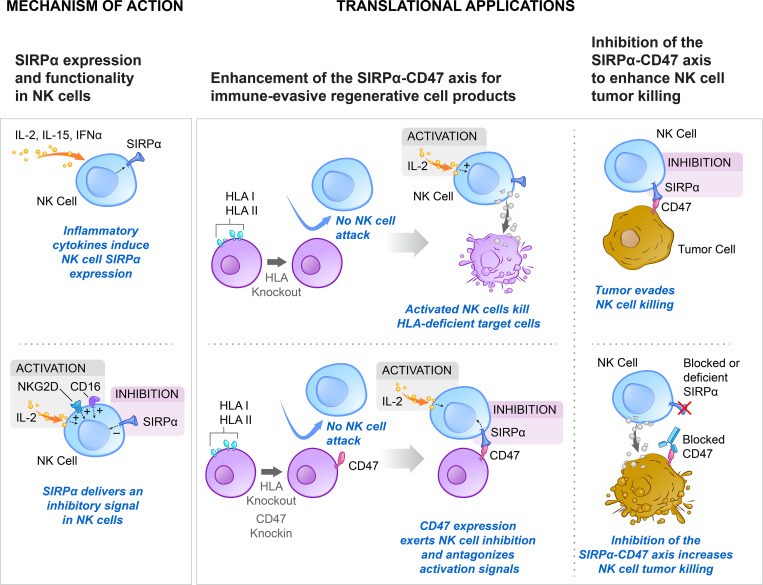
NK cells are able to upregulate SIRPα, which transmits a strong inhibitory signal upon CD47 engagement. Modulation of this immune checkpoint can enable the generation of universal allogeneic regenerative cell products or enhance NK cell–mediated tumor cell killing.
Abstract
Here we report on the existence and functionality of the immune checkpoint signal regulatory protein α (SIRPα) in NK cells and describe how it can be modulated for cell therapy. NK cell SIRPα is up-regulated upon IL-2 stimulation, interacts with target cell CD47 in a threshold-dependent manner, and counters other stimulatory signals, including IL-2, CD16, or NKG2D. Elevated expression of CD47 protected K562 tumor cells and mouse and human MHC class I–deficient target cells against SIRPα+ primary NK cells, but not against SIRPα− NKL or NK92 cells. SIRPα deficiency or antibody blockade increased the killing capacity of NK cells. Overexpression of rhesus monkey CD47 in human MHC-deficient cells prevented cytotoxicity by rhesus NK cells in a xenogeneic setting. The SIRPα–CD47 axis was found to be highly species specific. Together, the results demonstrate that disruption of the SIRPα–CD47 immune checkpoint may augment NK cell antitumor responses and that elevated expression of CD47 may prevent NK cell–mediated killing of allogeneic and xenogeneic tissues.
Graphical Abstract
Introduction
Natural killer (NK) cells express a large repertoire of activating and inhibitory receptors (Lanier, 2008) and function as key players for the elimination of cells that have undergone infection or malignant transformation (Vivier et al., 2008). To acquire functional competence, NK cells undergo an educational “licensing” process (Kim et al., 2012) to ensure that only those that express a cognate inhibitory receptor for self-MHC class I molecules become functionally mature. This central self-tolerance mechanism sets the triggering threshold, and the integration of all transmitted activating and inhibitory signals determines the outcome and magnitude of interactions with target cells (Raulet and Vance, 2006).
The emerging field of cancer immunotherapy explores novel methods for increasing NK cell antitumor immunity by using immune checkpoint inhibitors aimed at tilting the balance toward activation, many of which target inhibitory receptors shared by NK cells and T cells (Childs and Carlsten, 2015; Chiossone et al., 2018). Clinical trials exploring the efficacy of disruption of inhibitory killer cell Ig-like receptors (KIRs; Korde et al., 2014; Sola et al., 2009) and NKG2A (Andre et al., 2018) through mAb-mediated blockade and others are under development.
As next-generation cell replacement therapies are developed, there will be a need to generate universally compatible nonimmunogenic stem cells that evade immune rejection and can develop into functionally active somatic cells after transplantation. However, the disruption of MHC class I in allogeneic cells, necessary to prevent CD8+ T cell activation, increases the cells’ susceptibility to innate immune clearance (Gornalusse et al., 2017). Turning the concept of immune checkpoint inhibition on its head, we aimed to exploit cancer survival pathways for the silencing of innate immunity. The inhibitory ligand CD47 is an efficient immunomodulator (Advani et al., 2018) and was previously described to be up-regulated in cancer and to exclusively inhibit macrophage clearance (Willingham et al., 2012). Here, we report that CD47 transmits threshold-dependent, direct inhibitory signals to activated NK cells via SIRPα. CD47 overexpression thus emerges as a very effective checkpoint enhancer with implication for both cancer immunotherapy and regenerative medicine.
Results
Overexpression of mouse Cd47 prevents NK cell killing of MHC class I– and II–deficient target cells
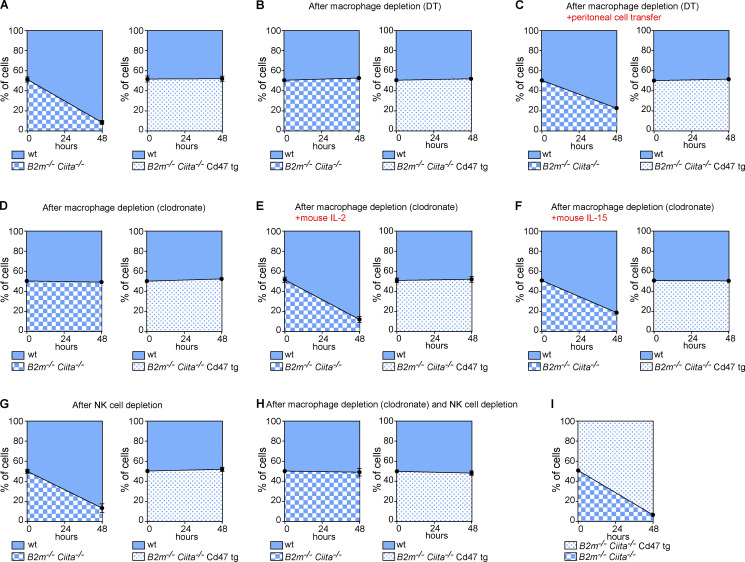
In C57BL/6 mouse induced pluripotent stem cells (miPSCs), the B2m and Ciita genes were targeted for disruption with Cas9 nuclease and guides to deplete both MHC class I and class II expression. According to the “missing-self” hypothesis, target cells lacking MHC class I fail to engage with inhibitory NK cell receptors and trigger cytotoxic killing (Kärre et al., 1986). KIRs in humans and the Ly49 receptors in mice bind MHC class I and mediate the inhibitory signaling via immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in their cytoplasmic tails. Although less well established, MHC class II molecules have also been suggested to regulate NK cell function and target cell recognition (Jiang et al., 1996; Johnson et al., 2018). The generated B2m−/−Ciita−/− miPSCs should thus exhibit increased NK cell susceptibility. To study their fate in vivo, a 1:1 mixture of CFSE-labeled WT miPSCs and B2m−/−Ciita−/− miPSCs was injected into the innate immune cell-rich peritoneum of syngeneic C57BL/6 mice (Fig. 1 A). After 48 h, CFSE-labeled peritoneal miPSCs were recovered and the percentages of both fractions assessed by flow cytometry. The vast majority of B2m−/−Ciita−/− miPSCs had been cleared, and mainly WT miPSCs could be retrieved. To assess an inhibitory function of up-regulated Cd47 against innate immune clearance, the mouse Cd47 cDNA sequence was synthesized and cloned into a lentiviral vector with blasticidin resistance, and B2m−/−Ciita−/− miPSC clones were transduced and antibiotic-selected pools of B2m−/−Ciita−/− Cd47 transgenic (tg) miPSCs were expanded. When a 1:1 mixture of WT miPSCs and B2m−/−Ciita−/− Cd47 tg miPSCs was injected, both fractions were equally recovered after 48 h. This demonstrated that Cd47 overexpression fully silenced all innate immune killing, despite complete lack of both MHC class I and II molecules. CD11b tg mice with a diphtheria toxin (DT)–inducible system that transiently depletes macrophages were used to selectively study NK cell–mediated rejection. DT-treated mice killed neither B2m−/−Ciita−/− nor B2m−/−Ciita−/− Cd47 tg miPSCs (Fig. 1 B). Only when the animals additionally received a peritoneal cell transfer from naive C57BL/6 mice were B2m−/−Ciita−/− miPSCs cleared (Fig. 1 C). This demonstrates that macrophages are required to activate NK cell killing, which is not induced by target cell MHC deficiency alone. In animals depleted of macrophages using clodronate, mouse IL-2 (a cytokine produced by lymphocytes) and mouse IL-15 (a cytokine produced by myeloid cells) were shown to deliver such activating signals (Fig. 1, D–F). When NK cells were depleted in mice using a mAb against NK1.1 to selectively study macrophages, B2m−/−Ciita−/−, but not B2m−/−Ciita−/−, Cd47 tg miPSCs were cleared without requiring another activating factor (Fig. 1 G). When both macrophages and NK cells were depleted, the B2m−/−Ciita−/− miPSCs were not eliminated, suggesting that no other immune cell populations contributed to miPSC clearance (Fig. 1 H). Animals clearing B2m−/−Ciita−/− miPSCs still spared concomitantly injected B2m−/−Ciita−/− Cd47 tg miPSCs, thus indicating that even in an environment of activated innate immune cells, Cd47 overexpression sufficiently suppressed cytotoxicity against MHC-deficient cells (Fig. 1 I).
Figure 1.
Cd47 overexpression protects MHC-deficient mouse iPSCs from killing by stimulated NK cells or macrophages in vivo. A 1:1 mixture of CFSE-labeled WT and either B2m−/−Ciita−/− or B2m−/−Ciita−/− Cd47 tg miPSCs was injected into the peritoneum of mice on a syngeneic C57BL/6 background. After 48 h, the ratio of recovered CFSE-positive miPSCs was determined (mean ± SD, triplicates in four animals per group). (A) Recipient WT C57BL/6 mice did not receive additional treatment. (B) A tg CD11b-DT receptor mouse on C57BL/6 background was used to selectively deplete macrophages. (C) The peritoneal cell populations in macrophage-depleted tg CD11b-DT receptor mice were restored by peritoneal cell transfer from WT C57BL/6 mice. (D) Macrophages were pharmacologically depleted in WT C57BL/6 mice using clodronate. (E) In macrophage-depleted mice, peritoneal NK cells were stimulated by peritoneal injections of mouse IL-2. (F) In macrophage-depleted mice, peritoneal NK cells were stimulated by injections of mouse IL-15. (G) In C57BL/6 mice, NK cells were depleted with an anti-NK1.1 depleting antibody. (H) In WT C57BL/6 mice, both macrophages and NK cells were depleted. (I) A 1:1 mixture of CFSE-labeled B2m−/−Ciita−/− and B2m−/−Ciita−/− Cd47 tg miPSCs was injected into WT C57BL/6 mice. wt, wild-type.
NK cells require an additional activation signal to kill MHC class I– and II–deficient target cells
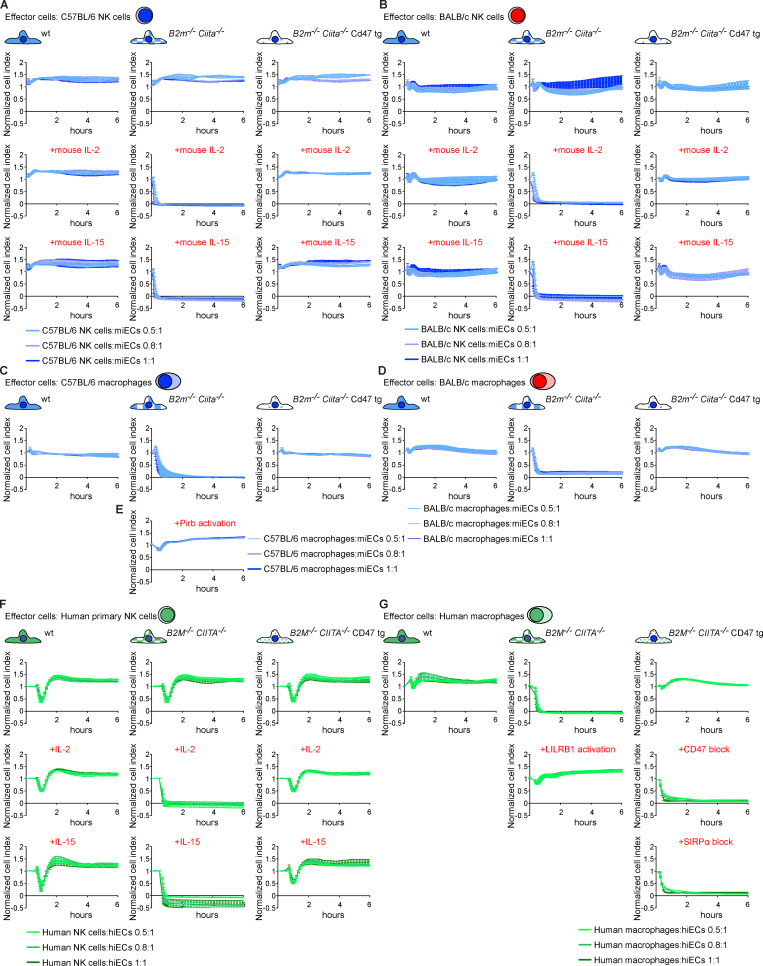
The kinetics of innate immune cell killing was assessed in vitro using a real-time cellular impedance assay, for which miPSCs were differentiated into mouse endothelial cells (miECs). No differences in the outcomes or kinetics of miEC killing were observed between syngeneic C57BL/6 or allogeneic BALB/c NK cells and macrophages (Fig. S1). WT iPSC-derived endothelial cells (iECs) were spared by all NK cells and macrophages. B2m−/−Ciita−/− miECs were ignored by unstimulated NK cells but rapidly killed if mouse IL-2 or mouse IL-15 was added, and the killing was faster when higher effector-to-target cell (E:T) ratios were used. Macrophages cleared B2m−/−Ciita−/− miECs without additional stimulation but were inhibited when their inhibitory MHC class I–binding receptor, Pirb (Kim et al., 2013), was cross-linked with an antibody. These findings suggest that mouse macrophages can sense and independently kill MHC class I–deficient cells using unknown activating receptors. B2m−/−Ciita−/− Cd47 tg miECs were resistant to all NK cells and macrophages.
Figure S1.
