTTAGAGAGCGTG-3′ BHQ (Patent CN103993101A).
SARS2-NSP14- Forward primer: 5′- TGGGGYTTTACRGGTAACCT-3’.
SARS2-NSP14- Reverse primer: 5′- AACRCGCTTAACAAAGCACTC-3′
SARS2-NSP14-Probe: FAM 5′- TAGTTGTGATGCWATCATGACTAG-3′ TAMRA. (Epithelix Sàrl)
2.6. HCoV-229E ALI assay
Human Small Airway Epithelial Cells (SAEC) (#CC-2547s) were purchased from Lonza. The cells were first expanded in PneumaCult™Ex Plus Medium (#05040, STEMCELL), followed by differentiation in PneumaCult™-ALI-S Medium (#05001, STEMCELL). On the day of infection, 100 μl of the HCoV-229E virus inoculum at the indicated MOI was added to the apical side of the cells, which were incubated for 1 h at 35 °C. Then, the virus inoculum were removed, and the cells were washed with 300 μl of PBS twice. The compounds were added to the basal medium at this time. At the indicated collection time point, 300 μl of medium was added to the apical side of each well and incubated for 5 min. The virus-containing media were collected and stored at −80 °C for virus titer measurements by TCID50 or viral RNA determination by real-time PCR. The slides were subjected to hematoxylin and eosin (H&E) staining to visualize the morphological changes of a possible CPE.
2.7. SARS-CoV-2 MucilAir™ assay
MucilAir™ reconstituted from human primary cells obtained from bronchial biopsies was provided by Epithelix SARL (Geneva, Switzerland) and maintained in ALI with MucilAir™ culture medium (EP04MM) in 24 well-plate. The coronavirus SARS-COV-2 was provided by Epithelix Sàrl and the study was performed in VirNext (BSL3 facilities, Lyon, France) (Pizzorno et al., 2020). 100 μL of SARS-COV-2 inoculum (MOI = 0.1) was applied to the apical side of the tissues for 1 h at 37 °C. Then the inoculum was removed and test compound was applied on the basal compartment. Test compound solution was renewed at 24 and 48 h. Apical washes were performed with 200 μL of OptiMEM™ culture medium for 10 min at 37 °C at the 48-h and 72-h time to collect samples for viral RNA quantification by Real time qPCR.
2.8. TEER assay
A total of 300 μL of culture medium was added to the apical compartment of the tissue cultures, resistance was measured across cultures with an EVOM2 volt-ohm-meter (World Precision Instruments, Sarasota, US). Resistance values (Ω) were converted to TEER (Ω*cm2) by using the following formula: TEER (Ω*cm2) = (resistance value (Ω) - 100(Ω)) x 0.33 (cm2), where 100 Ω is the resistance of the membrane and 0.33 cm2 is the total surface of the epithelium in the 24-well plate.
2.9. CCK-8 assay
At indicated time point, the CCK-8 assays were performed to quantify the cell viability. In brief, 10% CCK-8 reagent (Dojindo, Cat.CK04) in 100 μl cell culture medium were used to replace the old culture medium. The CCK-8 value were measured as absorbance at 450 nm after 2 h further incubation in 35° for MRC-5 cells.
2.10. Statistical analysis
Statistical analysis was performed using Prism software (GraphPad). The significance of differences between groups was assessed with a two-tailed Student's t-test. A value of P < 0.05 was considered statistically significant.
3. Results
3.1. The STING agonist diABZI exhibited potent anti HCoV-229E activity in MRC-5 cells
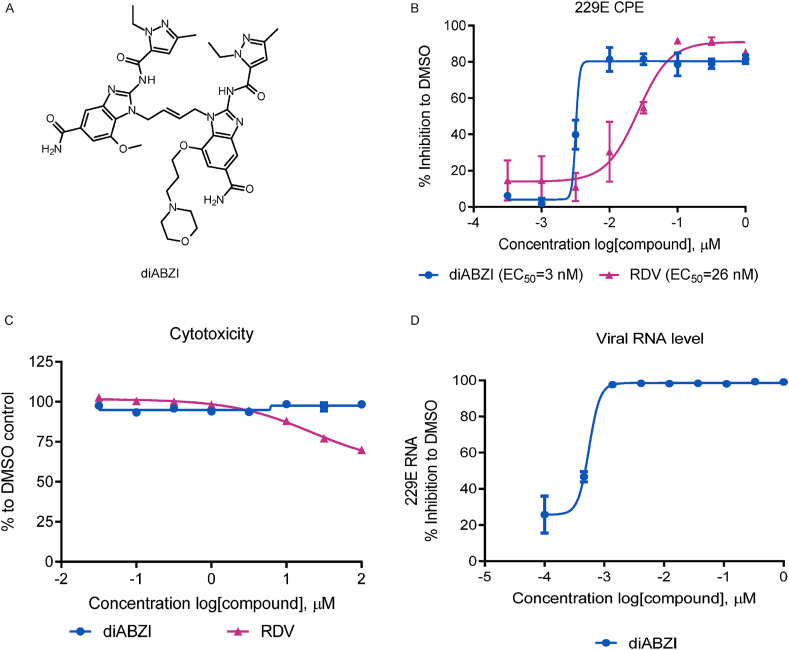
Dimeric ABZI (diABZI, Fig. 1 A), which exhibits an enhanced binding affinity and cellular functions in activating both human and murine STING, has elicited potent antiviral effects against PIV3 and HRV16 in previous study (Zhu et al., 2020b). Recently, Wu et al. suggested that the STING agonist cyclic dinucleotides could be adopted as a potential adjuvant for the SARS-CoV-2 vaccine (Wu et al., 2020); however, the antiviral potential of the STING agonist diABZI against coronavirus infections have yet to be explored. We therefore investigated the potential anti-coronavirus activity of diABZI. First the dose response of diABZI was examined using HCoV-229E (surrogate virus) CPE assay. MRC-5 cells infected with HCoV-229E at an MOI of 0.01 were treated with diABZI. After 72 h, the cell viability was measured via CCK-8 assay to quantify CPEs. Notably, diABZI dose dependently protected the cells from CPEs with an EC50 value of 3 nM, indicative of significant anti-HCoV-229E activity comparable to that of Remdesivir (RDV, EC50 = 26 nM) (Fig. 1B). Moreover, the half maximal cytotoxic concentration value (CC50) of diABZI was greater than 100 μM in MRC-5 cells, suggesting that the observed antiviral effects were not related to nonspecific cytotoxicity (Fig. 1C). There was a >80000-fold difference in the EC50 and CC50 values of diABZI, a higher selectivity index (SI) value compared to that of RDV (SI = 13074), indicating that diABZI has good therapeutic potential. To further confirm the anti-HCoV-229E ability of diABZI, MRC-5 cells infected with HCoV-229E at an MOI of 0.01 were treated with diABZI/DMSO for 48 h, and the cells were collected for RNA isolation followed by RNA quantification. The result in Fig. 1D demonstrated diABZI significant inhibited the HCoV-229E viral replication on MRC-5 cells.
