Abstract
Influenza B virus is a member of the Orthomyxoviridae family which can infect humans and causes influenza. Although it is not pandemic like influenza A virus, it nevertheless affects millions of people worldwide annually. MicroRNAs are small non-coding RNAs regulating gene expression at posttranscriptional level. They play various important roles in cellular processes including response to viral infection. MiRNA profiles from our previous study suggested that miR-30e-3p was one of the upregulated miRNAs that responded to influenza B virus infection. In this study, in silico prediction and in vitro investigation proved that this miRNA can directly target NA and NP genes of the influenza B virus and inhibit its replication. This finding might be useful for using miRNA as an alternative therapeutics for influenza virus infection.
Keywords: hsa-miR-30e-3p, microRNA, influenza B virus, NA, NP, inhibit
Impact statement
This is the first study to investigate the function of human miRNA in influenza B virus infection. The hsa-miR-30e-3p was computationally predicted and in vitro studies proved that this miRNA can directly target viral NA and NP genes and inhibit influenza B virus replication. Thus, it is the first report suggesting that human miRNA can directly target influenza B virus and inhibit viral replication. It also proved the concept that in response to viral infection, some host miRNAs are upregulated and miRNA can be used as a silencing strategy to inhibit viral replication at the early stage of viral infection.
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs with 20–22 nucleotide length.They are generally generated from 60 to 110 nucleotide stem-loop RNA precursor structures.1 The biogenesis of miRNAs requires members of the Argonaute protein family, Pol II-dependent transcription, and the RNase IIIs Drosha and Dicer.2,3 MiRNAs regulate gene expression through mRNA degradation or translational repression by pairing with their mRNA targets. The functions of miRNAs are involved in various biological processes, including development, differentiation, proliferation, apoptosis, and response to virus infection.3,4
The cellular miRNAs also evolved in the response to viral infection as shown in various studies. For example, the hsa-miR-32 was reported to reduce the accumulation of primate foamy virus type 1 (PFV-1).5 Three miRNAs including miR-323, miR-491, and miR-654 can directly target influenza A H1N1 (A/WSN/33) PB1 genes and inhibit viral replication.6 In addition, some studies using computational method to predict host miRNAs targeting viral genomes predicted that hsa-miR-489, hsa-miR-325, hsa-miR-876-3p, and hsa-miR-2117 target HA, PB2, MP, and NS of influenza A virus subtype H1N1 and inhibit viral replication.7 The computational prediction suggested that human miRNA hsa-miR-323, hsa-miR-491, and hsa-miR-654 target PB1 gene of H1N1 influenza A virus (A/WSN/33).8 The repression of PB1 which is a component of viral polymerase complex leads to inhibition of influenza A virus replication. The computational analysis also suggested that hsa-miR-3145 can target the PB1 gene of influenza A viruses three subtype including pH1N1, H5N1, and H3N2. The luciferase assay confirmed that the miR-3145 directly targeted the PB1 gene of influenza A viruses. And the in vitro transfection of miR-3145 expression vector confirmed that this miRNA can inhibit viral replication.9 Human let-7c was reported to be upregulated in influenza virus-infected A549 cells. The seed region of let-7c is perfectly complementary to the 3ʹ UTR of the viral M1 gene. In vitro study also confirmed that let-7c can downregulate viral M1 both in the RNA and protein levels.10
Influenza B virus is a member of the enveloped RNA virus in the family Orthomyxoviridae.11 Its genome contains eight negative single strands of RNA: polymerase basic-1(PB1), polymerase basic-2(PB2), polymerase acidic (PA), hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA), matrix (M), and nonstructural protein (NS).12 Influenza B virus causes influenza, which is a worldwide infectious problem. The clinical symptoms are generally similar to the infection of influenza A viruses including fever, muscle aching, headache, dry cough, fatigue, and nasal congestion.13 There are several differences between influenza B virus and influenza A virus such as no subtypes in influenza B, while influenza A has varieties of HA and NA genes which can be used for subtype classification.14 Genetic reassortment is a major process of evolution for the influenza B virus, producing new recombinant genomes supporting viral adaptation, which can cause epidemics and pandemics occasionally around the world.15 Currently, two subtypes of influenza A and one of influenza B lineage are included in the current trivalent seasonal influenza vaccines.