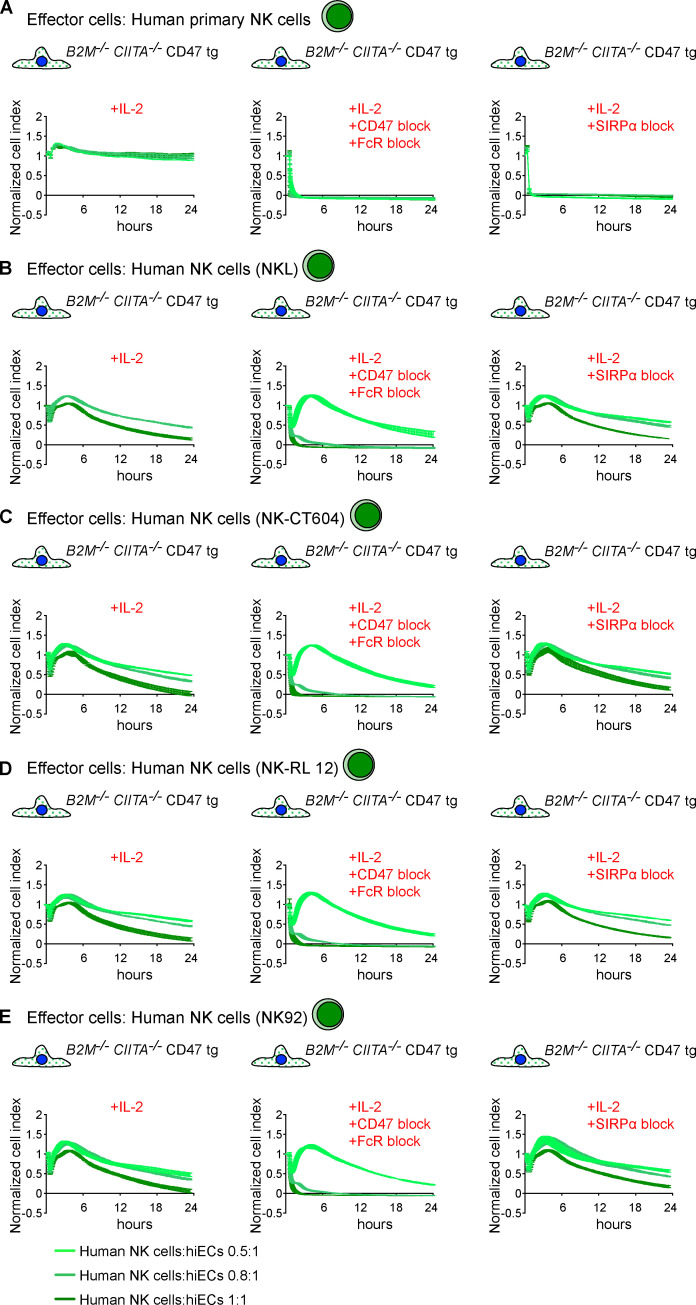
Cd47 protects MHC-deficient miECs and CD47 protects HLA-deficient hiECs from killing by NK cells and macrophages in vitro. (A–D) WT, B2m−/−Ciita−/−, and B2m−/−Ciita−/− Cd47 tg miECs were challenged with syngeneic C57BL/6 NK cells (A), allogeneic BALB/c NK cells (B), syngeneic C57BL/6 macrophages (C), or allogeneic BALB/c macrophages (D). Where indicated, NK cells were stimulated with mouse IL-2 or IL-15. Graphs show mean ± SD and three independent replicates per group and time point; three different E:T ratios are shown. (E) B2m−/−Ciita−/− miECs were incubated with syngeneic C57BL/6 macrophages treated with an activating anti-Pirb antibody (mean ± SD, three independent replicates per group and time point, and three different E:T ratios are shown). (F and G) WT, B2M−/−CIITA−/−, and B2M−/−CIITA−/− CD47 tg hiECs were challenged with allogeneic human primary NK cells (F) or allogeneic human macrophages (G). Where indicated, NK cells were stimulated with IL-2 or IL-15. Where indicated, an agonist anti-LILRB1 antibody (clone GHI/75) was added to the assay. A specific blocking antibody against CD47 (clone B6.H12) or a blocking peptide against SIRPα was used in some assays. Graphs show mean ± SD and three independent replicates per group and time point; three different E:T ratios are shown. wt, wild-type.
Cd47 inhibits NK cell activation via Sirpα ligation
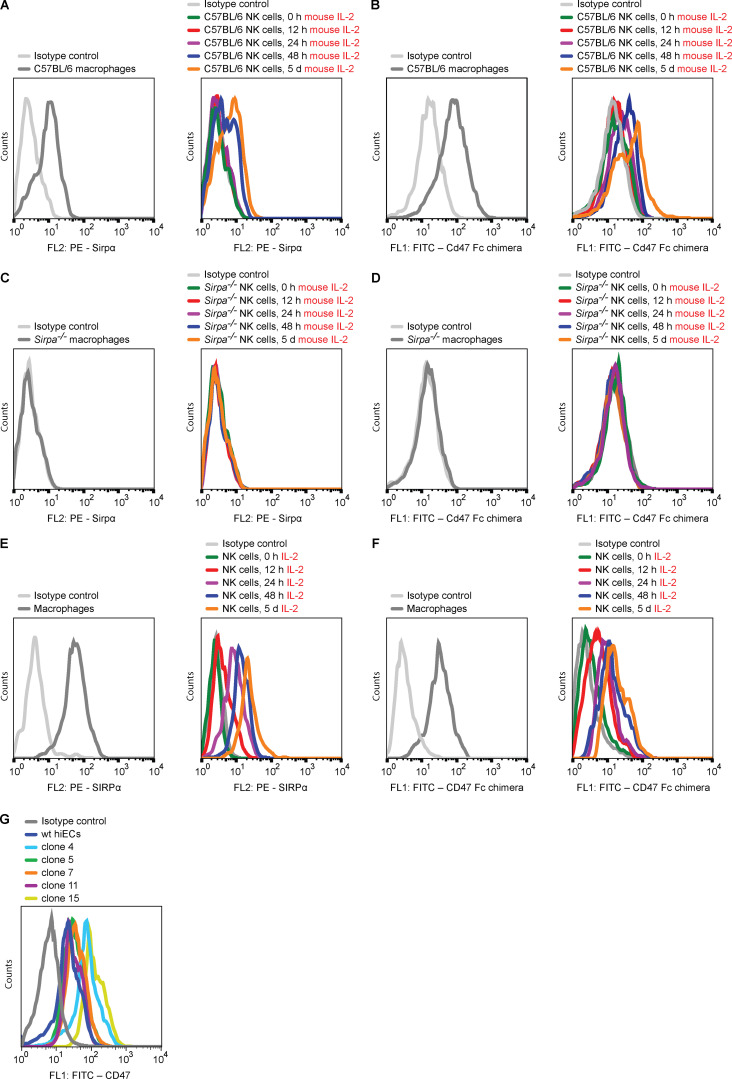
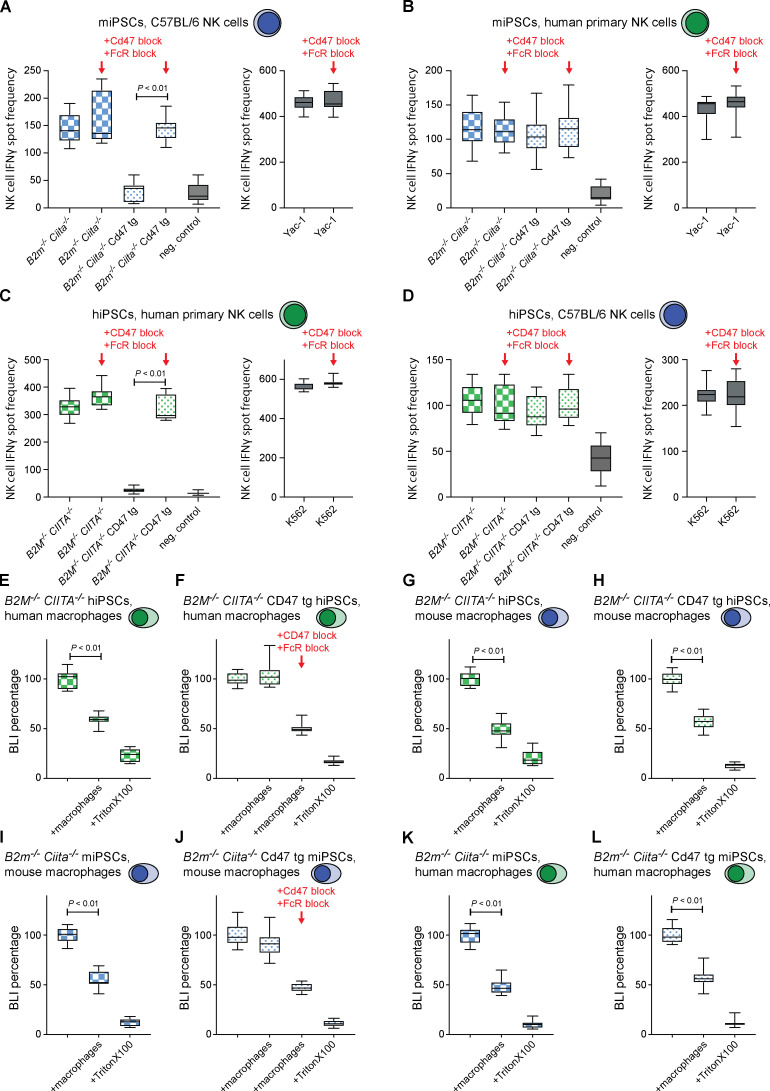
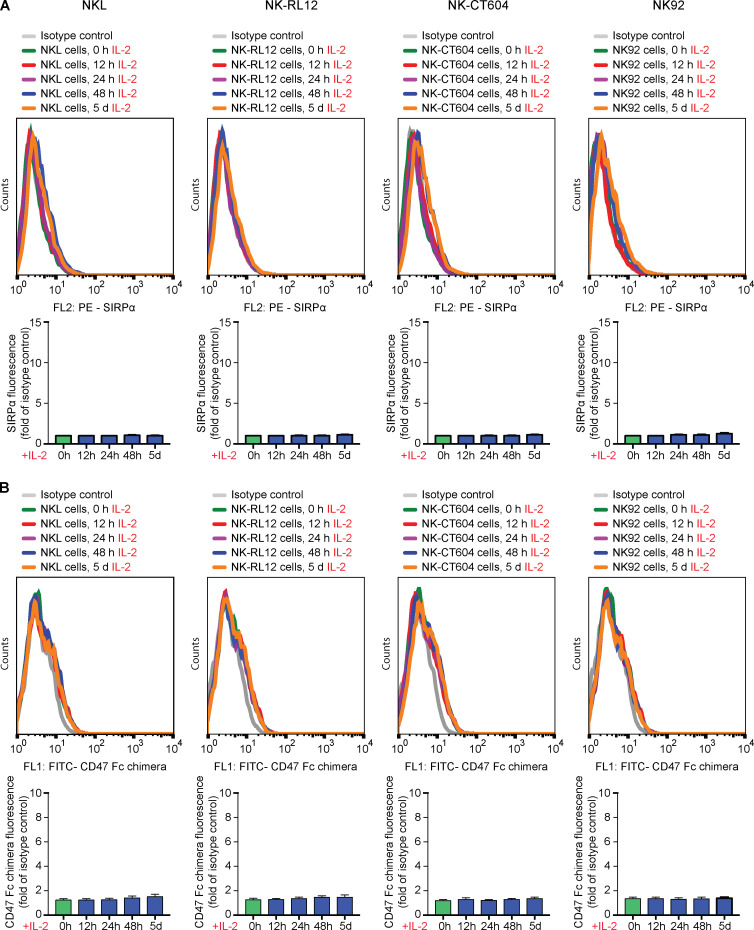
CD47 acts as a marker of “self” on red blood cells (Oldenborg et al., 2000), monocyte-derived dendritic cells (Dai et al., 2017), and other healthy cells (Bian et al., 2016). CD47 is a ligand for the signal regulatory protein α (SIRPα), binds SIRPγ with 10 times lower affinity, and has shown no detectable binding to SIRPβ (Hatherley et al., 2008; Nakaishi et al., 2008). We therefore assessed whether Sirpα is expressed on C57BL/6 NK cells and found only negligible expression compared with strong expression on macrophages (Fig. 2 A and Fig. S2 A). However, both NK cell Sirpα expression and Cd47 binding (Fig. 2 B and Fig. S2 B) significantly increased with prolonged mouse IL-2 stimulation over a 5-d period in vitro and with similar time kinetic. We then tested whether Sirpα expression similarly increases in our in vivo model for innate immune clearance and found that i.p. injection of mouse IL-2 up-regulated Sirpα expression on NK cells over 48 h (Fig. 2 C). We next assessed whether a Cd47–Sirpα interaction is responsible for NK cell inhibition. A blocking antibody for Cd47 was able to break the protection of B2m−/−Ciita−/− Cd47 tg miPSCs from macrophage clearance in vivo (Fig. 2 D). To prevent potential antibody-dependent cellular cytotoxicity (ADCC) against target cells by this anti-Cd47 blocking antibody, an Fc receptor (FcR) blocking solution was used. Cd47 blockade did not affect the susceptibility of B2m−/−Ciita−/− Cd47 tg miPSCs to nonactivated NK cells (Fig. 2 E), but led to the killing by activated NK cells (Fig. 2 F). Antibody blockade of the Cd47 receptor Sirpα similarly led to the killing of otherwise protected B2m−/−Ciita−/− Cd47 tg miPSCs by both macrophages and NK cells (Fig. 2, G–I). We also used Sirpa−/− mice on a C57BL/6 background to confirm the specificity of our results on Sirpα expression and Cd47 binding by NK cells. We found no Sirpα expression on (Fig. 2 J and Fig. S2 C) or Cd47 binding to Sirpa−/− macrophages or NK cells (Fig. 2 K and Fig. S2 D). Functionally, we saw that B2m−/−Ciita−/− Cd47 tg miPSCs were killed by both Sirpa−/− macrophages and activated Sirpa−/− NK cells (Fig. 2, L and M). Additional blocking of either Cd47 or Sirpα did not further affect the outcome (Fig. 2, N and O). In vitro impedance assays confirmed the killing of B2m−/−Ciita−/− Cd47 tg miECs by Sirpa−/− macrophages and activated Sirpa−/− NK cells (Fig. 2, P and Q). NK cell ELISpot assays showed that only Sirpa−/− NK cells, but not WT NK cells, released IFN-γ when encountering B2m−/−Ciita−/− Cd47 tg miECs (Fig. 2 R). The NK cell–sensitive mouse lymphoma cell line Yac-1, which does not express Cd47 and is low in MHC class I but expresses several activating NKG2D ligands, served as control for NK cell killing. Together, these data suggest that overexpressed Cd47 inhibits NK cell activation via Sirpα ligation and that Sirpα blockage or deficiency prevents this inhibition.