Fig. 1.
STING agonist inhibits HCoV-229E replication on MRC-5. (A) The chemical structures of ABZI-based STING receptor agonist diABZI. (B) MRC-5 cells were infected with HCoV-229E at MOI of 0.01 and treated with indicated concentrations of diABZI or RDV for 72 h. The CPE was determined by a CCK-8 assay and the percent inhibition relative to DMSO treatment was plotted. (C) The cell viability was assessed by measuring CCK-8 in non-infected MRC-5 in the presence of DMSO and indicated concentrations of diABZI or RDV. (D) The HCoV-229E viral RNA were isolated from infected MRC-5 at MOI of 0.01 for 48hrs treatment with indicated concentrations of diABZI or DMSO and measured by real-time PCR. The EC50 values were calculated using Prism software. The data are presented as the means ± SDs of triplicate samples from one experiment and are representative of at least three independent experiments.
In contrast to the strong antiviral activity of diABZI STING agonist, the natural STING ligand cGAMP exhibited only modest anti HCoV-229E activity with an EC50 of 120 μM in the presence of digitonin permeation (Supplementary Fig. 1A). Meanwhile, IFN-α could protect against HCoV-229E CPEs in MRC-5 cells with an EC50 of 0.12 μg/ml, indicating the presence of functional IFN-mediated antiviral response (Supplementary Fig. 1B).
Altogether, these results demonstrate that diABZI has potent anti HCoV-229E activity with a minimal effect on cellular cytotoxicity.
3.2. diABZI exhibited potent anti-HCoV-229E activity in human SAEC culture
Human reconstituted airway epithelial models have been broadly used in the area of respiratory diseases due to their high physiological relevance, as they faithfully recapitulate key characteristics of the in vivo airway. Here, we exploited the ALI model, which is convenient for viral inoculations because of the open apical area, to further characterize the anti-coronavirus effect of diABZI. Human SAECs (human small airway epithelial cells obtained from Lonza) were well differentiated under ALI conditions at 4 weeks after airlift, with the representative appearance of a pseudostratified ciliated columnar epithelium revealed by histology and microscopy (Supplementary Fig. 2A).
In parallel, we inoculated HCoV-229E on the apical surface at different MOIs to examine the replicative capacity of this viral strain in SAEC (Supplementary Fig. 2B). The characteristic features of HCoV-229E induced cell ultrastructure remodeling, epithelial damage and cell death, were easily distinguishable. In contrast, such a CPE was undetectable in the primary bronchial epithelial cell (PBEC) ALI model (data not shown).
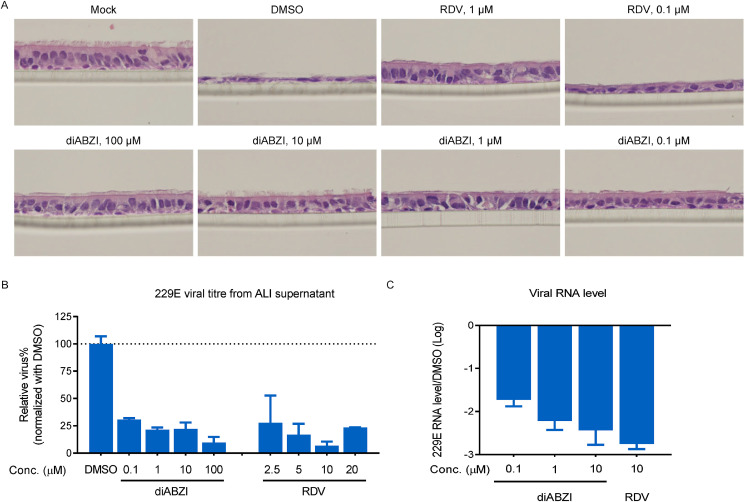
To explore the antiviral potential of diABZI in SAEC ALI, HCoV-229E was inoculated at an MOI of 0.01, followed by the addition of diABZI to the basal compartment which mimicked blood vessel drug transportation. Histological staining at 72 h post infection (hpi) showed that diABZI at concentrations as low as 0.1 μM successfully rescued the epithelium from CPEs, while RDV at the same dose had a modest protective effect (Fig. 2 A). Moreover, viral replication was monitored through repeated sampling and TCID50 titration on MRC-5 cells that underwent PBS washing from the apical surface. In parallel, comparative quantification of viral RNA in the supernatants from apical washes was performed by real-time PCR. Treatment with diABZI at concentration as low as 0.1 μM resulted in a 70% reduction in the HCoV-229E viral titer at 72 hpi, whereas RDV at 2.5 μM or higher exhibited a comparable antiviral viral ability (Fig. 2B). Similarly, diABZI administered basally with concentrations ranging from 0.1 to 10 μM showed a dose dependent inhibition, as observed by quantification of the viral RNA copy number in apical wash at 48 hpi (Fig. 2C). The amplitude of diABZI inhibition exceeded 1.5 log at 0.1 μM and reached 2 log at 1 μM, whereas obtaining a similar reduction by cGAMP required a minimum dose of 200 μM (Supplementary Fig. 3).
