Leukemia stem cells are heterogeneous and reside within the phenotypic hematopoietic stem cell and progenitor cell compartments in juvenile myelomonocytic leukemia.
Abstract
In this issue of JEM, Louka et al. (https://doi.org/10.1084/jem.20180853) report that leukemia stem cells lie within the phenotypic hematopoietic stem cell and progenitor cell compartments in juvenile myelomonocytic leukemia (JMML). Furthermore, they identify several candidate biomarker/therapeutic targets, such as CD96 and SLC2A1, that are of translational significance in JMML.
Despite recent research advancements establishing the genomic landscape in juvenile myelomonocytic leukemia (JMML; Caye et al., 2015), the identity of leukemia stem cells (LSCs) in this disease has remained largely unknown. In this study, Louka et al. address this gap in knowledge by investigating the origins of JMML using primary human patient specimens and report that the LSCs are heterogeneous and reside not only within the phenotypic hematopoietic stem cell (HSC) but also within the progenitor fractions. Additionally, the authors performed RNA profiling of LSCs and uncovered novel candidate biomarkers/therapeutic targets, such as CD96, SLC2A1, and STK24, in JMML (Louka et al., 2021).

Insights from Sriram Sundaravel and Ulrich Steidl.
The ability to initiate/propagate cancer is thought to reside within functionally defined cancer stem cells (CSCs), and the search for the identity of CSCs in various cancers has been ongoing for decades (Batlle and Clevers, 2017). While extensive research efforts have identified the CSCs in multiple cancers, the identity of CSCs/LSCs in rare cancers such as JMML has remained elusive, in part due to challenges in obtaining enough specimens to conduct detailed analyses. JMML is an aggressive pediatric hematopoietic disorder that is characterized by the presence of mutations in the RAS signaling pathway and exhibits hallmark features such as hyperproliferation and dysplasia in the monocyte and granulocyte lineages (Locatelli and Niemeyer, 2015). JMML is often fatal due to multiorgan failure or progression to acute myeloid leukemia (AML). Although allogeneic hematopoietic stem cell transplantation (HSCT) can be curative in this disease, ∼40–50% of the patients experience disease recurrence (Caye et al., 2020). Unraveling the cellular origins of JMML is of high importance and may provide crucial insights to manage and causatively treat this deadly disease.
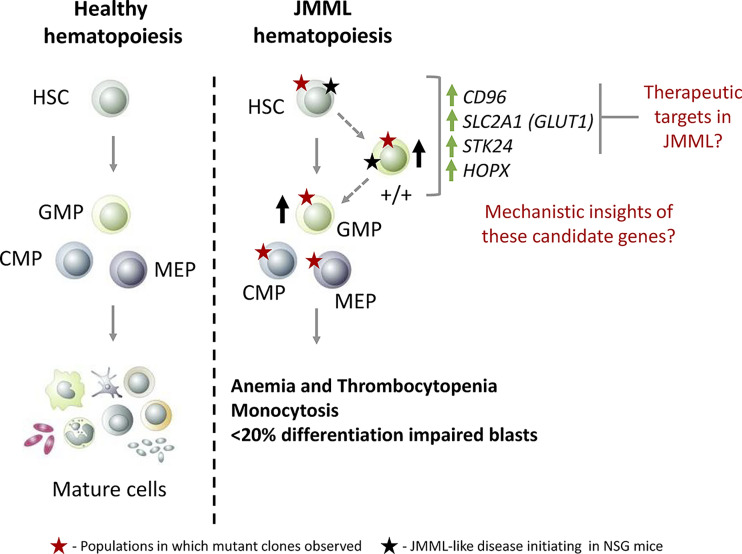
One of the hallmark features of cancer is intratumoral heterogeneity, and given the heterogeneous nature of LSCs in aggressive hematopoietic malignancies such as myelodysplastic syndromes (MDS) and AML (Chen et al., 2019; Shlush et al., 2017; Sarry et al., 2011; Yanagisawa et al., 2016), the authors hypothesized that the LSC landscape in JMML might be similarly heterogenous. Hence, the authors systematically examined hematopoietic stem/progenitor cell (HSPC) fractions in JMML to identify cell populations exhibiting LSC activity. pecifically, they focused on phenotypic HSCs (Lin−CD34+CD38−CD90+CD45RA−), granulocyte monocyte progenitors (GMPs; Lin−CD34+CD38+CD123+CD45RA+) and a novel double-positive (+/+; Lin−CD34+CD38−CD90+CD45RA+) population that was present only in JMML and had exclusive myeloid potential. They found that specifically the GMP and +/+ populations were aberrantly increased in JMML samples compared with healthy pediatric bone marrow samples, suggesting that they might contain LSCs. To functionally assess the LSC activity of the JMML immunophenotypic HSC, GMP, and +/+ populations, the authors performed a series of in vivo xenotransplantation experiments. They found that the JMML HSCs efficiently reconstituted the mice bone marrow and resulted in JMML-like disease. However, the engraftment potential of the +/+ and GMP populations exhibited marked interpatient heterogeneity, with only the +/+ populations resulting in JMML-like disease. Collectively, the authors demonstrated that cells with LSC activity in JMML resided not only within HSCs but also within the +/+ and GMP populations, highlighting the underlying heterogeneity (Fig. 1). These findings are consistent with the results reported by another recent study (Caye et al., 2020). Because identification of differentially regulated pathways in JMML HSCs, +/+, and GMPs could lead to uncovering novel candidate biomarkers/therapeutic targets, the authors performed RNA sequencing of these populations. Consequently, they identified pathways, such as proliferation and DNA repair, to be deregulated and identified candidate genes, including CD96, SLC2A1, HOPX, and STK24, that could serve as biomarkers/therapeutic targets in JMML (Fig. 1).
Figure 1. Proposed model of JMML hematopoiesis compared with normal hematopoiesis. Hematopoietic hierarchy is depicted by the use of solid gray arrows, and the proposed JMML hematopoietic hierarchy is represented by dotted gray arrows. The +/+ population exhibited shared expression profiles between HSC and GMP and is considered to be locked in an HSC-GMP transition state. HSPC populations such as +/+ and GMPs, which were aberrantly increased in JMML, are specified with a solid black up arrow. Various HSPC populations that harbored aberrant mutational clones are marked by a solid red star. Cell populations that were experimentally demonstrated to induce a JMML-like disease in immune-compromised mice are indicated by a solid black star. The authors demonstrated that all the HSPC subpopulations contained mutant clones; however, only the HSC and +/+ populations were able to induce JMML-like disease when transplanted to immune-compromised mice. It is noteworthy that xenotransplantation of the GMP population didn’t result in JMML-like disease despite successful engraftment, which raises the possibility that the GMPs might not harbor the potential to propagate JMML. Future investigations are needed to further clarify the role of GMP in JMML onset and/or relapse. The genes that are up-regulated in JMML HSC and +/+ populations are depicted using solid green up arrows. CMP, common myeloid progenitor; MEP, megakaryocyte erythroid progenitor.