Figure 2.
The protective effect of Cd47 overexpression against NK cell and macrophage killing is mediated through Sirpα. (A) Sirpα expression on C57BL/6 macrophages and time course of Sirpα expression on C57BL/6 NK cells stimulated with mouse IL-2 (mean ± SD, four independent experiments per group, ANOVA). (B) Cd47 binding to C57BL/6 macrophages and time course of Cd47 binding to C57BL/6 NK cells stimulated with mouse IL-2 (mean ± SD, four independent experiments per group, ANOVA). (C) Sirpα expression on naive C57BL/6 NK cells and 48 h after in vivo stimulation with i.p. mouse IL-2 (mean ± SD, six independent experiments per group, Student’s t test). (D–I) A 1:1 mixture of CFSE-labeled WT and B2m−/−Ciita−/− Cd47 tg miPSCs was injected into the peritoneum of syngeneic C57BL/6 mice and after 48 h, and the ratio of recovered CFSE-positive miPSCs was determined (mean ± SD, triplicates in four animals per group). Animals after NK cell depletion received an anti-Cd47 blocking antibody (clone BE0270) with the miPSC injection (D). Animals after macrophage depletion received an anti-Cd47 blocking antibody with the miPSC injection (E) and additionally mouse IL-2 to activate NK cells in vivo (F). Animals after NK cell depletion received an anti-Sirpα blocking antibody (clone P84; G). Animals after macrophage depletion received an anti-Sirpα blocking antibody (H) and additionally mouse IL-2 to activate NK cells in vivo (I). (J) Sirpα expression on Sirpa−/− macrophages and time course of Sirpα expression on Sirpa−/− NK cells stimulated with mouse IL-2 (mean ± SD, four independent experiments per group, ANOVA). (K) Cd47 binding to Sirpa−/− macrophages and time course of Cd47 binding to Sirpa−/− NK cells stimulated with mouse IL-2 (mean ± SD, four independent experiments per group, ANOVA). (L–O) In some Sirpa−/− mice, NK cells were depleted (L). In other Sirpa−/− mice, macrophages were depleted, and NK cells were stimulated with mouse IL-2 (M); some animals in addition received a blocking antibody for Sirpα (N) or Cd47 (O). Graphs show mean ± SD and triplicates in four animals per group. (P) WT, B2m−/−Ciita−/−, and B2m−/−Ciita−/− Cd47 tg miECs were challenged with Sirpa−/− macrophages (mean ± SD, three independent replicates per group and time point, and three different E:T ratios). (Q) B2m−/−Ciita−/− Cd47 tg miECs were challenged with mouse IL-2–stimulated C57BL/6 NK cells or Sirpa−/− NK cells. In some groups, an anti-Cd47 or anti-Sirpα blocking antibody was used (mean ± SD, three independent replicates per group and time point, and three different E:T ratios). (R) IFNγ ELISpot assays with B2m−/−Ciita−/− Cd47 tg miEC target cells and either mouse IL-2–stimulated C57BL/6 or Sirpa−/− NK cells. Yac-1 served as controls (boxes show 25th to 75th percentile with median, and whiskers show minimum to maximum; six independent samples per group, Student’s t test). wt, wild-type.
Figure S2.
Expression of SIRPα and CD47 binding of mouse and primary human NK cells. (A and B) The kinetics of Sirpα expression (A) and Cd47 binding (B) of C57BL/6 NK cells in the presence of mouse IL-2 was assessed by flow cytometry (representative histograms are shown of four independent experiments). (C and D) The kinetics of Sirpα expression (C) and Cd47 binding (D) of Sirpa−/− NK cells in the presence of mouse IL-2 was assessed by flow cytometry (representative histograms are shown of four independent experiments). (E and F) The kinetics of SIRPα expression (E) and CD47 binding (F) of human primary NK cells in the presence of IL-2 was assessed by flow cytometry (representative histograms are shown of four independent experiments). (G) The expression of CD47 on five B2M−/−CIITA−/− CD47 tg hiEC clones was assessed by flow cytometry (representative histograms of four independent experiments are shown). wt, wild-type.
The CD47–SIRPα interaction is highly species specific
Starting with a human episomal induced pluripotent stem cell (hiPSC) line derived from CD34+ umbilical cord blood, we simultaneously targeted the human B2M and CIITA genes for disruption with Cas9 nuclease and guides. Edited B2M−/−CIITA−/− hiPSCs were transduced with a lentiviral vector carrying a CD47 cDNA with an EF-1α short promotor to generate B2M−/−CIITA−/− CD47 tg hiPSCs. A comparison between the human and mouse CD47 proteins has been reported (Hatherley et al., 2008). We used IFN-γ ELISpot assays to assess NK cell activation and bioluminescence imaging (BLI) assays on firefly luciferase (Fluc)–expressing target cells to evaluate macrophage clearance in a set of cross-species in vitro experiments. C57BL/6 NK cells killed syngeneic B2m−/−Ciita−/− miPSCs and spared B2m−/−Ciita−/− Cd47 tg miPSCs, but Cd47 blockade broke this NK cell inhibition (Fig. 3 A). When primary human NK cells were used against the same mouse target iPSCs, mouse Cd47 tg expression did not protect from xenogeneic NK cell cytotoxicity (Fig. 3 B). Similarly, when human target cells were used, human CD47 expression protected from killing by human NK cells, but not by mouse NK cells (Fig. 3, C and D). The blocking of human CD47 prevented the inhibition of human NK cell–mediated killing. When macrophages were used in killing assays, CD47 expression sufficiently inhibited same-species effector cells but was unable to protect against xenogeneic effectors (Fig. 3, E–L). Together, these results show that CD47 was highly effective in inhibiting same-species primary NK cells and macrophages from killing MHC-deficient iPSCs but showed no effect on cross-species immune cells. This further supports the highly specific receptor–ligand interaction of the CD47–SIRPα axis (Matozaki et al., 2009).
Figure 3.
CD47 is species specific, with no cross-reactivity between mouse and human. (A and B) IFN-γ ELISpot assays were performed with B2m−/−Ciita−/− and B2m−/−Ciita−/− Cd47 tg miPSCs as target cells and syngeneic C57BL/6 (A) or xenogeneic human primary NK cells as effector cells (B). Cd47 antibody blockade (clone BE0270) was used in some groups, and Yac-1 was used as a control (boxes show 25th to 75th percentile with median, and whiskers show minimum to maximum; 12 independent experiments per miPSC group and 6 for Yac-1 groups, ANOVA with Bonferroni’s post hoc test). (C and D) IFN-γ ELISpot assays were performed with B2M−/−CIITA−/− and B2M−/−CIITA−/− CD47 tg hiPSCs as target cells and allogeneic human primary NK cells (C) or xenogeneic C57BL/6 NK cells as effector cells (D). CD47 antibody blockade (clone B6.H12) was used in some groups, and K562 was used as a control (boxes show 25th to 75th percentile with median, and whiskers show minimum to maximum, 12 independent experiments per hiPSC group and 6 for K562 groups, ANOVA with Bonferroni’s post hoc test). (E–H) Fluc+ B2M−/−CIITA−/− and B2M−/−CIITA−/− CD47 tg hiPSCs were incubated with allogeneic human macrophages (E and F) or xenogeneic C57BL/6 macrophages (G and H), and the BLI signal was quantified (boxes show 25th to 75th percentile with median, and whiskers show minimum to maximum; n = 16 [control], 20 [macrophages], and 9 [Triton X-100] independent samples, ANOVA with Bonferroni’s post hoc test). CD47 antibody blockade was used in some groups. (I–L) Fluc+ B2m−/−Ciita−/− and B2m−/−Ciita−/− Cd47 tg miPSCs were incubated with syngeneic C57BL/6 macrophages (I and J) or xenogeneic human macrophages (K and L), and the BLI signal was quantified (boxes show 25th to 75th percentile with median, and whiskers show minimum to maximum; n = 16 [control], 20 [macrophages], and 9 [Triton X-100] independent samples, ANOVA with Bonferroni’s post hoc test). Cd47 antibody blockade was used in some groups.
SIRPα transmits the inhibitory CD47 signal in human NK cells
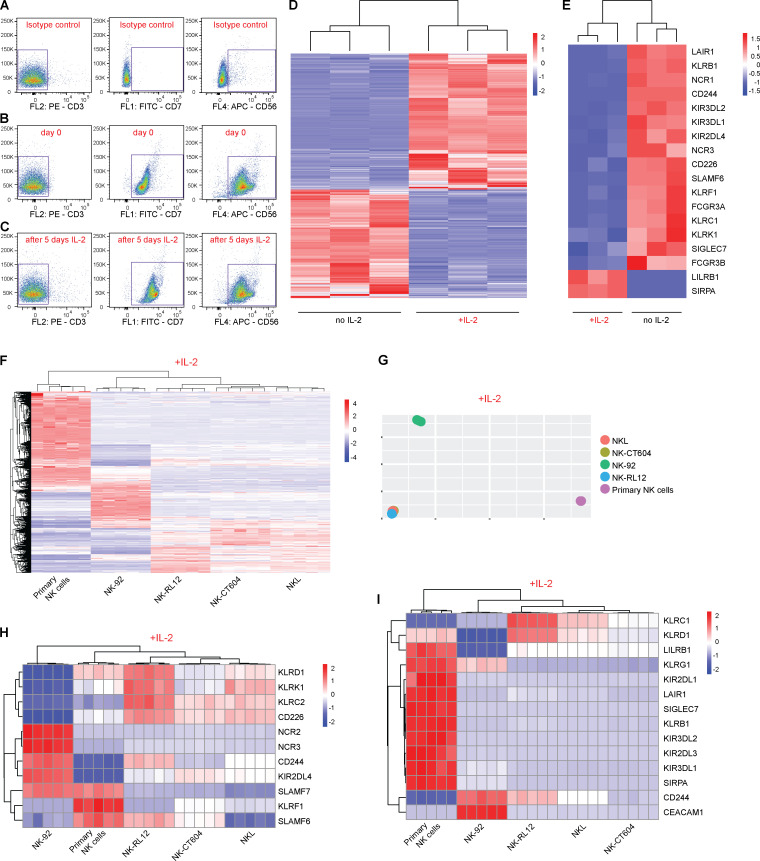
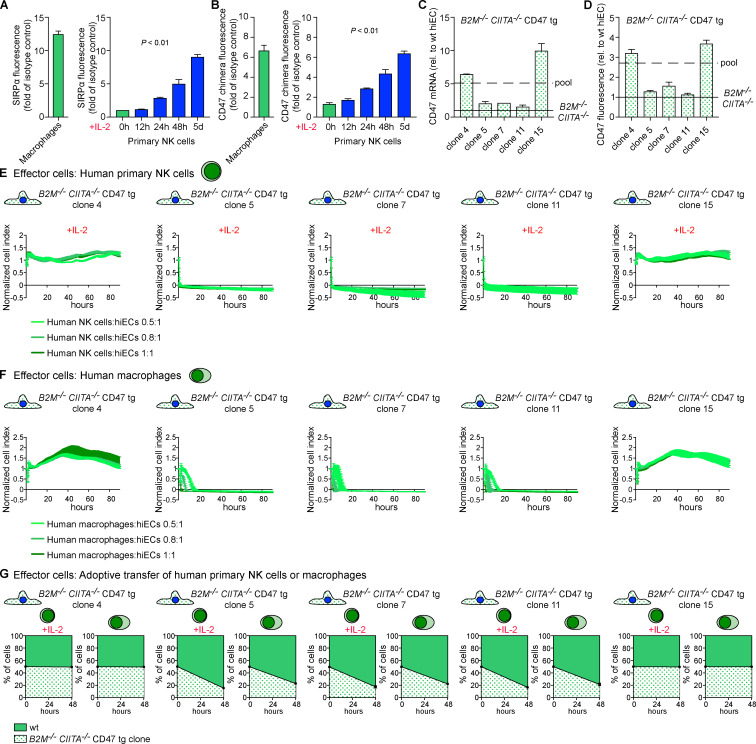
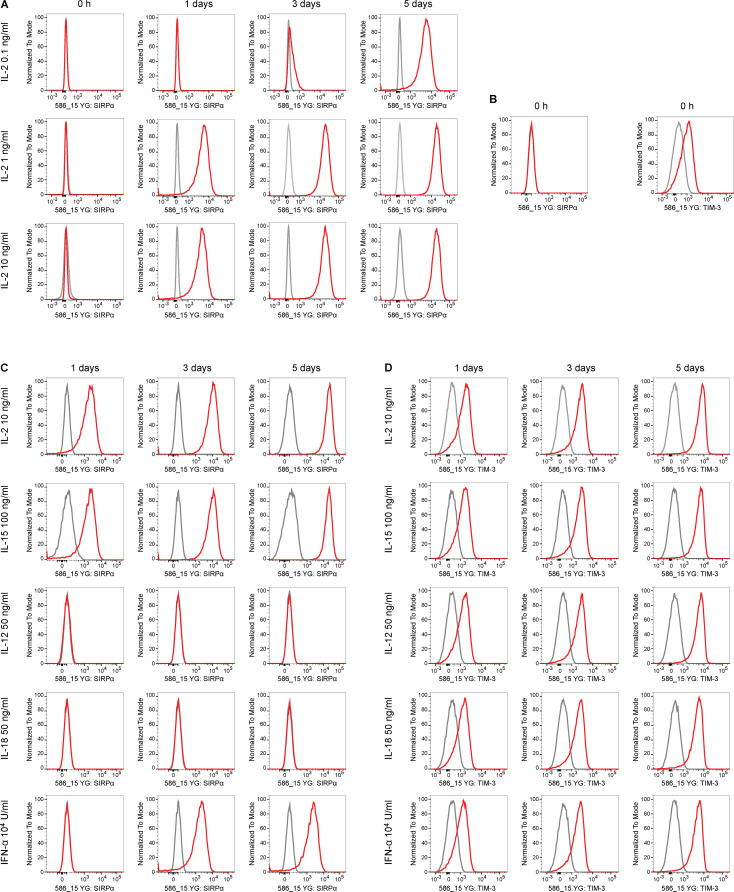
We assessed what factors or conditions were necessary to trigger the killing of HLA-deficient cells by human NK cells and macrophages and how they interact with each other. As we had seen in the mouse, only primary human NK cells activated with IL-2 or IL-15 killed HLA-deficient B2M−/−CIITA−/− hiPSCs, and CD47 overexpression completely prevented such NK cell killing (Fig. S1 E). Human macrophages detected and killed B2M−/−CIITA−/− hiPSCs, while activating the inhibitory HLA class I–sensing receptor LILRB1 with an antibody prevented cell killing (Fig. S1 G). CD47 or SIRPα blockade broke CD47-mediated protection. This supports the notion that HLA class I–LILRB1 and CD47–SIRPα pathways regulate macrophage function (Barkal et al., 2018). For subsequent transcriptome sequencing of human NK cells, a highly selected CD3−CD7+CD56+ population of primary NK cells was generated by flow cytometry sorting (Fig. 4, A and B). Coexpression of CD7 and CD56 has previously been shown to differentiate NK cells from CD56+ monocyte- or dendritic cell–like cells, which lack CD7 (Milush et al., 2009). Activation of primary human NK cells with IL-2 for 5 d led to the up-regulation of 3,270 genes and the down-regulation of 2,617 genes, respectively (Fig. 4 D). Among genes for inhibitory and stimulatory NK receptors (Lanier, 2008), SIRPA showed the highest IL-2–induced up-regulation (Fig. 4 E). SIRPα expression and capacity to bind CD47 was vastly higher on macrophages compared with unstimulated primary NK cells, but NK cell SIRPα expression and CD47 binding capacity were strongly up-regulated by IL-2 over time (Fig. 5, A and B; and Fig. S2, E and F). A 5-d course of IL-2 stimulation, however, did not affect their CD3−CD7+CD56+ phenotype (Fig. 4 C). The induction of NK cell SIRPα expression by IL-2 was found to be dose dependent (Fig. S3 A). We then tested whether other cytokines known to stimulate NK cell proliferation, cytotoxicity, and cytokine production would similarly induce SIRPα. While IL-2, IL-15, IL-12, IL-18, and IFN-α increased the expression of TIM-3, a marker for NK cell activation and maturation (Ndhlovu et al., 2012), only IL-2, IL-15, and IFN-α induced SIRPα (Fig. S3, B–D). To confirm our hypothesis that SIRPα is the inhibitory CD47 ligand on NK cells, we next aimed to identify SIRPα-deficient NK cell controls. Four IL-2–dependent human NK cell lines were sequenced and compared with IL-2–stimulated primary NK cells. Unsupervised clustering (Fig. 4 F) and principal-component analysis (Fig. 4 G) showed the distance of primary NK cells to the NK cell lines and the clustering of NKL with its sublines NK-RL12 and NK-CT604. We next assessed their expression of activating (Fig. 4 H) and inhibitory receptors (Fig. 4 I) and found that all four NK cell lines lacked SIRPA transcripts and SIRPα expression (Fig. S4 A) and showed no CD47 binding (Fig. S4 B). These four SIRPα-deficient NK cell lines were used in NK cell killing assays with WT and engineered human iECs (hiECs). We found that IL-2–activated primary NK cells and all four NK cell lines spared WT hiECs and rapidly killed B2M−/−CIITA−/− hiECs (Fig. S5). However, B2M−/−CIITA−/− CD47 tg hiECs were only spared by primary NK cells and killed by the SIRPα-negative NK cell lines (Fig. 6). SIRPα expression on NK cells was thus necessary to protect CD47-expressing but HLA-deficient target cells from killing. Blocking of either CD47 or SIRPα rendered B2M−/−CIITA−/− CD47 tg hiECs susceptible to primary NK cell cytotoxicity. We could thus confirm in the human setting that SIRPα was essential to transmit the inhibitory CD47-induced signal to NK cells.