Fig. 2.
STING agonist represses HCoV-229E amplification on human SAEC ALI. (A–C) Human SAEC ALI cells were differentiated and seeded in 24-well plates. The cells were infected with HCoV-229E at MOI of 0.01 on apical and treated with different doses of the indicated compounds in basal medium. (A) After 72 h, the slides were subjected to the H&E stains. (B) After 72 h, the infectious supernatant collected from apical cells by PBS wash were subjected to viral titration assay on MRC-5. (C) After 48 h, the viral RNAs were isolated from the infectious supernatant collected from apical cells by PBS wash and measured by real-time PCR. The data are presented as the means ± SDs of triplicate samples from one experiment and are representative of at least three independent experiments.
In summary, in both 2D culture (MRC-5) and 3D culture (SAEC ALI) infection models, HCoV-229E production was more sensitive to diABZI than RDV, with greater inhibition seen at the same treatment concentration, which is in agreement with the lower EC50 revealed by CPE assay.
3.3. The combination of diABZI and RDV had a synergistic antiviral effect against HCoV-229E
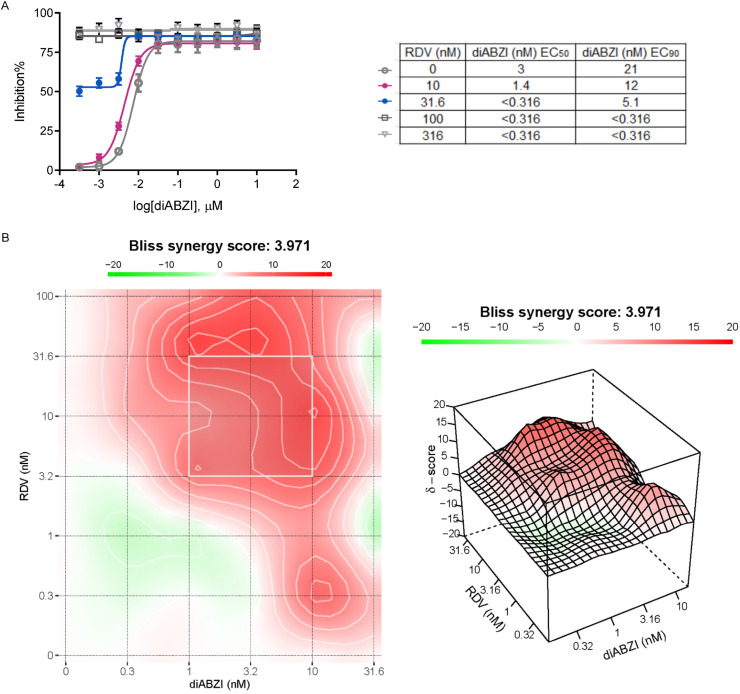
The combination of direct-acting antivirals and host directed immunomodulators could offer a two-pronged approach for the treatment of COVID-19. We therefore explored the antiviral potential of diABZI and RDV at several concentrations in a range of combination regimens against HCoV-229E in MRC-5 cells using a CPE assay (Fig. 3 A). For the single arm experiments, diABZI alone inhibited the HCoV-229E CPE with an average EC50 of 0.003 μM and EC90 of 0.021 μM, whereas RDV had an average EC50 of 0.026 μM and EC90 of 0.086 μM (Fig. 3A). The inhibitory effect of the combination at each dose was analyzed with SynergyFinder (https://synergyfinder.fimm.fi), which obtained a Bliss synergy score of 3.971 (Fig. 3B). This value indicates the synergistic interaction of diABZI and RDV, and this synergistic effect was not due to potential cytotoxicity of the combination because no noticeable cytotoxicity observed (data not shown). These results suggest that combining diABZI with RDV could potentially offer greater therapeutic efficacy than single usage alone.
Fig. 3.
The combination study for STING agonist diABZI with RDV. (A) (Top) Representative antiviral dose response curves of diABZI in combination with RDV against HCoV-229E. Serial dilutions of diABZI with a range of indicated concentration of RDV. (Bottom) In vitro antiviral activity shift in 50% and 90% of diABZI with fixed concentration of RDV. (B) A representative heat-map (Left) and 3-dimensional drug interaction landscape plotting synergy scores analyzed using SynergyFinder 2.0. The data are presented as the means ± SDs of triplicate samples from one experiment and are representative of at least three independent experiments.