To date, more research has been dedicated to influenza A than influenza B in terms of cell miRNA studies.16 Information concerning the biological process of miRNAs involved in influenza B virus infection is still sparse and unclear. Our previous studies investigated how human miRNA profiles responded to influenza A (subtype pH1N1, H5N1 and H3N2)17 and influenza B virus infection.18 Interestingly, hsa-miR-30e-3p was reported to be upregulated in both studies at 24-h post infection. Therefore, in this study, we aimed to investigate the role of hsa-miR-30e-3p in influenza B virus infection, especially focusing on viral gene regulation.
Materials and methods
Cell culture
Adenocarcinomic human alveolar basal epithelial cells (A549 cells) were used for an in vitro study. The A549 cells were cultured in high glucose containing Dulbecco’s modified Eagle medium (DMEM) with 1% (v/v) antibiotic/antimycotic and 10% fetal bovine serum (FBS). Cells were incubated at 37°C in a humidified atmosphere with 5% CO2. Culture media were replaced every three days.
Transfection of miRNA and viral infection
To study the effect of how hsa-miR-30e-3p expression was changed due to the infection of influenza B virus, the miRNA mimics, miRNA inhibitors, and mimic negative control were used for an in vitro study. From the preliminary study, the mimic transfection condition was optimized to upregulate the miR-30e-3p expression to the level found in our previous study (∼2 fold change). The 0.5 pmol of mimic per well transfection could upregulate the miRNA expression to the lowest fold changes (∼40 fold change) with reproducibility. The upregulated level of this miRNA prolonged up to 72 h post transfection. The 0.5 pmol miRNA mimics, 50 pmol miRNA inhibitors, and 0.5 pmol mimic negative control (mirVarna) were individually transfected into the A549 cells (3 × 105 per well) in six well-plates by Lipofectamine 2000 (Invitrogen) transfection reagent. In this experiment, each transfected group was performed in triplicates. After 24 h post transfection, the cells were harvested and the small RNA was extracted by miRNA extraction kit (Norgen). The expressions of miRNAs were quantified by poly-U polymerization followed by the stem-loop RT-qPCR relative quantification.19 The stem-loop RT-qPCR to quantify miRNA in every sample was performed in triplicates. The expression level of U6 snRNA was used as reference for normalization. The amount of miRNA was optimized to reach the miRNA expression level comparable to the expression level that responded to influenza B virus infection. The miRNA expression fold changes between each group were analyzed by Student’s t-test. The P ≤ 0.05 was considered as statistical significant.
Quantitation of viral production
To quantify the viral production, viral particles were extracted from 200 µL of the culture media by using QIAamp viral RNA mini kit (QIAGEN). Then the viral RNA was reverse transcribed with random hexamer primers. The viral M gene in the supernatant was used to represent the viral production. The qPCR of M gene was performed by using these components (primers; FluB-MF439 CTCTGTGCTTTRTGC GARAAAC 439–460 sense and FluB-MR CCTTCYCCATTCTTTTGACTTGC 671–649 antisense, probe; FluB-P135 Cy5-TCAGCAATGAACACAGCAA-BHQ3 541–559 sense).20 The qPCR of each sample was performed in triplicates. The viral M gene level was presented as % FluB in supernatant. The mimic negative control-transfected group was used as reference.
Cell viability assay
Cell proliferation was determined using the MTT Kit (Promega) according to the manufacturer’s instructions. For each sample, the MTT assay was performed in triplicates. In brief, 20 µL MTT (5 mg/mL) was added to the transfected cells. The supernatant was removed after 4 h incubation with MTT, and 200 µL DMSO was added into the plates to solubilize the formazan. The optical density at 490 nm was measured using a microplate reader (Thermo), representing the cell viability.
In silico prediction of viral target
Influenza B virus (B/Thailand/CU-B5522/2011) genome (accession no: JX513059.1- JX513066.1) was used for miRNA-targeted genes prediction. The seed region binding rule21 was used as criteria for target gene prediction. The hybridization pattern and binding energy between the candidate miRNA and its viral-targeted gene were predicted by RNAHybrid, (http://bibiserv.techfak.unibielefeld.de/rnahybrid).22
Vector construction
To validate the influenza B virus genes which can be targeted by miRNAs of interest, the 3′-UTR reporter assay was performed. The reporter target vector was constructed by ligating the miRNA target site of the viral gene into the 3′-UTR of the reporter gene: firefly luciferase (Luc). Oligonucleotides (Table 1) were designed containing the miRNA-target site region of viral genes connecting with restriction sites in both 5ʹ and 3ʹ which were similar to the reporter vector. The pmiRGLO (Promega) was used as a backbone vector. For the silencing positive control of reporter gene, the siLuc/Luc2 oligo nucleotides were ligated with pSilencer3.0-H1.