Figure 4.
IL-2 alters the expression of inhibitory and stimulatory receptors on primary human NK cells. (A–C) CD3−CD7+CD56+ primary human NK cells were sorted to avoid contamination with myeloid cells. Representative plots with side scatter vs. CD3 (left column), CD7 (middle column), or CD56 (right column) are shown. Rows show the isotype controls (A) and CD3−CD7+CD56+ NK cells before (B) and after 5 d of IL-2 stimulation (C). (D) Whole transcriptome sequencing of CD3−CD7+CD56+ primary NK cells was performed without IL-2 and after 5 d of IL-2 stimulation (heatmap shows row z-scores for the normalized expression of genes at least twofold differentially expressed among groups; FDR < 0.1, n = 3 per group). (E) Changes in gene expression of inhibitory and stimulatory NK receptors during 5 d of IL-2 stimulation (heatmap shows row z-scores for the normalized expression of selected NK cell receptors). All depicted genes are significantly (FDR < 0.1) differentially expressed. Changes are at least twofold, except for SIGLEC7 (log2 fold change = 0.84). (F) Whole transcriptome sequencing of primary human NK cells and NK cell lines were conducted after 24 h of IL-2 stimulation (heatmap shows z-scores for normalized expression of genes that were at least twofold differentially expressed in any four NK cell line compared with primary NK cells). (G) Principal-component analysis showing the distance of primary NK cells to all NK cell lines and the clustering of NKL with its sublines NK-RL12 and NK-CT604. (H and I) Gene expression of stimulatory NK cell receptors (H) and inhibitory NK cell receptors (I; heatmaps show z-scores for normalized expression).
Figure 5.
Threshold of CD47 expression for NK cell and macrophage inhibition. (A) SIRPα expression on macrophages and time course of SIRPα expression on highly selected CD3−CD7+CD56+ primary NK cells stimulated with IL-2 (mean ± SD, four independent experiments per group, ANOVA). (B) CD47 binding to macrophages and time course of CD47 binding to highly selected CD3−CD7+CD56+ primary NK cells stimulated with IL-2 (mean ± SD, four independent experiments per group, ANOVA). (C and D) From the pool of B2M−/−CIITA−/− CD47 tg hiECs, five clones with different levels of CD47 overexpression were selected. The CD47 expression on transduced cells was always higher than the basal expression level on WT hiECs but below or above the mean of the pool. CD47 was quantified by RT-PCR (C; mean ± SD, three independent experiments per group) and fluorescence (D; mean ± SD, four independent experiments per group). (E and F) The five B2M−/−CIITA−/− CD47 tg hiEC clones were challenged with IL-2–activated human primary NK cells (E) or human macrophages (F). Graphs show mean ± SD and three independent replicates per group and time point; three different E:T ratios are shown. (G) A 1:1 mixture of CFSE-labeled WT and one of the B2M−/−CIITA−/− CD47 tg hiEC clones was injected into the peritoneum of immunodeficient NSG mice. Additionally, either IL-2–activated primary NK cells or macrophages were coinjected. After 48 h, the ratio of recovered CFSE-positive hiECs was determined (mean ± SD, triplicates in four animals per group). wt, wild-type.
Figure S3.
Cytokine stimulation induces SIRPα expression on human NK cells. (A) Selected CD3−CD7+CD56+ primary NK cells showed a dose-dependent increase of SIRPα expression with IL-2 (representative histograms are shown of two independent experiments). (B) Unstimulated CD3−CD7+CD56+ primary NK cells did not express SIRPα but showed low-level expression of TIM-3 (representative histograms of two independent experiments are shown). (C and D) Five stimulatory cytokines for NK cells were assessed for their efficacy to induce SIRPα expression (C) and TIM-3 expression (B). While all five increased TIM-3, only IL-2, IL-15, and IFN-α increased NK cell SIRPα (representative histograms of two independent experiments are shown).
Figure S4.
Expression of SIRPα and CD47 binding of human NK cell lines. (A and B) The kinetics of SIRPα expression (A) and CD47 binding (B) of human NK cell lines after 24 h of IL-2 stimulation was assessed by flow cytometry (representative histograms are shown of four independent experiments, bar graphs show mean ± SD, four independent experiments per group, ANOVA).
Figure S5.
NK cell killing of hiECs in the presence and absence of CD47. (A–E) WT, B2M−/−CIITA−/−, and B2M−/−CIITA−/− CD47 tg hiECs were challenged with primary NK cells (A) or NK cells from a cell line (B–E) using in vitro impedance assays. Graphs show mean ± SD and three independent replicates per group and time point; three different E:T ratios are shown. wt, wild-type.
Figure 6.
Mechanistic interaction of CD47 with NK cell SIRPα. (A–E) B2M−/−CIITA−/− CD47 tg hiECs engaged with primary NK cells (A) or NK cells from a cell line (B–E) using in vitro impedance assays in the presence or absence of a specific blocking antibody against CD47 (clone B6.H12) or a blocking peptide against SIRPα. Graphs show mean ± SD and three independent replicates per group and time point; three different E:T ratios are shown.
CD47 delivers a threshold-dependent inhibitory signal in human NK cells
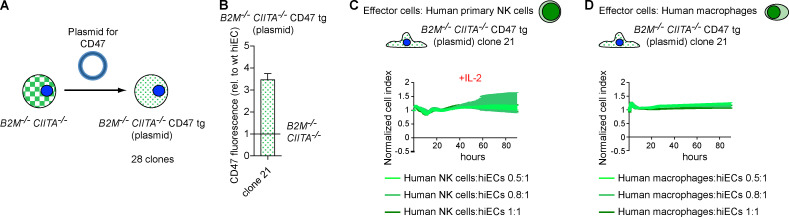
To determine the threshold of CD47 expression necessary to inhibit macrophages and NK cells from killing HLA-deficient hiEC target cells, single cells were collected from the pool of B2M−/−CIITA−/− CD47 tg hiECs and clones were expanded. CD47 expression levels by mRNA and flow cytometry were all above those of B2M−/−CIITA−/− hiECs but lower or higher than the mean of the B2M−/−CIITA−/− CD47 tg hiEC pool (Fig. 5, C and D; and Fig. S2 G). The five clones were then exposed to activated primary NK cells (Fig. 5 E). The two clones with CD47 expression levels above the average of the pool (clones 4 and 15) were resistant to primary NK cell killing, while the three clones with lower CD47 expression (clones 5, 7, and 11) were rapidly killed. When the five clones were exposed to macrophages, we observed similar results with the same two clones surviving and the same three clones vanishing (Fig. 5 F). An interesting observation is the on/off switch phenomenon either triggering rapid killing or achieving complete inhibition with no intermediate stages of NK cell or macrophage activation. Also unexpected was the similar threshold for CD47-mediated immune cell inhibition for both effector cell types. NK cell and macrophage killing of B2M−/−CIITA−/− CD47 tg hiEC clones was further assessed in vivo using adoptive transfer of purified human primary NK cells or macrophages into the peritoneum of immunocompromised NOD scid gamma (NSG) mice (Fig. 5 G). The results in this model recapitulated the in vitro results and showed survival or killing of the exact same clones. To further exclude that the lentiviral particles used to overexpress CD47 in the B2M−/−CIITA−/− CD47 tg hiEC pool had induced additional genomic alteration critical for NK cell and macrophage engagement, another B2M−/−CIITA−/− CD47 tg hiPSC pool was generated using plasmid vectors for CD47 transfection (Fig. 7 A). Out of 28 picked clones, hiPSC clone 21 showed the highest CD47 expression and was differentiated into hiECs, which then expressed CD47 at ∼3.5-fold of normal WT hiEC levels (Fig. 7 B). This CD47 expression was very similar to those in clones 4 and 15 of the lentiviral CD47-transduced cells. The B2M−/−CIITA−/− CD47 tg hiEC (plasmid) clone 21 showed resistance to human primary NK cells and macrophages (Fig. 7, C and D). Experiments with lentiviral transduction or plasmid transfection have thus consistently shown that the threshold for CD47 expression on hiECs to inhibit innate immune cells is ∼3.5-fold higher than the basal level.
Figure 7.
CD47 overexpression using plasmid vector transfection. (A) CD47 overexpression in B2M−/−CIITA−/− hiPSCs was achieved by plasmid vector transfection, and out of this B2M−/−CIITA−/− CD47 tg (plasmid) hiPSC pool, 28 clones were picked and expanded. Clone 21 showed the highest CD47 expression. (B) Clone 21 iPSCs were differentiated into hiECs, and the CD47 expression level was assessed by flow cytometry (mean ± SD, four independent experiments). (C and D) hiEC clone 21 was challenged with IL-2–activated human primary NK cells (C) or human macrophages (D). Graphs show mean ± SD and three independent replicates per group and time point; three different E:T ratios are shown.
SIRPα is a strong inhibitory receptor on NK cells
The functional role of SIRPα on NK cells was additionally assessed in a redirected antibody-dependent cytotoxicity assay against the FcR+ P815 mouse mastocytoma cell line. When agonist antibodies against activating or inhibitory receptors on NK cells or T cells engage the FcR on the P815 mouse mastocytoma cell line, they can increase or decrease, respectively, killing of this target cell line (Lanier et al., 1997). As has been shown previously, antibodies against CD16 initiate cytolytic NK cell activity, whereas control antibodies against other membrane molecules like CD56 do not (Lanier et al., 1988; Siliciano et al., 1985). By using an agonist antibody against SIRPα, this assay can mimic SIRPα-induced inhibition with high specificity, thus ruling out that an interaction between CD47 and another unknown receptor contributes to NK cell inhibition. Target cell killing of Fluc+ P815 was assessed by the drop of their BLI signal over 4 h. Approximately 50% of P815 cells were killed by IL-2–stimulated CD3−CD7+CD56+ NK cells. NK cell killing was further increased when CD16 was cross-linked by anti-CD16 and to a lesser degree by cross-linking NKG2D (Fig. 8). Concomitant engagement of SIRPα inhibited P815 lysis and offset both stimulatory signals. The inhibitory character of NK cell SIRPα was thus confirmed in a CD47-independent manner.
Figure 8.
Redirected antibody-dependent cytotoxicity assay against P815. (A) CD3−CD7+CD56+ NK cells were stimulated with IL-2 for 72 h, and SIRPα expression was assessed by flow cytometry (representative histogram of two independent experiments). (B and C) CD3−CD7+CD56+ NK cells were added to Fluc+ target P815 cells (green bar), or NK cells were added together with activating antibodies against CD16 (B) or NKG2D (C). The effect of anti-SIRPα to offset the activating signals was assessed. An antibody against CD56 served as control (mean ± SD, three independent experiments per group, ANOVA with Bonferroni’s post hoc test).