3.4. Time-of-drug-addition study to elucidate diABZI-mediated anti HCoV-229E activity
To delineate the antiviral mechanism of diABZI against HCoV-229E in more detail, we first carried out a time-of-addition experiment under three scenarios, to examine whether diABZI could inhibit viral replication after the virus enters the host cells and whether the STING-mediated antiviral response can be sustained after drug removal.
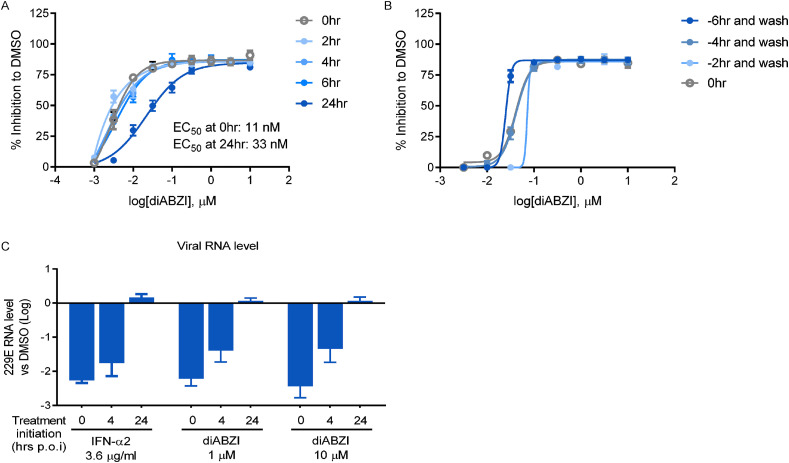
In brief, MRC-5 cells were inoculated with HCoV-229E and treated with diABZI at different time points, designated prior to infection (pre-treatment-wash, 2 h, 4 h, and 6 h), during infection (co-treatment, and 0 h), and post-infection (post-treatment, 2 h, 4 h, 6 h, and 24 h). Antiviral effects were determined based on the percentage of CPE inhibition. As shown in Fig. 4 A, the anti-HCoV-229E activity of diABZI was negligibly affected by delayed treatment initiation within the first 6 h; interestingly, 24 h post-treatment condition also inhibited HCoV-229E infection, shifting the EC50 value to 33 nM, threefold higher than that observed during co-treatment (11 nM).
Fig. 4.
Time-of-drug-addition study for STING agonist diABZI. (A) MRC-5 cells were infected with HCoV-229E at an MOI of 0.01 and treated with different doses of diABZI at the indicated times. After 72 h, the CPE was determined by a CCK-8 assay and the percent inhibition relative to DMSO treatment was plotted. (B) MRC-5 cells were pre-treated with different doses of diABZI at the indicated times, followed by infection with HCoV-229E at an MOI of 0.01 after PBS wash to remove the residue drug. After 72 h, the CPE was determined by a CCK-8 assay and the percent inhibition relative to DMSO treatment was plotted. (C) SAEC ALI were infected with HCoV-229E on apical cell at an MOI of 0.01 and treated with the indicated concentrations of compounds at different time points. After 48 h, the viral RNAs were isolated from the infectious supernatant collected from apical cells by PBS wash and measured by real-time PCR. The CPE was determined by a CCK-8 assay and the percent inhibition relative to DMSO treatment was plotted. The data are presented as the means ± SDs of triplicate samples from one experiment and are representative of at least three independent experiments.
For the pre-treatment-wash regimen, MRC-5 cells were incubated with diABZI for 2 h, 4 h, or 6 h prior to PBS washing, and infected with HCoV-229E in the absence of the drug. It was interesting to note that the dose response curves showed little difference between the pre-treat-wash and co-treatment conditions, suggesting that diABZI may be used as prophylactics to protect the host from viral infection (Fig. 4B).
We also performed the post-treatment experiment in an HCoV-229E infected primary SAEC ALI model (Fig. 4C). Notably, when diABZI treatment was delayed to 4 hpi, the antiviral response was impaired as evidenced by the decreased extent of viral load reduction in comparison to that upon co-treatment. Moreover, post-treatment with diABZI for 24 h, even at a dose of 10 μM, did not inhibit RNA replication. Determining why the treatment of primary SAECs was more dependent on the time of treatment requires further investigation by future study or investigation. In any case, the time-of-addition studies confirmed the potent antiviral activity of diABZI against HCoV-229E, providing important insight of diABZI into the STING-mediated induction of type I IFNs and subsequent antiviral signaling pathways responsible for the inhibition of HCoV-229E infection and replication.