Table 1.
Sequences of oligo nucleotides for vector construction.
| Oligo name | Sequence (5ʹ-3ʹ) |
|---|---|
| Backbone vector: pmiRGLO | |
| NEP-428-top | CTAGCATGTTGATGGCCCAACTGAAATC |
| NEP-449-bottom | TCGAGATTTCAGTTGGGCCATCAACATG |
| NB&NA-1123-top | CTAGCCGATGTCTAAAACTGAAAGGC |
| NB&NA-1142-bottom | TCGAGCCTTTCAGTTTTAGACATCGG |
| NP-23-top | CTAGCAGTAAAAGAACTGAAAAC |
| NP-39-bottom | TCGAGTTTTCAGTTCTTTTACTG |
| PB2-1406-top | CTAGCTGAATGCATCTGACTATACACTGAAAGGC |
| PB2-1433-bottom | TCGAGCCTTTCAGTGTATAGTCAGATGCATTCAG |
| PB2-1451-top | CTAGCATGTAATTGACGACTTTAGCTCTACTGAAACC |
| PB2-1481-bottom | TCGAGGTTTCAGTAGAGCTAAAGTCGTCAATTACATG |
| PB1-956-top | CTAGCTGGCTATGACTGAAAGAC |
| PB1-972-bottom | TCGAGTCTTTCAGTCATAGCCAG |
| Backbone vector: pSilencer3.0-H1 | |
| siLuc/Luc2-TS | GATCCCACCCCAACATCTTCGACGTTCAAGAGACGTCGAAGATGTTGGGGTGTTTTTTGGAAA |
| siLuc/Luc2-BS | AGCTTTTCCAAAAAACACCCCAACATCTTCGACGTCTCTTGAACGTCGAAGATGTTGGGGTGG |
| miR-30e-3p-TS | GATCCGCTACTGTAAACATCCTTGACTGGAAGCTGTAAGGTGTTCAGAGGAGCTTTCAGTCGGATGTTTACAGCGGTTTTTTGGAAA |
| miR-30e-3p-BS | AGCTTTTCCAAAAAACCGCTGTAAACATCCGACTGAAAGCTCCTCTGAACACCTTACAGCTTCCAGTCAAGGATGTTTACAGTAGCG |
| miR-30e-3p_mut-TS | GATCCGCTACTGTAAACATCCTTGACTGGAAGCTGTAAGGTGTTCAGAGGAGCCCCACTCCGGATGTTTACAGCGGTTTTTTGGAAA |
| miR-30e-3p_mut-BS | AGCTTTTCCAAAAAACCGCTGTAAACATCCGGAGTGGGGCTCCTCTGAACACCTTACAGCTTCCAGTCAAGGATGTTTACAGTAGCG |
Luciferase assay
The A549 cells were divided into four groups. The first group was transfected with miRNA expression vector. The second group was transfected with pSilencer-siLuc/Luc2. The third group was transfected with pSilencer-scramble and the last group was mock. Then all four groups were cotransfected with the reporter target vector (pmiRGLO). For each group, the transfection was performed biological triplicated. After 48 h post transfection, cells were harvested and the luciferase activities were measured by Dual luciferase assay (Promega). For each sample, the luciferase assay was performed in triplicate. The relative luciferase activity was analyzed to determine the expression of reporter gene using the following formula: Relative Luciferase activity = (RLuc activity)/(Luc activity). The relative luciferase activity between each group was analyzed by Student’s t-test. The P ≤ 0.05 was considered as statistical significant.
Results
Effect of miR-30e-3p transfection
Our previous studies suggested that miR-30e-3p was upregulated in human cells infected with influenza A and influenza B viruses. However, the role played by this miRNA in influenza B virus infection was limited. Therefore, miR-30e-3p was selected as the candidate to study the effect of miRNA on influenza B virus production.