CD47 in tumor biology
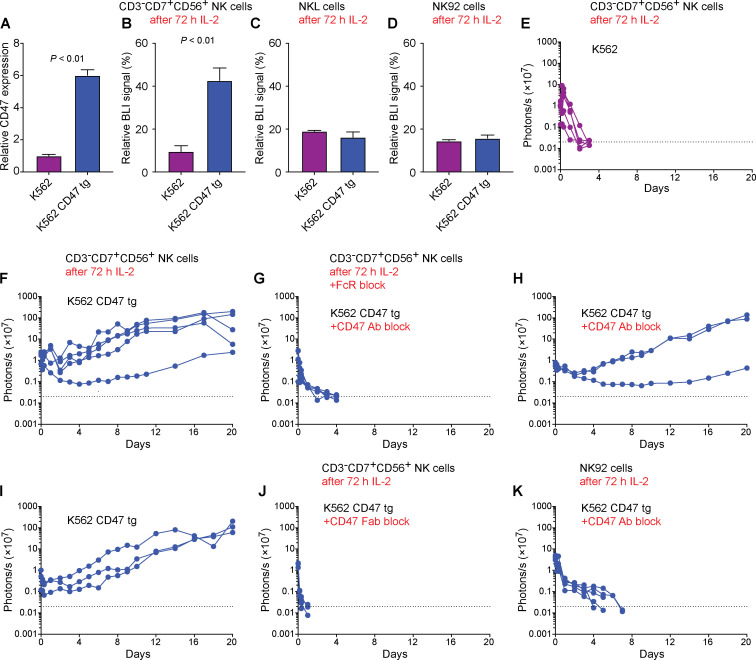
The NK-sensitive erythroleukemia cell line K562 was transduced to overexpress CD47 to evaluate whether it exerts inhibitory function against tumor cell killing. Using lentiviral particles, we found that CD47 expression was increased approximately sixfold compared to K562 baseline levels (Fig. 9 A). This CD47 overexpression markedly alleviated K562 killing by IL-2–stimulated CD3−CD7+CD56+ primary NK cells (Fig. 9 B). However, K562 killing by IL-2–stimulated SIRPα-negative NKL or NK92 was not affected by CD47 overexpression (Fig. 9, C and D). We next assessed K562 survival in vivo after transplantation into NSG mice, which received adoptively transferred NK cells. K562 were rapidly eliminated by IL-2–stimulated CD3−CD7+CD56+ primary NK cells (Fig. 9 E). However, CD47-overexpressing K562 cells escaped killing by the same IL-2–stimulated CD3−CD7+CD56+ primary NK cells and progressed to form tumors in all animals (Fig. 9 F). We then performed different experiments blocking the CD47 signal. Before they were injected, the K562 CD47 tg cells were incubated with the CD47 blocking antibody used throughout this study. The IL-2–stimulated CD3−CD7+CD56+ primary NK cells were treated with FcR blocking agent to mitigate their ADCC capacity. We observed rapid killing of CD47-overexpressing K562 cells when CD47 was blocked (Fig. 9 G). This killing must have been executed by the transferred IL-2–stimulated CD3−CD7+CD56+ primary NK cells, because the K562 CD47 tg cells were not killed by native NSG immune cells with (Fig. 9 H) or without (Fig. 9 I) the addition of the blocking CD47 antibody. To further rule out that ADCC contributed to the target cell killing, we used Fab (antigen-binding fragment) fragments of the CD47 blocking antibody, which cannot engage effector cell FcRs. We again observed rapid killing of K562 CD47 tg cells (Fig. 9 J). Last, we used CD16-deficient NK92 cells, which thus cannot kill via ADCC, in conjunction with the anti-CD47 antibody and again observed rapid killing of K562 CD47 tg targets (Fig. 9 K). Our results therefore indicate that CD47 delivers an inhibitory signal to NK cells and that increased CD47 expression can be used by tumor cells to mitigate NK cell killing as long as the effector cells express SIRPα.
Figure 9.
CD47 overexpression in K562 cells. (A) CD47 overexpression was achieved with lentiviral particles and the relative surface expression on K562 CD47 tg was assessed in flow cytometry (mean ± SD, three independent samples per group, Student’s t test). (B–D) CD3−CD7+CD56+ primary NK (B), NKL (C), or NK92 (D) cells were stimulated with IL-2 for 72 h before they were added to Fluc+ K562 or K562 CD47 tg, and target cell killing was assessed in BLI assays (mean ± SD, three independent samples per group, Student’s t test). (E) Fluc+ K562 cells were transplanted into NSG mice, which received adoptive transfer of IL-2–treated CD3−CD7+CD56+ primary NK cells. Cell survival was longitudinally monitored using BLI (five mice; each line represents one mouse). (F–G) Fluc+ K562 CD47 tg cells were transplanted into NSG mice, and cell survival was longitudinally monitored using BLI (five mice per graph; each line represents one mouse). IL-2–treated CD3−CD7+CD56+ primary NK cells were used as effector cells without blocking CD47 (F) or with CD47 antibody block and FcR block (G; clone B6.H12). (H and I) Fluc+ K562 CD47 tg cells were transplanted into NSG mice with (H) or without (I) CD47 antibody block but without transfer of human NK cells (three mice per graph; each line represents one mouse). (J) Fluc+ K562 CD47 tg cells were transplanted into NSG mice that received IL-2–treated CD3−CD7+CD56+ primary NK cells (five mice; each line represents one mouse). Free Fab fragments of the CD47 antibody (clone B6.H12; H) were used to block CD47 interactions. (K) IL-2–treated NK92 effector cells were transferred in conjunction with the CD47 blocking antibody in mice transplanted with Fluc+ K562 CD47 tg cells (five mice; each line represents one mouse).
CD47 engineering for xenogeneic cell products
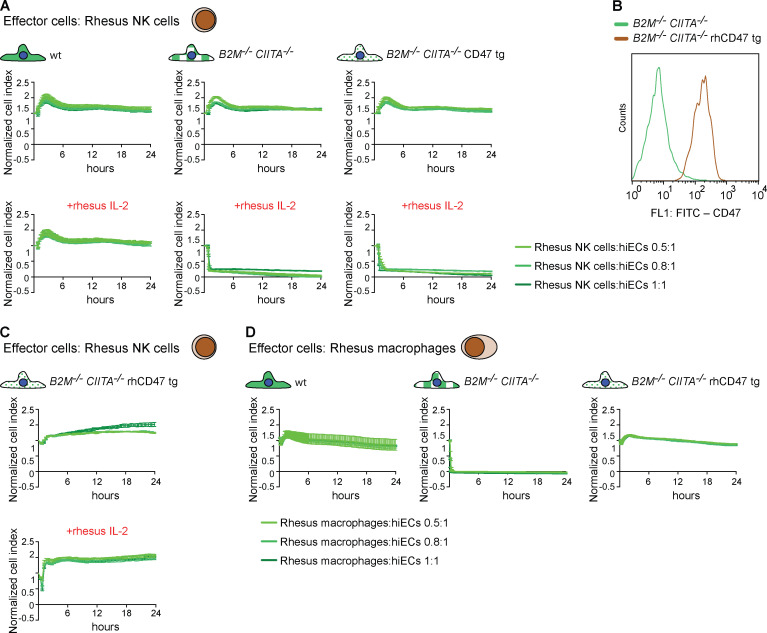
We assessed the cellular immune response against human B2M−/−CIITA−/− CD47 tg hiECs by innate immune cells from rhesus monkeys, a species relevant for preclinical testing of cell products. We intentionally selected endothelial cells for these xenogeneic experiments, because this is a highly immunogenic cell type. Rhesus NK cells (activated with rhesus IL-2) rapidly killed human B2M−/−CIITA−/− CD47 tg hiECs in the in vitro screening assay with the same kinetics as B2M−/−CIITA−/− hiECs (Fig. 10 A). This shows the limitations of predictive modeling when crossing species. We then cloned the rhesus monkey rhCD47 sequence into a lentivirus and transduced human B2M−/−CIITA−/− hiPSCs to generate B2M−/−CIITA−/− rhCD47 tg hiPSCs, which were then differentiated into hiECs. The expression of rhCD47 was absent in B2M−/−CIITA−/− hiECs but very highly expressed in B2M−/−CIITA−/− rhCD47 hiECs (Fig. 10 B). Human B2M−/−CIITA−/− rhCD47 tg hiECs were spared by both rhesus NK cells (Fig. 10 C) and rhesus macrophages in vitro (Fig. 10 D). These results demonstrate that rhCD47 expression inhibits rhesus monkey innate immune cells even in a xenogeneic setting.
Figure 10.
Inhibition of rhesus monkey NK cells and macrophages by B2M−/−CIITA−/− hiECs overexpressing rhesus CD47. (A) WT, B2M−/−CIITA−/−, and B2M−/−CIITA−/− CD47 tg hiECs were challenged with rhesus NK cells using in vitro impedance assays. Rhesus NK cells were used either unstimulated or stimulated with rhesus IL-2. Graphs show mean ± SD and three independent replicates per group and time point; three different E:T ratios are shown. (B) The rhesus CD47 tg was expressed in B2M−/−CIITA−/− rhCD47 hiECs, and expression by fluorescence is compared with primary rhesus ECs (representative histogram of two independent experiments). (C) B2M−/−CIITA−/− rhCD47 tg hiECs were challenged with unstimulated or rhesus IL-2–stimulated rhesus NK cells. Graphs show mean ± SD and three independent replicates per group and time point; three different E:T ratios are shown. (D) WT, B2M−/−CIITA−/−, and B2M−/−CIITA−/− rhCD47 tg hiECs were challenged with rhesus macrophages. Graphs show mean ± SD and three independent replicates per group and time point; three different E:T ratios are shown. wt, wild-type.
Discussion
Previous approaches to inhibit NK cell responses against HLA class I–deficient cells included overexpression of single-chain HLA-E fused with B2M (Gornalusse et al., 2017) or retention of HLA-C (Xu et al., 2019). However, HLA-E will not inhibit CD94-NKG2A–negative NK cell populations, which may be suppressed though HLA-C by KIR or HLA-G by LILRB1. HLA-C retention, on the other side, may allow alloantigen presentation to T cells with subsequent immune activation. This study now adds CD47 to the list of inhibitory NK cell checkpoint pathways. The CD47–SIRPα axis delivers strong inhibitory signals that are capable of overriding the stimulatory signals provided by HLA deficiency and IL-2 or IL-15 activation of NK cell without the above shortcomings.
Our data complement the growing knowledge about the complex CD47–SIRPα axis in immune cells. In macrophages, SIRPα was found to be an inhibitory receptor for cytotoxicity and phagocytosis (Barclay and Van den Berg, 2014) and a key modulator of the innate allorecognition response (Dai et al., 2017; Oberbarnscheidt et al., 2014). The macrophage antitumor functions, however, are modulated by the carbon metabolism and CpG-stimulated alterations in metabolism, which allow circumventing inhibitory signals mediated by CD47 (Liu et al., 2019). In polymorphonuclear leukocytes, it has been reported that SIRPα cytoplasmic signaling ITIMs are cleaved during active inflammation and that the loss of SIRPα ITIMs enhances the polymorphonuclear leukocyte inflammatory response (Zen et al., 2013). While SIRPα had primarily been viewed as an inhibitory receptor on myeloid cells only, recent reports showed interaction of CD47 with certain T cell subsets (Myers et al., 2019) or NK cells (Deuse et al., 2019), hinting at their SIRPα expression. In both cell populations, SIRPα is not expressed in the naive state but is up-regulated upon stimulation.
Crystallographic analysis revealed how the convoluted interacting face of CD47 intercalates with the corresponding regions in SIRPα and explained the specificity for SIRPα in atomic detail (Hatherley et al., 2008). The failure of human SIRPβ to bind CD47 was explained by a subtle structural rearrangement at the bottom of the binding domain (Hatherley et al., 2008; Seiffert et al., 2001). Human SIRPγ can interact with CD47, albeit with 10 times lower affinity than SIRPα (Brooke et al., 2004; Hatherley et al., 2008), but has no known motifs for recruiting signaling proteins (Barclay and Van den Berg, 2014) and is therefore thought not to signal, although it might contribute to T cell adhesion (Piccio et al., 2005). While SIRPα was reported not to be a positive or negative regulator of functionality in T cells (Myers et al., 2019), we herein found that SIRPα is a strong inhibitor of NK cell–mediated cytotoxicity. Our data show that SIRPα expression on NK cells was required for CD47 to inhibit NK cell killing and that NK cell lines lacking SIRPα were not susceptible to CD47-mediated inhibition of iEC and tumor cell killing. Engineering of NK cells to delete SIRPA for adoptive therapy in cancer patients might enhance their antitumor function and efficacy.
NK cell killing or quiescence is governed by the balance of stimulatory and inhibitory signals (Chiossone et al., 2018). We observed that MHC deficiency alone in the engineered iECs did not provide a sufficient stimulatory signal to trigger NK cell activation and that additional stimulatory signals, including IL-2 and IL-15, were necessary. We found that a threshold for CD47 overexpression of at least ∼3.5-fold above basal levels in iECs was required to inhibit stimulated NK cells from killing HLA-deficient targets. This threshold for activated NK cells was found to be similar to that required to silence macrophages. The increase of NK cell SIRPα expression by cytokine stimulation to levels similar to constitutive SIRPα expression on macrophages might explain this finding. Broadly, the CD47 expression threshold against HLA-deficient cells described herein is very similar to the changes in CD47 expression observed under physiological conditions and in cancer biology. CD47 reportedly is up-regulated approximately fourfold by hematopoietic stem and progenitor cells in response to an insult that induces mobilization and protects from phagocytosis during migration to the periphery (Jaiswal et al., 2009). Mouse and human myeloid leukemic cells were found to show CD47 up-regulation of this magnitude, and CD47 overexpression again provided a survival benefit in immunodeficient mouse models and in vitro phagocytosis assays (Jaiswal et al., 2009). While the level of CD47 overexpression was reported to directly correlate with cell survival (Jaiswal et al., 2009), we observed more of a binary outcome in our specific immune assays. That may be due to the fact that a mere slowing of innate cell clearance still results in cell death and only complete innate silencing allows durable engraftment. For the engineering of hypoimmunogenic cells, this minimum threshold for CD47 expression should be respected.
To date, few regenerative human cell products have progressed to preclinical testing in nonhuman primates, and primarily human embryonic stem cell–derived cardiomyocytes (Liu et al., 2018) and retinal tissue (Shirai et al., 2016) have been evaluated. We show that CD47 is highly species specific without cross-reactivity between the mouse, human, and rhesus monkey isoforms tested herein, despite high predicted homology between humans and rhesus. This requires the engineered cells to overexpress rhCD47 for xenotransplantation studies using human cells in rhesus. Our engineering strategy was effective to completely silence all rhesus monkey NK cell and macrophage responses against human B2M−/−CIITA−/− rhesus CD47 tg hiECs. Previous studies to suppress immune activation against WT human embryonic stem cell–derived grafts in macaque hearts (Liu et al., 2018) and eyes (Shirai et al., 2016) used high levels of immunosuppression. Exploitation of the CD47–SIRPα immune checkpoint may advance the immune silencing technology and enable the generation of universal regenerative human cell products.
Materials and methods
Mouse assays
Mouse models
BALB/cJ (000651), C57BL/6J (000664), CD11b-DTR (B6.FVB-Tg(ITGAM-DTR/EGFP)34Lan/J; 006000), and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (005557, all 6–12 wk old) mice were purchased from the Jackson Laboratories and used as immune cell donors or graft recipients for different assays. Sirpa−/− C57BL/6 mice were generated by Y. Liu as previously described (Bian et al., 2016). Animals were randomly assigned to experimental groups. The number of animals per experimental group is presented in each figure. Mice received humane care in compliance with the University of California, San Francisco Institutional Animal Care and Use Committee and performed according to local guidelines.