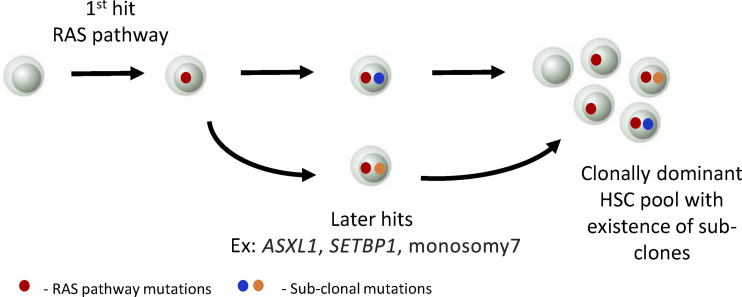
Next, to analyze the clonal landscape in JMML, the authors performed single-cell index sorting of Lin−CD34+ HSPCs followed by colony genotyping. In doing so, they found that the JMML HSPC compartment was not only clonally dominant with RAS pathway mutations but also contained subclones with other canonical leukemia mutations, such as ASXL1, SETBP1, and monosomy7 (Fig. 2). In addition, the results from these studies support the existence of both linear and branching clonal evolution patterns in JMML, which is similar to evolution patterns recently observed in other myeloid malignancies (Chen et al., 2019; Shlush et al., 2017; Miles et al., 2020). Collectively, these results further reinforce the notion that JMML HSPCs are indeed heterogeneous at phenotypic and clonal levels. Since prior work has implicated subclones in contributing to relapse in other myeloid malignancies, and given the presence of subclones in JMML, it will be interesting to assess the subclonal dynamics of JMML HSPCs during disease progression, HSCT, remission, and relapse in future studies.
Figure 2. Proposed model of HSC clonal evolution in JMML. JMML HSCs are clonally dominant with mutations in the RAS pathway genes (KRAS, NRAS, PTPN11, NF1, CBL), which are indicated by a solid red circle. The results also support the presence of subclones, which are indicated by solid blue/orange circles. The authors present evidence supporting linear and/or early branching clonal evolution patterns in JMML.
One likely explanation for the increased relapse rates in JMML could be incomplete eradication/persistence of LSCs. A growing body of literature has established paradigms where LSCs are present during clinical/morphological remission and act as precursors to leukemia relapse (Will et al., 2012). Consistent with this notion, the authors observed the presence of mutant HSPCs in post-HSCT samples of JMML patients. Because LSCs are postulated to drive disease relapse after remission/treatment, the authors wanted to examine the dynamics of JMML LSCs during diagnosis, after HSCT and relapse using longitudinal samples from individual patients. They conducted phenotypic analyses of the HSPC compartment and noticed that the aberrant +/+ population exhibited striking dynamics. They observed that the +/+ population that was present during diagnosis was undetectable by flow cytometry after HSCT. However, when the patients relapsed, there was a marked increase in the frequency of the +/+ population, suggesting it as a potential relapse driver. It could thus be informative to perform RNA profiling on +/+ populations at diagnosis and relapse to uncover specific pathways that facilitated disease relapse in future studies.
Another striking finding from this study is that CD96, a cell surface receptor that belongs to the Ig gene superfamily, was up-regulated in JMML LSCs. This finding is particularly interesting from a translational aspect because CD96 has been previously demonstrated to be an LSC marker/target in adult AML (Hosen et al., 2007). This provided the authors a rationale to more closely examine CD96 as an LSC marker in JMML. When they performed cell surface phenotypic analysis, they found that CD96 was indeed dramatically increased within the stem cell populations in JMML compared with respective cord blood counterparts. Interestingly, they also observed that CD96 expression was detected in stem cell populations of two JMML patients after HSCT during clinical remission who eventually relapsed. This finding is particularly interesting because it underscores the potential utility of CD96 as a JMML biomarker in disease monitoring and possibly as a predictive biomarker for relapse. Subsequently, the authors performed a series of xenotransplantation experiments to assess the LSC activity in CD96-positive and CD96-negative JMML LSCs. They found that JMML LSCs expressing CD96 engrafted rapidly and developed JMML-like disease in a shorter time span compared with CD96-negative JMML LSCs. Collectively, these findings establish CD96 as a “true” LSC marker in JMML with multiple translational implications. One can envision the development of antibodies and/or chimeric antigen receptor T cells against CD96 to target LSCs in JMML in the future.