3.5. diABZI mediated STING activation inhibited HCoV-229E via TBK1-IRF pathway
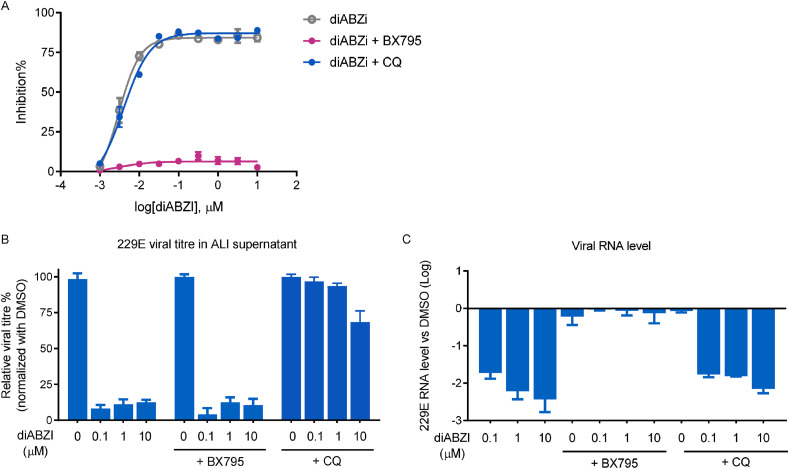
In addition to its role in the induction of type I interferon responses through TANK-binding kinase 1 (TBK1), STING activation has been shown to induce autophagy via ATG5 (Gui et al., 2019). Our previous report demonstrated that diABZI-mediated activation of STING inhibited the replication of PIV3 and HRV16 through TBK1 and autophagy, respectively. Here, to explore which signaling pathway plays a major role in the anti-coronavirus activity of diABZI, the TBK1 inhibitor BX795 or the autophagy inhibitor chloroquine (CQ) was applied during diABZI treatment upon HCoV-229E infection in both MRC-5 cells and the SAEC ALI model. As shown in Fig. 5 A and Fig. 5B, BX795 completely abolished the antiviral activity of diABZI in HCoV-229E infected MRC-5 cells and SAECs. 229E viral RNA quantification revealed a similar pattern: only BX795 but not CQ eradicated the diABZI antiviral ability in HCoV-229E infected SAECs (Fig. 5C). In contrast, the protection against the CPE and reduction in viral titer were barely affected by the addition of CQ, while CQ or BX795 alone at the tested concentration had no effect on HCoV-229E replication (data not shown). These results suggest that diABZI mediated anti HCoV-229E activity is independent of autophagy but largely requires TBK1-IRF activation and the type I IFN response.
Fig. 5.
The mechanism of action study for STING agonist diABZI on MRC5 and reconstituted human ALI system. (A) MRC-5 cells were infected with HCoV-229E at an MOI of 0.01 and treated with the indicated concentrations of diABZI in the presence or absence of 5 μM TBKi (BX795) or 10 μM CQ for 72 h. (B) SAEC ALI were infected with HCoV-229E on apical cell at an MOI of 0.01 and treated with the indicated concentrations of diABZI in the presence or absence of 10 μM TBKi (BX795) or 10 μM CQ for 72 h in basal medium. The infectious supernatants collected from apical cells by PBS wash were subjected to viral titration assay on MRC-5. (C) SAEC ALI were infected with HCoV-229E on apical cell at an MOI of 0.01 and treated with the indicated concentrations of diABZI in the presence or absence of 10 μM TBKi (BX795) or 10 μM CQ for 72 h in basal medium. After 48 h, the viral RNAs were isolated from the infectious supernatant collected from apical cells by PBS wash and measured by real-time PCR. The data are presented as the means ± SDs of triplicate samples from one experiment and are representative of at least three independent experiments. *P < 0.05, **P < 0.01.
3.6. diABZI exhibited potent anti-SARS-CoV-2 activity in reconstituted human bronchial airway epithelia culture
To establish the potential use of diABZI in treating human coronaviruses, especially highly pathogenic strains, we next determined the anti-SARS-CoV-2 activity of diABZI in MucilAir bronchial HAE reconstituted from human primary cells obtained from bronchial biopsies provided by Epithelix SARL (Geneva, Switzerland) and maintained at the ALI Similar to SAECs, the MucilAir bronchial HAE recapitulates much of the complexity and architecture of the respiratory epithelium and is known to be one of the most physiologically relevant cell culture systems in which to study respiratory pathogens. Vero E6 is a workhorse cell line used for in vitro studies of SARS-CoV and permissive to SARS-CoV-2 infection, however, diABZI lost its STING dependent anti-PIV3 ability on VERO cells, indicating the deficiency of Vero E6 cell in STING signaling (Supplementary Fig. 4). Considering the deficiency of STING signaling on Vero E6 cells, the MucilAir bronchial HAE model is an ideal tool to determine the effect of diABZI on SARS-CoV-2 infection (Langereis et al., 2015; Matsuyama et al., 2020; Osada et al., 2014; Pizzorno et al., 2020).
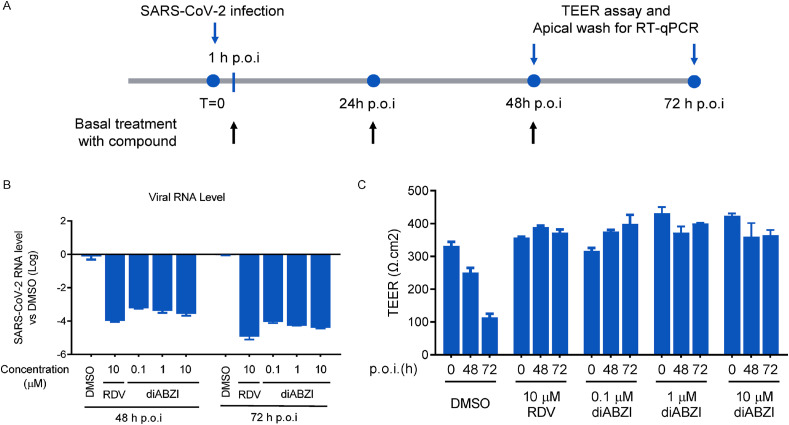
First the test compound (0.1, 1, 10 μM diABZI, or RDV, or DMSO as a control) was added to the basolateral chamber of the ALI, followed by apical challenge with SARS-CoV-2 (Beta CoV/France/IDF0571/2020 strain) challenge apically. The viral RNA collected from apical washes at 48 h and 72 h hpi was quantified, and disruption of the epithelial layer was measured as TEER (Fig. 6 A). As shown in Fig. 6B, diABZI in all tested concentration range potently decreased the level of SARS-CoV-2 viral RNA in a dose-dependent manner. Strikingly, treatment with diABZI at the concentration as low as 0.1 μM fully protected the integrity of the epithelial layer in the ALI model from SARS-CoV-2 challenge (Fig. 6C). Meanwhile, neither diABZI nor RDV had a negative impact on TEER over time, unlike the decrease in TEER indicative epithelial damage observed in the DMSO group (Fig. 6C). Taken together, these results demonstrate that diABZI could potently inhibit SARS-CoV-2 infection in a model of the HAE and has a favorable cytotoxicity profile.