Generation and gene editing of mouse iPSCs
miPSC generation and gene editing were performed as described previously (Deuse et al., 2019). Briefly, C57BL/6 miPSCs were generated from mouse tail tip fibroblasts using a mini-intronic plasmid carrying sequences of Oct4, Klf4, Sox2, and c-Myc. These miPSCs underwent CRISPR-Cas9 inactivation of the mouse B2m and Ciita genes. Cd47 overexpression was achieved by lentiviral transduction in a subsequent step. Cell pools of WT and engineered B2m−/−Ciita−/− miPSCs and B2m−/−Ciita−/− Cd47 tg miPSCs were used for different assays.
miPSC culture and transduction to express Fluc
After the MEF feeder cells attached and were 100% confluent, miPSCs were grown on MEF in KO DMEM 10829 with 15% KO serum replacement, 1% glutamine, 1% MEM with nonessential amino acids, 1% penicillin/streptomycin (pen/strep; all Gibco), 0.2% β-mercaptoethanol, and 100 U leukemia inhibitory factor (both Millipore). Cells were maintained in 10-cm dishes, medium was changed daily, and the cells were passaged every 2–3 d using 0.05% trypsin-EDTA (Gibco). For luciferase expression, 105 miPSCs were plated in one gelatin-coated 6-well plate and incubated overnight at 37°C at 5% CO2. The next day, medium was changed, and one vial of Fluc lentiviral particles expressing the luciferase II gene under the reengineered EF1a promotor (Gen Target) was added to 1.5 ml medium. After 36 h, 1 ml cell medium was added. After an additional 24 h, a complete medium change was performed. After 2 d, luciferase expression was confirmed by adding D-luciferin (Promega). Signals were quantified with Ami HT (Spectral Instruments Imaging) in maximum photons per second per centimeter square per steradian (p/s/cm2/sr).
Endothelial cell differentiation of miPSCs
miPSCs were plated on gelatin in 6-well plates and maintained in mouse iPSC medium. After the cells reached 60% confluency, the differentiation was initiated and medium was changed to RPMI-1640 containing 2% B-27 minus insulin (both Gibco) and 5 µM CHIR-99021 (Selleckchem). On day 2, the medium was changed to reduced medium of RPMI-1640 containing 2% B-27 minus insulin (both Gibco) and 2 µM CHIR-99021 (Selleckchem). From days 4–7, cells were exposed to RPMI-1640 endothelial cell (EC) medium of RPMI-1640 containing 2% B-27 minus insulin plus 50 ng/ml mouse vascular endothelial growth factor (mouse VEGF; R&D Systems), 10 ng/ml mouse fibroblast growth factor basic (mouse FGFb; R&D Systems), 10 µM Y-27632 (Sigma-Aldrich), and 1 µM SB 431542 (Sigma-Aldrich). Endothelial cell clusters were visible from day 7, and cells were maintained in Endothelial Cell Basal Medium 2 (PromoCell) plus supplements, 10% heat-inactivated FCS (FCS hi; Gibco), 1% pen/strep, 25 ng/ml VEGF, 2 ng/ml FGFb, 10 µM Y-27632 (Sigma-Aldrich), and 1 µM SB 431542 (Sigma-Aldrich). The differentiation process was completed after 21 d, and undifferentiated cells detached during the differentiation process. For purification, cells went through magnetic-activated cell sorting (MACS) purification according the manufacturer’s protocol using anti-CD15 mAb-coated magnetic microbeads (Miltenyi) for negative selection. The highly purified miECs in the flowthrough were cultured in miEC medium as described above. TrypLE was used for passaging the cells 1:3 every 3 to 4 d.
Mouse NK cell isolation
Mouse NK cells were isolated from fresh BALB/c, C57BL/6, or C57BL/6 Sirpa−/− spleens 18 h after poly I:C injection (100 µg i.p.; Sigma-Aldrich). After red cell lysis, NK cells were purified with MagniSort Mouse NK cell Enrichment Kit (Invitrogen), followed by CD49b MACS (Miltenyi). This cell population was highly selected for NK cells with a purity of >99%.
Mouse macrophage differentiation from splenocytes
Mouse splenocytes were isolated from fresh mouse spleens and were resuspended in RPMI-1640 with 10% FCS hi and 1% pen/strep (all Gibco). Cells were plated in 24-well plates at a concentration of 106 cells per ml and stimulated with 10 ng/ml mouse M-CSF. Medium was changed every second day until day 6. Then, 1 µg/ml mouse IL-2 (both PeproTech) was added 24 h before the cells were used in assays.
Flow cytometry
Sirpα expression (PE-conjugated antibody, clone P84, IgG1,κ; BD Biosciences) and CD47 binding (recombinant mouse Cd47 protein Fc chimera [catalog no. 1866-CD-050; R&D Systems] with FITC-conjugated anti-human IgG1 secondary antibody) were assessed by flow cytometry. Results were expressed as mean fluorescent intensity fold change to isotype-matched control Ig staining or secondary antibody staining.
Mouse in vivo innate cytotoxicity assay
Five million WT miECs and five million B2m−/−Ciita−/− miECs or B2m−/−Ciita−/− Cd47 tg miECs were mixed and stained with 5 µM CFSE (Thermo Fisher). Cells in saline were injected i.p. into syngeneic C57BL/6 mice, CD11b-DTR mice or Sirpa−/− mice. Some mice received a coinjection i.p. with 1 µg mouse IL-2 or mouse IL-15 (PeproTech). After 48 h, cells were collected from the abdomen and stained with PerCP-eFlour710–labeled anti-MHC class I (clone AF6-88.5.5.3, mouse IgG2a,κ; eBioscience) mAb for 45 min at 4°C. The CFSE-positive and MHC class I–negative population was analyzed by flow cytometry (FACSCalibur; BD Biosciences) and compared between the WT and the engineered miEC group. All animals were pretreated 18 h with poly I:C injection (100 µg in sterile PBS i.p.; Sigma-Aldrich) before miEC injection. Some animals were pretreated with clodronate (200 µl i.p. 3 d before the experiment; Liposoma) to eliminate macrophages and make the assay more specific for NK cells. Some animals were pretreated with anti-NK1.1 (clone PK136, 200 µl i.p. 3 d before the experiment; BD Biosciences) to eliminate NK cells for macrophage-specific experiments. Some animals received clodronate and anti-NK1.1 for cell depletion. Some of the CD11b-DTR mice were pretreated with DT (Lystlab) 3 d and 1 d before the experiment at a concentration of 25 ng/g mouse weight in 100 µl saline i.p. For peritoneal transfer, 106 peritoneal cells from naive C57BL/6 mice were injected on day 0 with the target miECs. Some animals were pretreated with an anti-Cd47 blocking antibody (clone MIAP301, rat IgG2a,κ; BioXCell; 100 µg i.p., 2 d before implantation of the miEC). Some animals were pretreated with an anti-Sirpα blocking antibody (clone P84, rat IgG1,κ; BioLegend; 100 µg i.p. 2 d before implantation of the miEC). To investigate mouse in vivo innate killing of B2m−/−Ciita−/− miECs and B2m−/−Ciita−/− Cd47 tg miECs, 5 × 106 of both cells were injected after staining with DiO and DiD, respectively according to the manufacturer’s protocol (Vybrant Multicolor cell labeling kit; Invitrogen). Syngeneic C57BL/6 mice were pretreated 18 h with poly I:C (100 µg i.p.; Sigma-Aldrich) in saline before cell injection. After 48 h, cells were collected from the peritoneum and analyzed by flow cytometry (FACSCalibur; BD Bioscience).
NK cell killing and macrophage killing assay by XCelligence
NK cell killing and macrophage killing assays were performed on the XCelligence SP platform and MP platform (ACEA Biosciences). Special 96-well E-plates (ACEA Biosciences) were coated with collagen (Sigma-Aldrich) and 4 × 105 WT, B2m−/−Ciita−/−, or B2m−/−Ciita−/− Cd47 tg miECs and plated in 100 µl cell-specific medium. After the cell index value reached 0.7, BALB/c, WT C57BL/6, or Sirpa−/− C57BL/6 NK cells or macrophages were added at an E:T ratio of 0.5:1, 0.8:1, or 1:1 with or without 1 ng/ml mouse IL-2 or 1 ng/ml mouse IL-15 (both PeproTech). As a negative control, cells were treated with 2% Triton X-100 in cell-specific media (data not shown). Some wells were treated with rabbit anti-mouse Pirb antibody (polyclonal, catalog no. MBS1489822; MyBioSource) at a concentration of 50 ng/ml. In some experiments, anti-mouse Cd47 blocking antibody (clone MIAP301, rat IgG2a,κ; BioXCell) and mouse FcR block (catalog no. 130–092-575, concentration 1:5; Miltenyi) or anti-Sirpα blocking antibody (clone P84, rat IgG1,κ; BioLegend) was used. Data were standardized and analyzed with RTCA software (ACEA Biosciences).
Macrophage killing by BLI
Luciferase-expressing B2m−/−Ciita−/− miPSCs or B2m−/−Ciita−/− Cd47 tg miPSCs were counted and plated at a concentration of 105 cells per 24 wells. After 16 h, mouse WT or Sirpa−/− macrophages or human macrophages were added to the miPSCs at an E:T ratio of 1:1. After 120 min, luciferase expression was detected by adding D-luciferin (Promega). As controls, target cells were nontreated or treated with 2% Triton X-100 in cell-specific media. In some conditions, target cells were pretreated with blocking anti-mouse Cd47 antibody (1 µg/ml, clone MIAP410; BioXCell) 2 h before the addition of target cells. Signals were quantified with Ami HT (Spectral Instruments Imaging) in p/s/cm2/sr.
NK cell ELISpot assays
For NK cell–specific ELISpot assays, NK cells from WT and Sirpa−/− C57BL/6 mice or human primary NK cells were co-cultured with mouse B2m−/−Ciita−/− or B2m−/−Ciita−/− Cd47 tg miPSCs, and their IFN-γ release (BD Biosciences) was measured. Yac-1 cells (Sigma-Aldrich) served as positive control. Mitomycin-treated (50 µg/ml for 30 min) stimulator cells were incubated with NK cells (1:1) for 24 h, and IFN-γ spot frequencies were enumerated using an ELISpot plate reader. In some cases, cells were stimulated with mouse IL-2 (1 ng/ml; PeproTech) or stimulator cells were pretreated with anti-mouse Cd47 blocking antibody (clone MIAP301, rat IgG2a,κ; BioXCell) and effector cells with mouse FcR block (catalog no. 130–092-575, concentration 1:5; Miltenyi) for 120 min at 37°C.
Human cells and assays
Generation and gene editing of hiPSCs
A human episomal iPSC line derived from CD34+ umbilical cord blood using a three-plasmid, seven-factor (SOKMNLT; SOX2, OCT4 (POU5F1), KLF4, MYC, NANOG, LIN28, and SV40L T antigen) Epstein-Barr nuclear antigen-1–based episomal system was used (Thermo Fisher). Using CRISPR-Cas9 technology, the B2M and CIITA genes were disrupted to generate B2M−/−CIITA−/− hiPSCs as described previously (Deuse et al., 2019). To achieve CD47 overexpression, the human CD47 cDNA gene sequence was synthetized in frame with puromycin and separated with a P2A cleaving peptide to allow functional expression of both the CD47 and puromycin. The synthetized CD47-P2A-puro was then subcloned into a pLenti vector, which drives the CD47-P2A-puro cassette under control of a EF-1α short promoter. B2M−/−CIITA−/− hiPSCs were transfected to obtain a pool of B2M−/−CIITA−/− CD47 tg hiPSC. Alternatively, cells were transfected with the CD47 expression plasmid using lipofectamine to generate a B2M−/−CIITA−/− CD47 tg (plasmid) hiPSC pool. For single-cell cloning, cultures were dissociated using TrypLE. Dissociated cells were then stained with Tra1-60 Alexa 488 and propidium iodide. Stained hiPSCs were sorted using a FACSAria II cell sorter (BD Biosciences). Single cells were seeded into 96-well plates. Doublets and debris were excluded from seeding by selective gating on forward and side light scatter emission, and viable pluripotent cells were selected based on the absence of propidium iodide and the presence of Tra1-60 Alexa 488 staining. Single cells were then expanded into full-size colonies, after which the colonies were consolidated, banked, and tested for CD47 expression. Five clones with different levels of CD47 expression from pLenti-derived B2M−/−CIITA−/− CD47 tg hiPSCs, as well as one clone from the B2M−/−CIITA−/− CD47 tg (plasmid) hiPSC pool, were selected for further analyses.
Human iPSC culture and transduction to express Fluc
Human iPSCs were cultured on diluted feeder-free Matrigel (hESC qualified; BD Biosciences)–coated 10-cm dishes in Essential 8 Flex medium (Thermo Fisher). Medium was changed every 24 h, and Versene (Gibco) was used for cell passaging at a ratio of 1:6. For luciferase transduction, 105 hiPSCs were plated in one 6-well and incubated overnight at 37°C with 5% CO2. The next day, medium was changed, and one vial of Fluc lentiviral particles expressing luciferase II gene under reengineered EF1a promotor (Gen Target) was added to 1.5 ml medium. After 36 h, 1 ml cell medium was added. After 24 h, complete medium change was performed. After 2 d, luciferase expression was confirmed by adding D-luciferin (Promega). Signals were quantified in p/s/cm2/sr.
Endothelial cell differentiation from hiPSCs
The differentiation protocol was initiated at 60% confluency, and medium was changed to RPMI-1640 containing 2% B-27 minus insulin (both Gibco) and 5 µM CHIR-99021 (Selleckchem). On day 2, the medium was changed to reduced medium of RPMI-1640 containing 2% B-27 minus insulin (Gibco) and 2 µM CHIR-99021 (Selleckchem). From culture days 4–7, cells were exposed to RPMI-1640 EC medium, RPMI-1640 containing 2% B-27 minus insulin plus 50 ng/ml human VEGF (R&D Systems), 10 ng/ml human FGFb (R&D Systems), 10 µM Y-27632 (Sigma-Aldrich), and 1 µM SB 431542 (Sigma-Aldrich). Endothelial cell clusters were visible from day 7, and cells were maintained in Endothelial Cell Basal Medium 2 (PromoCell) plus supplements, 10% FCS hi (Gibco), 1% pen/strep, 25 ng/ml VEGF, 2 ng/ml FGFb, 10 µM Y-27632 (Sigma-Aldrich), and 1 µM SB 431542 (Sigma-Aldrich). The differentiation protocol was completed after 14 d; undifferentiated cells detached during the differentiation process. TrypLE Express (Gibco) was used for passaging the cells 1:3 every 3–4 d.
NK cell culture
Human primary NK cells were purchased from StemCell Technologies (70036) and cultured in RPMI-1640 plus 10% FCS hi and 1% pen/strep before performing the assays.