In this study, A549 cells were used as models for the in vitro study. These cells were transfected with the miR-30e-3p mimic, chemically modified double-stranded RNAs that mimic endogenous miRNAs. After 24 h post transfection, the amount of hsa-miR-30e-3p was investigated by using a stem-loop RT-qPCR system. The results revealed that in cells transfected with miR-30e-3p mimic, the amount of this miRNA was significantly increased when compared with cells transfected with negative control RNA and mock (transfection reagent only) (Figure 1(a)). The cell viability was examined in all transfected cell groups. The cell viability percentages of mock cells were not different from untreated cells, indicating that the amount of transfection reagent did not affect the cell viability. Further, the cell viability of miR-30e-3p mimic-transfected cells was also not different in comparison to mock and negative control-transfected cells (Figure 1(b)).
Figure 1.
The miR-30e-3p expression levels in A549 cells transfected with miR-30e-3p mimic, inhibitor, and negative control (a). The cell viability of transfected cells (b). Error bars represent standard deviation and the (*) represents statistical significance at P ≤ 0.05.
miR-30e-3p inhibits influenza B virus production
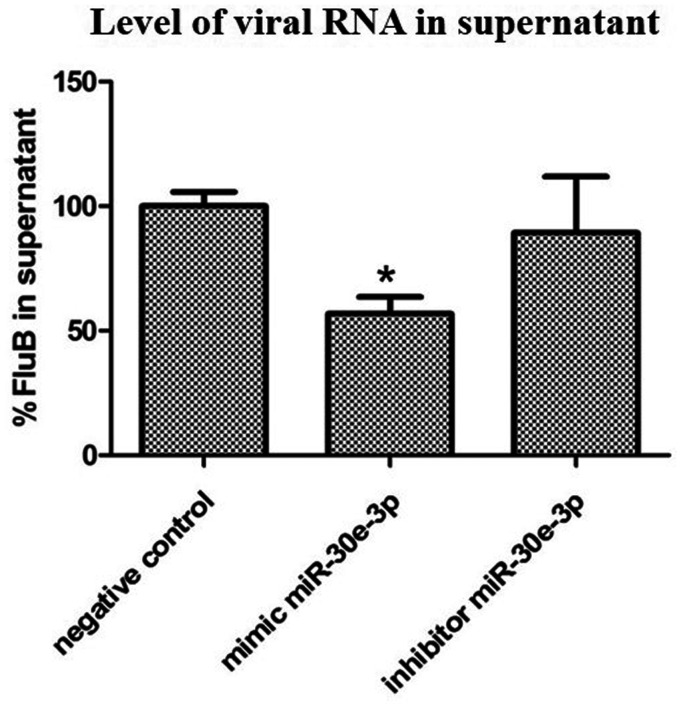
The effect of miR-30e-3p on influenza B virus production was studied by using the transfection of miR-30e-3p mimic, miR-30e-3p inhibitor, and negative control to the A549 cells. At 24 h post-transfection, cells were infected with influenza B virus at MOI = 0.5. The infection of influenza B virus in each group was biologically triplicated. After 24 h post-infection, supernatant was collected and extracted for total viral RNAs. Reverse transcription was performed by using random hexamer and the cDNA of influenza B virus was determined by using the TaqMan probe. The results, as shown in Figure 2, indicated that the upregulated miR-30e-3p significantly reduced influenza B virus production.
Figure 2.
Level of influenza B virus RNA in supernatant from A549 cells transfected with miR-30e-3p mimic, miR-30e-3p inhibitor, and negative control. Error bars represent standard deviation and the (*) represents statistical significance at P ≤ 0.05.