Fig. 6.
The STING agonist exhibits anti-SARS-CoV-2 ability on MucilAir™. (A) The diagram of assay procedure. (B–C) MucilAir™ were infected with SARS-CoV-2 at an MOI 0.1 at apical cell, followed by treatment with indicated concentrations of compounds for 48 h and 72 h. At indicated time point, (B) SARS-CoV-2 viral RNA quantification and (C) transepithelial electrical resistance (TEER) assay were performed for treatment of diABZI or RDV. The data are presented as the means ± SD of triplicate samples from one experiment and are representative of at least three independent experiments. *P < 0.05, **P < 0.01.
4. Discussion
In the face of the global public health crisis caused by coronavirus infection, an effective treatment is urgently needed. However, the recently released interim results from one of the world's largest trials of COVID-19 therapies indicate that drugs repurposed for the treatment of COVID-19, including Remdesivir, hydroxychloroquine, lopinavir, and interferon regimens, appeared to have little or no effect on hospitalized COVID-19 patients (Pan et al., 2020b). IFNs are pleotropic cytokines with broad spectrum antiviral activity. Previous studies in cell culture systems have shown the antiviral effects of IFNs against MERS and SARS (Hart et al., 2014; Scagnolari et al., 2004). In addition, pre-treatment with IFN completely prevented HCoV-229E replication, CPEs and massive cell death in monocyte-derived dendritic cells (Mesel-Lemoine et al., 2012). Recently, recombinant interferons were reported to exhibit anti-SARS-CoV-2 activity in Vero and primary human bronchial epithelial cells (Busnadiego et al., 2020; Mantlo et al., 2020; Yuan et al., 2020). However, a recent study also suggested that IFNs may have pro-viral functions through enhanced expression of the viral entry receptor angiotensin-converting enzyme 2 (ACE2) (Ziegler et al., 2020). Together with the unsatisfactory efficacy results obtained from the COVID-19 clinical trials so far, substantial optimization is needed to make IFN-based treatment an effective therapy for COVID-19 patients and there is clearly a need for additional therapeutic options.
Serving as the first line of defense against viral infections, STING is required for the induction of innate immunity signals during infection from a broad range of viruses. Thus, STING agonists have the potential to be developed as prophylactic or therapeutic treatments against a broad range of viral infections. In a previous study, using diABZI as a tool, we demonstrated its potent antiviral activity against the representative respiratory RNA viruses, PIV3 and HRV16, upon STING activation (Zhu et al., 2020b). In the current study, its potent anti-coronavirus activity against HCoV-229E and SARS-CoV-2 was also demonstrated. We also confirmed that this antiviral activity is dependent on TBK1, which is located downstream of STING and responsible for the induction of a broad range of ISGs and production of IFNs (Barber, 2015). These encouraging results support the accelerated further optimization and advancement of novel STING agonists to the clinic to address the current huge unmet medical need.
Substantial progress towards the discovery of novel STING agonists with improved properties for potential therapeutic applications has been made over the years. The prokaryotic cyclic di-(3’:5′)-guanosine monophosphate (c-di-GMP) and eukaryotic 2′, 3′-cGAMP CDNs are both naturally-occurring STING agonists with potent immunostimulatory effects (Deng et al., 2014; Ohkuri et al., 2017). However, the CDN type molecules derived from natural STING ligands have disadvantages in terms of stability and permeability that limit their therapeutic applications. 5, 6-dimethylxanthenone-4-acetic acid (DMXAA) was the first identified non-CDN STING agonist with murine STING activity, but failed to bind and activate human STING (Conlon et al., 2013). In 2018, diABZI was discovered as a new class of synthetic small molecule STING receptor agonist with much higher potency than CDNs and suitable for systemic administration (Ramanjulu et al., 2018). Recently, an orally available non-nucleotide STING agonist, MSA-2, was reported to have favorable activity and tolerability profiles (Pan et al., 2020a). Further optimization of these molecules for improved safety margins may still be needed for their clinical application against coronavirus infections.
Our results also indicate that synergistic effects may be achieved through combination therapy consisting of immunomodulators and direct antiviral agents. This would allow immunomodulators to be administered at lower doses than alone. STING agonists, such as diABZI, trigger potent innate immune activation inside the host upon application. Cations must be taken to minimize the side effects and potential immunopathogenesis caused by STING stimulation. It may be essential to develop a pro- or lung-specific STING agonist to prevent coronaviruses infection, as well as intranasal delivery approach (Kumaki et al., 2017).