Macrophage differentiation from peripheral blood mononuclear cells (PBMCs)
PBMCs were isolated by Ficoll separation from fresh blood and were resuspended in RPMI-1640 with 10% FCS hi and 1% pen/strep (all Gibco). Cells were plated in 24-well plates at a concentration of 106 cells/ml and 10 ng/ml human M-CSF (PeproTech). Medium was changed every second day. From day 6 onward, 1 µg/ml human IL-2 (PeproTech) was added to the medium for 24 h before performing assays.
NK cell and macrophage killing assay by XCelligence
NK cell killing assays and macrophage killing assays were performed on the XCelligence SP platform and MP platform (ACEA Biosciences). Special 96-well E-plates (ACEA Biosciences) were coated with collagen (Sigma-Aldrich) and 4 × 105 WT, B2M−/−CIITA−/−, or B2M−/−CIITA−/− CD47 tg (pooled or single clones) hiECs were plated in 100 µl cell-specific medium. After the cell index value reached 0.7, human NK cells or human macrophages were added at an E:T ratio of 0.5:1, 0.8:1, or 1:1 with or without 1 ng/ml human IL-2 or human IL-15 (both from PeproTech). In some cases, NK cells were pretreated with human FcR block (catalog no. 130–059-901, concentration 1:5; Miltenyi) for 4 h before addition of target cells. As a negative control, cells were treated with 2% Triton X-100 in cell-specific media (data not shown).
Some wells were pretreated with an anti-CD47 blocking antibody (10 µg/ml, clone B6.H12, mouse IgG1,κ; BioXCell) for 2 h and during the assay or with an anti-SIRPα blocking peptide (2 µg/ml, catalog no. MBS822365; MyBioSource) for 2 h and during the assay. In some cases, cells were treated with an activating anti-LILRB1 antibody (clone GHI/75, mouse IgG2b; Abcam) at a concentration of 1 µg/ml. Data were standardized and analyzed with the RTCA software (ACEA Biosciences).
Transcriptome sequencing and data analysis
For the sequencing of CD3−CD7+CD56+ primary human NK cells, trimmomatic (Bolger et al., 2014) was employed to remove sequencing adapters and for quality control. Low-quality bases (Phred quality score <20) were trimmed off the 3′ end, and three bases were removed from the 5′ end of each sequencing read. Thereafter, reads shorter than 32 bases were removed. The remaining sequencing reads were aligned to the human reference assembly (GRCh38.98) using STAR (v2.7.3a; Dobin et al., 2013) with default parameters. Differential expression was assessed with DESeq2 (Love et al., 2014).
Sequencing of human NK cell lines was performed on a BGISeq-500 platform in single-end mode with 50-bp read length. For the primary NK cells and the four NK cell lines, between 25.4 and 25.7 million reads were sequenced per sample; for the other samples between 31.4 and 33.7 million reads were sequenced. Sequence reads were aligned to the human reference assembly (GRCh38.95) using STAR (v2.6.1c). Differential expression analysis was performed with DESeq2. Genes were considered differentially expressed when the absolute log2 fold change was ≥1 and the false discovery rate (FDR) was ≤0.1. Sequence data have been submitted to the European Nucleotide Archive, and the corresponding accession no. is PRJEB36016.
RT-PCR
For RT-PCR, RNA was extracted using the RNeasy Plus Mini Kit (Qiagen). Genomic DNA contamination was removed using the gDNA spin column. cDNA was generated using Applied Biosystems High-Capacity cDNA Reverse Transcription Kit. Human CD47 primers (Sinobiological) were used to amplify target sequences. GAPDH primers (Santa Cruz Biotechnology) were used as a housekeeping gene control. PCR reactions were performed with TopTaq DNA Polymerase (Qiagen) on Mastercycler nexus (Eppendorf AG) and visualized on 2% agarose gels. Real-time RT-PCR was performed on the QuantStudio6 (Agilent Technologies) with SYBR Premix Ex Taq II (Takara) according to the manufacturer’s instructions. The RNA fold change was estimated according to the comparative Ct method with the 2−ΔΔCt formula.
Flow cytometry
Human iECs were harvested and labeled with FITC-conjugated anti-CD47 antibody (clone CC2C6, mouse IgG1,κ; BioLegend) and FITC-conjugated mouse IgG1,κ isotype-matched control antibody (MOPC-21; BioLegend). SIRPα expression (PE-conjugated antibody, clone 15–414, IgG2a,κ; BioLegend) on CD3−CD7+CD56+ primary human NK cells and CD47 binding (recombinant CD47 protein Fc chimera, ab216207; Abcam; with FITC-conjugated anti-human IgG1 secondary antibody) were assessed by flow cytometry. Results were expressed as mean fluorescent intensity fold change to isotype-matched control Ig staining or secondary antibody staining.
In vivo cytotoxicity assay with adoptive transfer
Five million WT hiECs and five million B2M−/−CIITA−/− CD47 tg hiECs were mixed and stained with 5 µM CFSE (Thermo Fisher). Cells in saline with human IL-2 (1 µg; PeproTech) and 2.5 × 106 human primary NK cells (StemCell Technologies) or 2.5 × 106 human macrophages (differentiated from PBMCs) were injected i.p. into immunodeficient NSG mice. Human primary NK cells were pretreated with human IL-2 (1 ng/ml; PeproTech) in vitro 12 h before injection. After 48 h, cells were collected from the peritoneum and stained with allophycocyanin-conjugated anti-HLA-A, anti-HLA-B, and anti-HLA-C antibody (clone G46_2.6, BD Biosciences) for 45 min at 4°C. The CFSE-positive and HLA-A/HLA-B/HLA-C–negative population was analyzed by flow cytometry (FACSCalibur; BD Bioscience) and compared between the WT and engineered hiEC group for different clones of B2M−/−CIITA−/− CD47 tg hiECs.
In vivo K562 killing assay with adoptive transfer
One million K562 or K562 CD47 tg targets were injected s.c. into immunodeficient NSG mice. Human CD3−CD7+CD56+ primary NK cells or NK92 were pretreated with human IL-2 (1 ng/ml; PeproTech) in vitro 72 h before being s.c. injected together with the target cells. In some experiments, NK cells were pretreated with human FcR block (catalog no. 130–059-901, concentration 1:5; Miltenyi) for 1 h. In some experiments, anti-CD47 blocking antibody (10 µg/ml, clone B6.H12, mouse IgG1,κ; BioXCell) or anti-CD47 blocking Fab (10 µg/ml, clone B6.H12, catalog no. HPAB-0840-CN-F(E); Creative Biolabs) was used. Survival of Fluc+ K562 or K562 CD47 tg was longitudinally assessed by BLI.
Macrophage killing by BLI
Luciferase-expressing B2M−/−CIITA−/− or B2M−/−CIITA−/− CD47 tg hiPSCs were counted and plated at a concentration of 105 cells per 24 wells. After 16 h, human macrophages or mouse C57BL/6 macrophages were added to the hiPSCs at an E:T ratio of 1:1. After 120 min, luciferase expression was detected by adding D-luciferin. As controls, target cells were nontreated or treated with 2% Triton X-100 in cell-specific media. In some conditions, target cells were pretreated with anti-CD47 blocking antibody (10 µg/ml) 2 h before addition of target cells. Signals were quantified with Ami HT (Spectral Instruments Imaging) in p/s/cm2/sr.
NK cell ELISpot assays
For NK cell–specific ELISpot assays, human primary NK cells or mouse C57BL/6 NK cells were co-cultured with B2M−/−CIITA−/− or B2M−/−CIITA−/− CD47 tg hiPSCs and their IFN-γ release (BD Biosciences) was measured. K562 cells (Sigma-Aldrich) served as positive control. Mitomycin-treated (50 µg/ml for 30 min) stimulator cells were incubated with NK cells at an E:T ratio of 1:1 for 24 h, and IFN-γ spot frequencies were enumerated using an ELISpot plate reader. In some cases, NK cells were stimulated with human IL-2 (1 ng/ml; PeproTech) and in some conditions, responder cells were pretreated with anti-CD47 blocking antibody (for 120 min at 37°C).
P815 BLI killing assay
Fluc+ P815 cells were counted and plated at a concentration of 103 cells per 96 wells and mixed with CD3−CD7+CD56+ primary human NK cells at an E:T ratio of 10:1. All NK cells were preincubated with human IL-2 (Life Technologies) at a concentration of 1 µg/ml for 72 h. After 4 h in the BLI killing assay, luciferase expression was detected by adding D-luciferin (Promega). As controls, target cells were left untreated or treated with 2% Triton X-100 in cell-specific media. In some conditions, target cells were treated with anti-CD16 antibody (clone 3G8, mouse IgG1,κ, 10 µg/ml; BioLegend), anti-NKG2D antibody (clone 149810, mouse IgG1, 10 µg/ml; R&D Systems), anti-SIRPα (clone 2H7E2, mouse IgG1, 10 µg/ml; antibodies-online), or anti-CD56 (clone NCAM1/784, mouse IgG1, Abcam, 10 µg/ml). Signals were quantified with Ami HT (Spectral Instruments Imaging) in p/s/cm2/sr.
K562 BLI killing assay
Fluc+ K562 or K562 CD47 tg cells were counted and plated at a concentration of 105 cells per 24 wells and mixed with CD3−CD7+CD56+ primary human NK, NKL, or NK92 cells at an E:T ratio of 1:1. After 2 h, luciferase expression was detected by adding D-luciferin (Promega). All NK cells were preincubated with human IL-2 (Life Technologies) at a concentration of 1 µg/ml for 72 h. Signals were quantified with Ami HT (Spectral Instruments Imaging) in p/s/cm2/sr.
Nonhuman primate assays
Gene editing of human iPSCs overexpressing rhesus CD47
Human B2M−/−CIITA−/− iPSCs overexpressing rhesus CD47 (rhCD47) were cultured on diluted feeder-free Matrigel (hESC qualified, BD Biosciences)–coated 6-well plates in Essential 8 Flex medium (Thermo Fisher) and transduced to express Fluc as described above.
Endothelial cell differentiation from B2M−/−CIITA−/− rhCD47 tg hiPSCs
Differentiation was initiated with B2M−/−CIITA−/− rhCD47 tg hiPSC at 60% confluency, and medium was changed to RPMI-1640 containing 2% B-27 minus insulin (both Gibco) and 5 µM CHIR-99021 (Selleckchem). On day 2, the medium was changed to reduced medium: RPMI-1640 containing 2% B-27 minus insulin (Gibco) and 2 µM CHIR-99021 (Selleckchem). From days 4–7, cells were cultured in RPMI-1640 EC medium of RPMI-1640 containing 2% B-27 minus insulin plus 50 ng/ml human VEGF, 10 ng/ml human FGFb; R&D Systems), 10 µM Y-27632 (Sigma-Aldrich), and 1 µM SB 431542 (Sigma-Aldrich). Endothelial cell clusters were visible from day 7 and cells were maintained in Endothelial Cell Basal Medium 2 (PromoCell) plus 10% FCS hi (Gibco), 1% pen/strep, 25 ng/ml VEGF, 2 ng/ml FGFb, 10 µM Y-27632 (Sigma-Aldrich), and 1 µM SB 431542 (Sigma-Aldrich). The differentiation process was completed after 14 d and undifferentiated cells detached during the differentiation process. TrypLE Express (Gibco) was used for passaging the cells 1:3 every 3–4 d.
Rhesus NK cell isolation
Rhesus PBMCs were isolated from fresh blood by Ficoll separation or purchased from Human Cells Biosciences. Cells were sorted on the FACSAria Fusion using FITC-conjugated anti-CD8 (ab28010, 1:5; Abcam) and PE-conjugated anti-NKG2A (130–114-092, 1:50; Miltenyi) antibodies to select a CD8+ NKG2A+ NK cell population as previously reported (Pomplun et al., 2015).
Macrophage differentiation from rhesus PBMCs
PBMCs were isolated by Ficoll separation from fresh blood and resuspended in RPMI-1640 with 10% FCS hi and 1% pen/strep (all from Gibco). Cells were plated in 24-well plates at a concentration of 106 cells/ml and 10 ng/ml rhesus M-CSF (Biorhyt). Medium was changed every second day until day 6. Macrophages were stimulated from day 6 with 1 µg/ml rhesus IL-2 (MyBioSource) for 24 h before the cells were used for assays.
Rhesus NK cell and macrophage killing by XCelligence
NK cell killing assays and macrophage killing assays were performed on the XCelligence SP platform and MP platform (ACEA Biosciences). Special 96-well E-plates (ACEA Biosciences) were coated with collagen (Sigma-Aldrich), and 4 × 105 WT, B2M−/−CIITA−/−, B2M−/−CIITA−/− CD47 tg, and B2M−/−CIITA−/− rhCD47 tg hiECs were plated in 100 µl cell-specific medium. After the cell index value reached 0.7, rhesus NK cells or rhesus macrophages were added with an E:T ratio of 0.5:1, 0.8:1, or 1:1 with or without 1 ng/ml rhesus IL-2 (MyBioSource). As a negative control, cells were treated with 2% Triton X-100 in cell-specific media (data not shown). Data were standardized and analyzed with the RTCA software (ACEA Biosciences).
Flow cytometry
Rhesus iECs were plated in 6-well plates in medium containing 100 ng/ml IFN-γ (Prospec). Cells were harvested and labeled with FITC-conjugated anti-CD47 antibody (clone CC2C6; BioLegend) and FITC-conjugated mouse IgG1k isotype-matched control antibody (MOPC-21; BioLegend). Results were expressed as mean fluorescence intensity fold change to isotype-matched control Ig.
ELISpot assays
For unidirectional ELISpot assays, recipient PBMCs were isolated from rhesus monkeys before (day 0) or at >7 d after cell injection. T cells were purified by CD3 MACS (Miltenyi) and used as responder cells. Donor cells were mitomycin treated (50 µg/ml for 30 min; Sigma-Aldrich) and used as stimulator cells. 100,000 stimulator cells were incubated with 5 × 105 recipient responder T cells for 36 h, and IFN-γ and IL-5 spot frequencies (UCyTech BioSciences) were enumerated using an ELISpot plate reader.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 software. Analysis of differences between two groups was performed using an unpaired two-tailed Student’s t test. Statistical analyses of three or more groups were performed using a one-way ANOVA followed by the post hoc test stated in the figure legends.