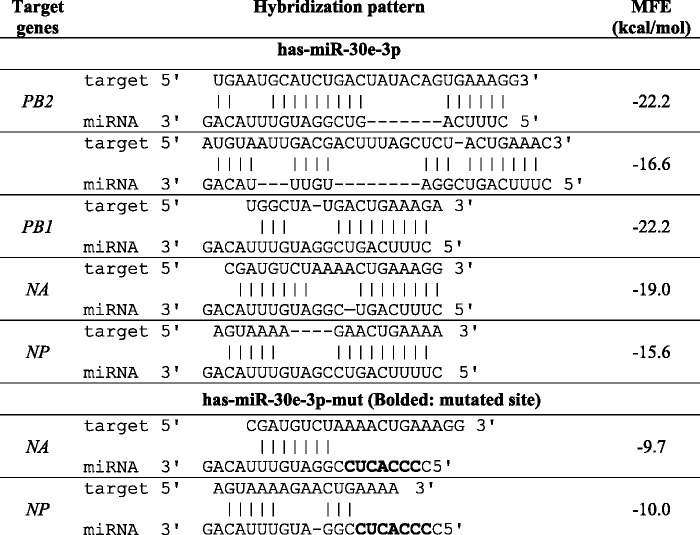
NA and NP are the viral targets of miR-30e-3p
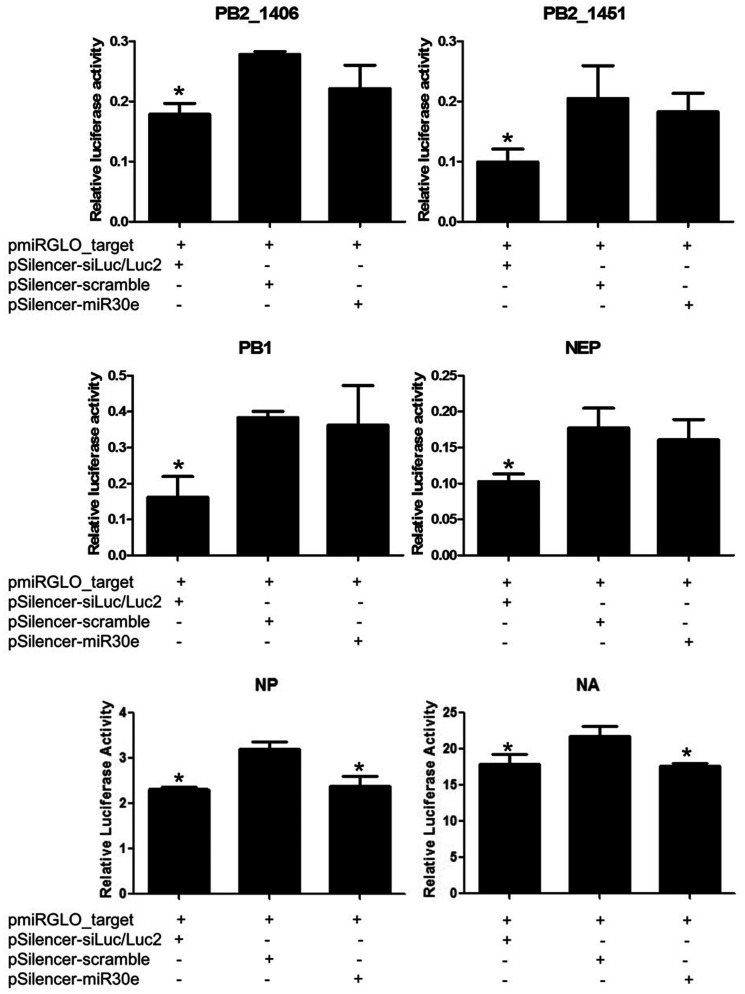
In silico prediction for viral targets of miR-30e-3p suggested that the miRNA may target five regions of the influenza B virus genome including two positions on PB2 and one position on PB1, NA, and NP. The predicted hybridization and minimum free energy (MFE) of each pair between hsa-miR-30e-3p and its targets are shown in Table 2. To validate the effectiveness of miR-30e-3p to silence the expression of these viral genes, 3ʹ-UTR reporter assay was performed. The sequences of predicted viral target sites were used to construct reporter vector by using pmiRGLO as backbone. For miRNA expression vector, the sequence of duplex miR-30e was used with the backbone, pSilencer3.0-H1. For silencing positive control, pSilencer_siLuc2 was constructed to silence the expression of Luc2 gene which is the reporter gene in pmiRGLO. All constructed vectors were confirmed by Sanger sequencing. Luciferase assay was performed after 48 h post-transfection and the relative luciferase activity was calculated. As shown in Figure 3, the relative luciferase activities of cells transfected with pSilencer-30e and the pmiRGLO_target of PB2 and PB1 were not statistically different when compared with the scramble group. In contrast, the relative luciferase activities of the miR-30e-3p overexpressing group were significantly different from the scramble group in NA and NP genes. Thus, it can be inferred that the miR-30e-3p can directly target NA and NP of influenza B virus and silence their expressions.
Table 2.
Predicted influenza B viral targets of miR-30e-3p and miR-30e-3p-mut.
|
Figure 3.