In addition to CD96, the authors also evaluated another candidate they identified, SLC2A1 (GLUT1), as a therapeutic target in JMML. GLUT1 is a membrane protein that is known for its role in glucose transport. Previous studies have shown that inhibiting glucose transport by targeting GLUT1 exerts cytotoxic effect on cancer cells (Liu et al., 2012). Louka et al. (2021) examined GLUT1 as a therapeutic target by testing whether JMML HSPCs are vulnerable to pharmacological inhibition of GLUT1. Interestingly, the authors found that GLUT1 inhibitors preferentially exerted cytotoxic activity on JMML HSPCs compared with cord blood HSPCs. Moreover, the fact that mitogen-activated protein kinase kinase (MEK) inhibitors are in clinical development for JMML provided the rationale for the authors to test a combination therapy along with GLUT1 inhibitors. They found synergism between GLUT1 and MEK inhibitors, with the combination treatment resulting in the maximum cytotoxic effect compared with individual treatments. These findings are exciting from a therapeutic standpoint, and it will be interesting to further test GLUT1 inhibitors alone or in combination with MEK inhibitors in preclinical in vivo JMML xenotransplantation models to further develop this as a potential therapeutic strategy in JMML.
One of the questions that arise from this study is, what are the molecular mechanisms by which JMML LSCs lead to disease progression and relapse? The newly identified immunophenotypic markers provide a novel tool to pursue this and other questions on LSCs in JMML in the future. In addition to CD96 and SLC2A1, several additional candidate pathogenic genes have been identified in this study; however, functional roles of many of these genes (SLC2A1, HOPX, STK24) are unknown in the context of JMML. For example, higher HOPX expression is associated with poor prognosis in AML (Lin et al., 2017), suggesting that HOPX status could potentially also be relevant in JMML patients. Furthermore, previous studies have implicated aberrant glucose metabolism in leukemia (Chen et al., 2014); however, the underlying mechanisms are still only partially understood. Future mechanistic and translational investigations geared toward understanding how SLC2A1, HOPX, and STK24 facilitate JMML progression and relapse could be very informative in that regard.
In conclusion, the work by Louka et al. (2021) represents a significant advance in the field of JMML research by not only identifying the cell of origin of this disease but also uncovering multiple actionable candidate targets with translational significance. Additionally, this study may serve as a road map for investigating cellular origins of other rare cancers in the future.
Acknowledgments
S. Sundaravel is supported by the National Cancer Institute (K00CA223044) and the Harry Eagle Scholar award.
References
- Batlle, E., and Clevers H.. 2017. Nat. Med. 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- Caye, A., et al. 2015. Nat. Genet. 10.1038/ng.3420 [DOI] [Google Scholar]
- Caye, A., et al. 2020. Leukemia. 10.1038/s41375-019-0662-y [DOI] [Google Scholar]
- Chen, W.L., et al. 2014. Blood. 10.1182/blood-2014-02-554204 [DOI] [Google Scholar]
- Chen, J., et al. 2019. Nat. Med. 10.1038/s41591-018-0267-4 [DOI] [Google Scholar]
- Hosen, N., et al. 2007. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.0704271104 [DOI] [Google Scholar]
- Lin, C.C., et al. 2017. Haematologica. 10.3324/haematol.2016.161257 [DOI] [Google Scholar]
- Liu, Y., et al. 2012. Mol. Cancer Ther. 10.1158/1535-7163.MCT-12-0131 [DOI] [Google Scholar]
- Locatelli, F., and Niemeyer C.M.. 2015. Blood. 10.1182/blood-2014-08-550483 [DOI] [PubMed] [Google Scholar]
- Louka, E., et al. 2021. J. Exp. Med. 10.1084/jem.20180853 [DOI] [Google Scholar]
- Miles, L.A., et al. 2020. Nature. 10.1038/s41586-020-2864-x [DOI] [Google Scholar]
- Sarry, J.E., et al. 2011. J. Clin. Invest. 10.1172/JCI41495 [DOI] [Google Scholar]
- Shlush, L.I., et al. 2017. Nature. 10.1038/nature22993 [DOI] [Google Scholar]
- Will, B., et al. 2012. Blood. 10.1182/blood-2011-12-399683 [DOI] [Google Scholar]
- Yanagisawa, B., et al. 2016. Exp. Hematol. 10.1016/j.exphem.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]