Online supplemental material
Fig. S1 shows that CD47 protects MHC-deficient target cells from killing by syngeneic or allogeneic NK cells and macrophages. Fig. S2 shows flow cytometry data on the expression of SIRPα on mouse and primary human NK cells and SIRPα binding to target cell CD47. Fig. S3 shows the impact of the five stimulatory cytokines (IL-2, IL-15, IL-12, IL-18, and IFN-α) on NK cell SIRPα expression. Fig. S4 shows that the four human NK cell lines (NKL, NK-RL12, NK-CT604, and NK92) are deficient in SIRPα and show no CD47 binding. Fig. S5 shows the killing efficacy of primary human NK cells and the four human NK cell lines (NKL, NK-RL12, NK-CT604, and NK92) on WT, B2M−/−CIITA−/−, and B2M−/−CIITA−/− CD47 tg hiECs in an IL-2 environment.
Acknowledgments
We thank C. Pahrmann for her overall assistance. We thank R. Nelson and K. Copeland with Spectral Instruments Imaging for their support with in vitro imaging and M. Lewis with Agilent for his support with the XCelligence experiments.
S. Schrepfer, T. Deuse, and L.L. Lanier received funding from the National Heart, Lung, and Blood Institute of the National Institutes of Health under award number R01HL140236. L.L. Lanier is supported in part by the Parker Institute for Cancer Immunotherapy. We acknowledge the University of California San Francisco Parnassus Flow Core (RRID:SCR_018206), supported in part by the National Institutes of Health (grant P30 DK063720 and S10 instrumentation grant S10 1S10OD021822-01).
Author contributions: T. Deuse L.L. Lanier, and S. Schrepfer designed the experiments, supervised the project, and wrote the manuscript. X. Hu performed the immunobiology experiments, molecular biology, imaging studies, and cell culture work and analyzed the data. S. Agbor-Enoh and M.K. Jang performed transcriptome sequencing. M. Alawi and C. Saygi performed bioinformatics analyses. A. Gravina performed the K562 cell culture and related immune assays. G. Tediashvili and V.Q. Nguyen performed in vivo BLI and flow cytometry experiments, respectively. Y. Liu generated the knockout mice. H. Valantine supervised the sequencing and gave conceptual advice. All authors contributed to editing the manuscript.
References
- Advani, R., Flinn I., Popplewell L., Forero A., Bartlett N.L., Ghosh N., Kline J., Roschewski M., LaCasce A., Collins G.P., et al. 2018. CD47 Blockade by Hu5F9-G4 and Rituximab in Non-Hodgkin’s Lymphoma. N. Engl. J. Med. 379:1711–1721. 10.1056/NEJMoa1807315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- André, P., Denis C., Soulas C., Bourbon-Caillet C., Lopez J., Arnoux T., Bléry M., Bonnafous C., Gauthier L., Morel A., et al. 2018. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell. 175:1731–1743.e13. 10.1016/j.cell.2018.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay, A.N., and Van den Berg T.K.. 2014. The interaction between signal regulatory protein alpha (SIRPα) and CD47: structure, function, and therapeutic target. Annu. Rev. Immunol. 32:25–50. 10.1146/annurev-immunol-032713-120142 [DOI] [PubMed] [Google Scholar]
- Barkal, A.A., Weiskopf K., Kao K.S., Gordon S.R., Rosental B., Yiu Y.Y., George B.M., Markovic M., Ring N.G., Tsai J.M., et al. 2018. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat. Immunol. 19:76–84. 10.1038/s41590-017-0004-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, Z., Shi L., Guo Y.L., Lv Z., Tang C., Niu S., Tremblay A., Venkataramani M., Culpepper C., Li L., et al. 2016. Cd47-Sirpα interaction and IL-10 constrain inflammation-induced macrophage phagocytosis of healthy self-cells. Proc. Natl. Acad. Sci. USA. 113:E5434–E5443. 10.1073/pnas.1521069113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M., Lohse M., and Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke, G., Holbrook J.D., Brown M.H., and Barclay A.N.. 2004. Human lymphocytes interact directly with CD47 through a novel member of the signal regulatory protein (SIRP) family. J. Immunol. 173:2562–2570. 10.4049/jimmunol.173.4.2562 [DOI] [PubMed] [Google Scholar]
- Childs, R.W., and Carlsten M.. 2015. Therapeutic approaches to enhance natural killer cell cytotoxicity against cancer: the force awakens. Nat. Rev. Drug Discov. 14:487–498. 10.1038/nrd4506 [DOI] [PubMed] [Google Scholar]
- Chiossone, L., Dumas P.Y., Vienne M., and Vivier E.. 2018. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 18:671–688. 10.1038/s41577-018-0061-z [DOI] [PubMed] [Google Scholar]
- Dai, H., Friday A.J., Abou-Daya K.I., Williams A.L., Mortin-Toth S., Nicotra M.L., Rothstein D.M., Shlomchik W.D., Matozaki T., Isenberg J.S., et al. 2017. Donor SIRPα polymorphism modulates the innate immune response to allogeneic grafts. Sci. Immunol. 2:eaam6202. 10.1126/sciimmunol.aam6202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuse, T., Hu X., Gravina A., Wang D., Tediashvili G., De C., Thayer W.O., Wahl A., Garcia J.V., Reichenspurner H., et al. 2019. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat. Biotechnol. 37:252–258. 10.1038/s41587-019-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornalusse, G.G., Hirata R.K., Funk S.E., Riolobos L., Lopes V.S., Manske G., Prunkard D., Colunga A.G., Hanafi L.A., Clegg D.O., et al. 2017. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat. Biotechnol. 35:765–772. 10.1038/nbt.3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatherley, D., Graham S.C., Turner J., Harlos K., Stuart D.I., and Barclay A.N.. 2008. Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47. Mol. Cell. 31:266–277. 10.1016/j.molcel.2008.05.026 [DOI] [PubMed] [Google Scholar]
- Jaiswal, S., Jamieson C.H., Pang W.W., Park C.Y., Chao M.P., Majeti R., Traver D., van Rooijen N., and Weissman I.L.. 2009. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 138:271–285. 10.1016/j.cell.2009.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y.Z., Couriel D., Mavroudis D.A., Lewalle P., Malkovska V., Hensel N.F., Dermime S., Molldrem J., and Barrett A.J.. 1996. Interaction of natural killer cells with MHC class II: reversal of HLA-DR1-mediated protection of K562 transfectant from natural killer cell-mediated cytolysis by brefeldin-A. Immunology. 87:481–486. 10.1046/j.1365-2567.1996.483556.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D.B., Nixon M.J., Wang Y., Wang D.Y., Castellanos E., Estrada M.V., Ericsson-Gonzalez P.I., Cote C.H., Salgado R., Sanchez V., et al. 2018. Tumor-specific MHC-II expression drives a unique pattern of resistance to immunotherapy via LAG-3/FCRL6 engagement. JCI Insight. 3:120360. 10.1172/jci.insight.120360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärre, K., Ljunggren H.G., Piontek G., and Kiessling R.. 1986. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 319:675–678. 10.1038/319675a0 [DOI] [PubMed] [Google Scholar]
- Kim, Y., Ponomarenko J., Zhu Z., Tamang D., Wang P., Greenbaum J., Lundegaard C., Sette A., Lund O., Bourne P.E., et al. 2012. Immune epitope database analysis resource. Nucleic Acids Res. 40(Web Server issue, W1):W525-30. 10.1093/nar/gks438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T., Vidal G.S., Djurisic M., William C.M., Birnbaum M.E., Garcia K.C., Hyman B.T., and Shatz C.J.. 2013. Human LilrB2 is a β-amyloid receptor and its murine homolog PirB regulates synaptic plasticity in an Alzheimer’s model. Science. 341:1399–1404. 10.1126/science.1242077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korde, N., Carlsten M., Lee M.J., Minter A., Tan E., Kwok M., Manasanch E., Bhutani M., Tageja N., Roschewski M., et al. 2014. A phase II trial of pan-KIR2D blockade with IPH2101 in smoldering multiple myeloma. Haematologica. 99:e81–e83. 10.3324/haematol.2013.103085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier, L.L. 2008. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9:495–502. 10.1038/ni1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier, L.L., Ruitenberg J.J., and Phillips J.H.. 1988. Functional and biochemical analysis of CD16 antigen on natural killer cells and granulocytes. J. Immunol. 141:3478–3485. [PubMed] [Google Scholar]
- Lanier, L.L., Corliss B., and Phillips J.H.. 1997. Arousal and inhibition of human NK cells. Immunol. Rev. 155:145–154. 10.1111/j.1600-065X.1997.tb00947.x [DOI] [PubMed] [Google Scholar]
- Liu, Y.W., Chen B., Yang X., Fugate J.A., Kalucki F.A., Futakuchi-Tsuchida A., Couture L., Vogel K.W., Astley C.A., Baldessari A., et al. 2018. Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat. Biotechnol. 36:597–605. 10.1038/nbt.4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M., O’Connor R.S., Trefely S., Graham K., Snyder N.W., and Beatty G.L.. 2019. Metabolic rewiring of macrophages by CpG potentiates clearance of cancer cells and overcomes tumor-expressed CD47-mediated ‘don’t-eat-me’ signal. Nat. Immunol. 20:265–275. 10.1038/s41590-018-0292-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matozaki, T., Murata Y., Okazawa H., and Ohnishi H.. 2009. Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway. Trends Cell Biol. 19:72–80. 10.1016/j.tcb.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Milush, J.M., Long B.R., Snyder-Cappione J.E., Cappione A.J. III, York V.A., Ndhlovu L.C., Lanier L.L., Michaëlsson J., and Nixon D.F.. 2009. Functionally distinct subsets of human NK cells and monocyte/DC-like cells identified by coexpression of CD56, CD7, and CD4. Blood. 114:4823–4831. 10.1182/blood-2009-04-216374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, L.M., Tal M.C., Torrez Dulgeroff L.B., Carmody A.B., Messer R.J., Gulati G., Yiu Y.Y., Staron M.M., Angel C.L., Sinha R., et al. 2019. A functional subset of CD8+ T cells during chronic exhaustion is defined by SIRPα expression. Nat. Commun. 10:794. 10.1038/s41467-019-08637-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaishi, A., Hirose M., Yoshimura M., Oneyama C., Saito K., Kuki N., Matsuda M., Honma N., Ohnishi H., Matozaki T., et al. 2008. Structural insight into the specific interaction between murine SHPS-1/SIRP alpha and its ligand CD47. J. Mol. Biol. 375:650–660. 10.1016/j.jmb.2007.10.085 [DOI] [PubMed] [Google Scholar]
- Ndhlovu, L.C., Lopez-Vergès S., Barbour J.D., Jones R.B., Jha A.R., Long B.R., Schoeffler E.C., Fujita T., Nixon D.F., and Lanier L.L.. 2012. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood. 119:3734–3743. 10.1182/blood-2011-11-392951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbarnscheidt, M.H., Zeng Q., Li Q., Dai H., Williams A.L., Shlomchik W.D., Rothstein D.M., and Lakkis F.G.. 2014. Non-self recognition by monocytes initiates allograft rejection. J. Clin. Invest. 124:3579–3589. 10.1172/JCI74370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenborg, P.A., Zheleznyak A., Fang Y.F., Lagenaur C.F., Gresham H.D., and Lindberg F.P.. 2000. Role of CD47 as a marker of self on red blood cells. Science. 288:2051–2054. 10.1126/science.288.5473.2051 [DOI] [PubMed] [Google Scholar]
- Piccio, L., Vermi W., Boles K.S., Fuchs A., Strader C.A., Facchetti F., Cella M., and Colonna M.. 2005. Adhesion of human T cells to antigen-presenting cells through SIRPbeta2-CD47 interaction costimulates T-cell proliferation. Blood. 105:2421–2427. 10.1182/blood-2004-07-2823 [DOI] [PubMed] [Google Scholar]
- Pomplun, N., Weisgrau K.L., Evans D.T., and Rakasz E.G.. 2015. OMIP-028: activation panel for Rhesus macaque NK cell subsets. Cytometry A. 87:890–893. 10.1002/cyto.a.22727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet, D.H., and Vance R.E.. 2006. Self-tolerance of natural killer cells. Nat. Rev. Immunol. 6:520–531. 10.1038/nri1863 [DOI] [PubMed] [Google Scholar]
- Seiffert, M., Brossart P., Cant C., Cella M., Colonna M., Brugger W., Kanz L., Ullrich A., and Bühring H.J.. 2001. Signal-regulatory protein alpha (SIRPalpha) but not SIRPbeta is involved in T-cell activation, binds to CD47 with high affinity, and is expressed on immature CD34(+)CD38(-) hematopoietic cells. Blood. 97:2741–2749. 10.1182/blood.V97.9.2741 [DOI] [PubMed] [Google Scholar]
- Shirai, H., Mandai M., Matsushita K., Kuwahara A., Yonemura S., Nakano T., Assawachananont J., Kimura T., Saito K., Terasaki H., et al. 2016. Transplantation of human embryonic stem cell-derived retinal tissue in two primate models of retinal degeneration. Proc. Natl. Acad. Sci. USA. 113:E81–E90. 10.1073/pnas.1512590113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano, R.F., Pratt J.C., Schmidt R.E., Ritz J., and Reinherz E.L.. 1985. Activation of cytolytic T lymphocyte and natural killer cell function through the T11 sheep erythrocyte binding protein. Nature. 317:428–430. 10.1038/317428a0 [DOI] [PubMed] [Google Scholar]
- Sola, C., André P., Lemmers C., Fuseri N., Bonnafous C., Bléry M., Wagtmann N.R., Romagné F., Vivier E., and Ugolini S.. 2009. Genetic and antibody-mediated reprogramming of natural killer cell missing-self recognition in vivo. Proc. Natl. Acad. Sci. USA. 106:12879–12884. 10.1073/pnas.0901653106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier, E., Tomasello E., Baratin M., Walzer T., and Ugolini S.. 2008. Functions of natural killer cells. Nat. Immunol. 9:503–510. 10.1038/ni1582 [DOI] [PubMed] [Google Scholar]
- Willingham, S.B., Volkmer J.P., Gentles A.J., Sahoo D., Dalerba P., Mitra S.S., Wang J., Contreras-Trujillo H., Martin R., Cohen J.D., et al. 2012. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. USA. 109:6662–6667. 10.1073/pnas.1121623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., Wang B., Ono M., Kagita A., Fujii K., Sasakawa N., Ueda T., Gee P., Nishikawa M., Nomura M., et al. 2019. Targeted Disruption of HLA Genes via CRISPR-Cas9 Generates iPSCs with Enhanced Immune Compatibility. Cell Stem Cell. 24:566–578.e7. 10.1016/j.stem.2019.02.005 [DOI] [PubMed] [Google Scholar]
- Zen, K., Guo Y., Bian Z., Lv Z., Zhu D., Ohnishi H., Matozaki T., and Liu Y.. 2013. Inflammation-induced proteolytic processing of the SIRPα cytoplasmic ITIM in neutrophils propagates a proinflammatory state. Nat. Commun. 4:2436. 10.1038/ncomms3436 [DOI] [PMC free article] [PubMed] [Google Scholar]