In severe COVID-19 patients, the overproduction of inflammatory cytokines, such as IL-6 and TNF, and low levels of IFNs are often observed, which might be the cause of sustained viral replication and disease progression (Zhou et al., 2020). The Glucocorticoids dexamethasone, which exerts anti-inflammation effect in different pathways (Zhu et al., 2010), could reduce mortality in only severe COVID-19 patients who require invasive mechanical ventilation or oxygen (41.4%–29.3%), but increased the death rate in patients who received no respiratory support (14.0%–17.3%) (Group et al., 2020). These results suggest that the timing of dexamethasone treatment is critical. On the other hand, during the early stage of disease, suppressing viral replication and increasing the IFN response via immune modulator drugs, such as STING agonist, may be a viable approach to maintain a balanced immune response and prevent disease progression. In addition, it is also very intriguing to know whether the combination treatment of STING agonists with the cytokine storm inhibitors could relieve the COVID-19 disease progression in severe/late stage patients in future investigation.
Conflict of interest statement
The authors are employees of F. Hoffman-La Roche Ltd.
Financial support
This work was supported by F. Hoffman-La Roche Ltd.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2021.105015.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahn J., Barber G.N. STING signaling and host defense against microbial infection. Exp. Mol. Med. 2019;51:1–10. doi: 10.1038/s12276-019-0333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber G.N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fatkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C., Members A.-S.G. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busnadiego I., Fernbach S., Pohl M.O., Karakus U., Huber M., Trkola A., Stertz S., Hale B.G. Antiviral activity of type I, II, and III interferons Counterbalances ACE2 inducibility and restricts SARS-CoV-2. mBio. 2020:11. doi: 10.1128/mBio.01928-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi N., Ferrarese R., Criscuolo E., Diotti R.A., Castelli M., Scagnolari C., Burioni R., Antonelli G., Clementi M., Mancini N. Interferon-beta 1a inhibits SARS-CoV-2 in vitro when administered after virus infection. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiaa350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon J., Burdette D.L., Sharma S., Bhat N., Thompson M., Jiang Z., Rathinam V.A., Monks B., Jin T., Xiao T.S., Vogel S.N., Vance R.E., Fitzgerald K.A. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J. Immunol. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoudi-Monfared E., Rahmani H., Khalili H., Hajiabdolbaghi M., Salehi M., Abbasian L., Kazemzadeh H., Yekaninejad M.S. A randomized clinical trial of the efficacy and safety of interferon beta-1a in treatment of severe COVID-19. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.01061-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Liang H., Xu M., Yang X., Burnette B., Arina A., Li X.D., Mauceri H., Beckett M., Darga T., Huang X., Gajewski T.F., Chen Z.J., Fu Y.X., Weichselbaum R.R. STING-dependent Cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group R.C., Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., Prudon B., Green C., Felton T., Chadwick D., Rege K., Fegan C., Chappell L.C., Faust S.N., Jaki T., Jeffery K., Montgomery A., Rowan K., Juszczak E., Baillie J.K., Haynes R., Landray M.J. Dexamethasone in hospitalized patients with Covid-19 - preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui X., Yang H., Li T., Tan X., Shi P., Li M., Du F., Chen Z.J. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart B.J., Dyall J., Postnikova E., Zhou H., Kindrachuk J., Johnson R.F., Olinger G.G., Frieman M.B., Holbrook M.R., Jahrling P.B., Hensley L. Interferon-beta and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 2014;95:571–577. doi: 10.1099/vir.0.061911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y., Salazar A.M., Wandersee M.K., Barnard D.L. Prophylactic and therapeutic intranasal administration with an immunomodulator, Hiltonol((R)) (Poly IC:LC), in a lethal SARS-CoV-infected BALB/c mouse model. Antivir. Res. 2017;139:1–12. doi: 10.1016/j.antiviral.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis M.A., Rabouw H.H., Holwerda M., Visser L.J., van Kuppeveld F.J. Knockout of cGAS and STING rescues virus infection of plasmid DNA-transfected cells. J. Virol. 2015;89:11169–11173. doi: 10.1128/JVI.01781-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., de Vries M., Valero-Jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V.D. bioRxiv; 2020. SARS-CoV-2 Is Sensitive to Type I Interferon Pretreatment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesel-Lemoine M., Millet J., Vidalain P.O., Law H., Vabret A., Lorin V., Escriou N., Albert M.L., Nal B., Tangy F. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J. Virol. 2012;86:7577–7587. doi: 10.1128/JVI.00269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk, P.D., Marsden, R.J., Tear, V.J., Brookes, J., Batten, T.N., Mankowski, M., Gabbay, F.J., Davies, D.E., Holgate, S.T., Ho, L.-P., Clark, T., Djukanovic, R., Wilkinson, T.M.A., Crooks, M.G., Dosanjh, D.P.S., Siddiqui, S., Rahman, N.M., Smith, J.A., Horsley, A., Harrison, T.W., Saralaya, D., McGarvey, L., Watson, A., Foster, E., Fleet, A., Singh, D., Hemmings, S., Aitken, S., Dudley, S., Beegan, R., Thompson, A., Rodrigues, P.M.B., Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. The Lancet Respiratory Medicine. [DOI] [PMC free article] [PubMed]
- Ohkuri T., Kosaka A., Ishibashi K., Kumai T., Hirata Y., Ohara K., Nagato T., Oikawa K., Aoki N., Harabuchi Y., Celis E., Kobayashi H. Intratumoral administration of cGAMP transiently accumulates potent macrophages for anti-tumor immunity at a mouse tumor site. Cancer Immunol. Immunother. 2017;66:705–716. doi: 10.1007/s00262-017-1975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada N., Kohara A., Yamaji T., Hirayama N., Kasai F., Sekizuka T., Kuroda M., Hanada K. The genome landscape of the african green monkey kidney-derived vero cell line. DNA Res. 2014;21:673–683. doi: 10.1093/dnares/dsu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B.S., Perera S.A., Piesvaux J.A., Presland J.P., Schroeder G.K., Cumming J.N., Trotter B.W., Altman M.D., Buevich A.V., Cash B., Cemerski S., Chang W., Chen Y., Dandliker P.J., Feng G., Haidle A., Henderson T., Jewell J., Kariv I., Knemeyer I., Kopinja J., Lacey B.M., Laskey J., Lesburg C.A., Liang R., Long B.J., Lu M., Ma Y., Minnihan E.C., O'Donnell G., Otte R., Price L., Rakhilina L., Sauvagnat B., Sharma S., Tyagarajan S., Woo H., Wyss D.F., Xu S., Bennett D.J., Addona G.H. An orally available non-nucleotide STING agonist with antitumor activity. Science. 2020;369 doi: 10.1126/science.aba6098. [DOI] [PubMed] [Google Scholar]
- Pan H., Peto R., Karim Q.A., Alejandria M., Henao-Restrepo A.M., García C.H., Kieny M.-P., Malekzadeh R., Murthy S., Preziosi M.-P., Reddy S., Periago M.R., Sathiyamoorthy V., Røttingen J.-A., Swaminathan S. medRxiv; 2020. Repurposed Antiviral Drugs for COVID-19 –interim WHO SOLIDARITY Trial Results. 2020.2010.2015.20209817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections-more than just the common cold. J. Am. Med. Assoc. 2020;323:707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- Pizzorno A., Padey B., Julien T., Trouillet-Assant S., Traversier A., Errazuriz-Cerda E., Fouret J., Dubois J., Gaymard A., Lescure F.X., Duliere V., Brun P., Constant S., Poissy J., Lina B., Yazdanpanah Y., Terrier O., Rosa-Calatrava M. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep Med. 2020;1:100059. doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prytherch Z., Job C., Marshall H., Oreffo V., Foster M., BeruBe K. Tissue-Specific stem cell differentiation in an in vitro airway model. Macromol. Biosci. 2011;11:1467–1477. doi: 10.1002/mabi.201100181. [DOI] [PubMed] [Google Scholar]
- Ramanjulu J.M., Pesiridis G.S., Yang J., Concha N., Singhaus R., Zhang S.Y., Tran J.L., Moore P., Lehmann S., Eberl H.C., Muelbaier M., Schneck J.L., Clemens J., Adam M., Mehlmann J., Romano J., Morales A., Kang J., Leister L., Graybill T.L., Charnley A.K., Ye G., Nevins N., Behnia K., Wolf A.I., Kasparcova V., Nurse K., Wang L., Puhl A.C., Li Y., Klein M., Hopson C.B., Guss J., Bantscheff M., Bergamini G., Reilly M.A., Lian Y., Duffy K.J., Adams J., Foley K.P., Gough P.J., Marquis R.W., Smothers J., Hoos A., Bertin J. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature. 2018;564:439–443. doi: 10.1038/s41586-018-0705-y. [DOI] [PubMed] [Google Scholar]
- Scagnolari C., Vicenzi E., Bellomi F., Stillitano M.G., Pinna D., Poli G., Clementi M., Dianzani F., Antonelli G. Increased sensitivity of SARS-coronavirus to a combination of human type I and type II interferons. Antivir. Ther. 2004;9:1003–1011. [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.X., Fish E.N. Global virus outbreaks: interferons as 1st responders. Semin. Immunol. 2019;43:101300. doi: 10.1016/j.smim.2019.101300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-J., Chen Y.-X., Li Y.-M. bioRxiv; 2020. Adopting STING Agonist Cyclic Dinucleotides as a Potential Adjuvant for SARS-CoV-2 Vaccine. 2020.2007.2024.217570. [Google Scholar]
- Yuan S., Chan C.C., Chik K.K., Tsang J.O., Liang R., Cao J., Tang K., Cai J.P., Ye Z.W., Yin F., To K.K., Chu H., Jin D.Y., Hung I.F., Yuen K.Y., Chan J.F. Broad-spectrum host-based antivirals targeting the interferon and lipogenesis pathways as potential treatment options for the pandemic coronavirus disease 2019 (COVID-19) Viruses. 2020:12. doi: 10.3390/v12060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Yang J., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890. doi: 10.1016/j.chom.2020.04.017. e882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Hu H., Liu H., Shen H., Yan Z., Gao L. A synthetic STING agonist inhibits the replication of Human Parainfluenza Virus 3 and Rhinovirus 16 through distinct mechanisms. Antivir. Res. 2020:104933. doi: 10.1016/j.antiviral.2020.104933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.Y., Liu Q., Chen J.X., Lan K., Ge B.X. MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation of MAPKs in macrophages. J. Immunol. 2010;185:7435–7442. doi: 10.4049/jimmunol.1000798. [DOI] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., 2nd, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.E., Barbry P., Leslie A., Kiem H.P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J., lung-network@humancellatlas.org, H.C.A.L.B.N.E.a., Network, H.C.A.L.B. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.