Luciferase assay determined the direct target of miR-30e-3p to predicted regions of influenza B including PB2, PB1, NA, and NP genes. Error bars represent standard deviation and the (*) represents statistical significance at P ≤ 0.05.
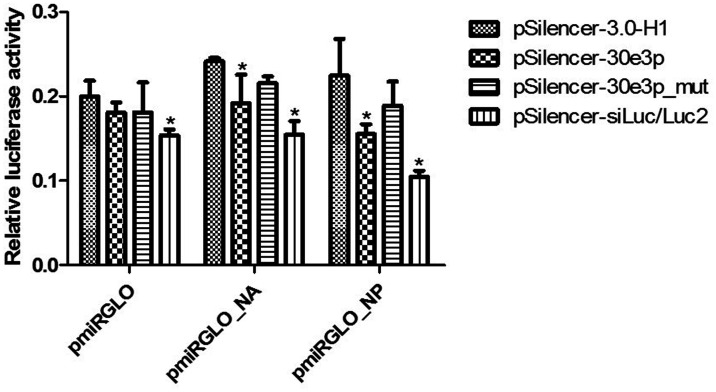
To confirm that hsa-miR-30e-3p can directly target NA and NP of the influenza B virus, the pmiRGLO_target of NA and NP was mutated in miR-30e-3p’s seed region binding sites as shown in the sequences and hybridization patterns in Table 2. The 3ʹ-UTR reporter assay was performed as described above. The results showed that the relative luciferase activities of mutated pmiRGLO_target of NA and NP were not significantly different from cells transfected with empty vector, pSilencer-3.0-H (Figure 4). This suggests that the hsa-miR-30e-3p can directly target NA and NP genes of influenza B virus and cause gene silencing.
Figure 4.
Luciferase assay determined the direct target of miR-30e-3p to NP and NA genes of influenza B virus. Error bars represent standard deviation and the (*) represents statistical significance at P ≤ 0.05.
Discussion
This study is the first investigation of human miRNA directly targeting influenza B virus infection. There are two co-circulating lineages of influenza B viruses; Victoria and Yamagata. These two lineages are different in hemagglutinin surface glycoprotein. In this study, we used influenza B virus B/Thailand/CU-B5522/2011 representing the Victoria lineage. The predicted viral target of these two lineages may be different. Even though the viral targets reported in this study may not be involved with the Yamagata lineage, these data might be useful when compared to the reported miRNA responses to influenza A viruses and future Yamagata lineage studies.
On the other hand, the hsa-miR-30e-3p which is the mature miRNA from the opposite strand of miR-30e-5p (miR-30e) is also reported as being upregulated in the cells infected with dengue virus.23 The hsa-miR-30e-3p was reported to target IĸBα, and upregulated IFN-β, leading to the upregulation of downstream IFN-stimulated genes (ISGs) including OAS1, MxA, and IFITM1 and inhibited dengue virus replication. Interestingly, the hsa-miR-30e-3p was found to be upregulated at 24 h post infection in three subtypes of influenza A viruses (pH1N1, H3N2, and H5N1)17 and influenza B virus infection.18 However, the function of how miR-30e-3p related to influenza virus replication was not clear Thus, this study is the first report suggesting the role played by hsa-miR-30e-3p in influenza B virus replication.
In addition, the hsa-miR-30 family has been suggested to be relevant in lung repair.24 This may indicate the role of miRNAs in the host response to viral infection. Five members of the hsa-miR-30 family, including hsa-miR-30a, hsa-miR-30b, and hsa-miR-30c, hsa-miR-30 d, and hsa-miR-30e, were reported to be downregulated during the infection of influenza A virus (H5N1). Overexpression of these miRNAs can suppress host SOCS1, SOCS3, and NEDD4 genes. This regulation positively regulates the type I IFN signaling pathway and inhibits influenza A virus infection. It was also suggested that the influenza A virus could disrupt the host antiviral immune response by downregulating miR-30 expression.
Previous studies suggested that miR-30e-3p is related to the maintenance of physiological conditions such as adipogenesis,25 heart development,26 and pathogenesis in many diseases, including neural tube defects,27 cancer,28,29 and trauma.29 Human miR-30e-3p is also related to autophagy inhibition. The miR-30e-3p found to be downregulated in Coronary microembolization (CME) at 6,9,12 h but not in CME at 1 and 3 h. The expression of miR-30e-3p was down regulated and accompanied by the inhibition of autophagy and decreased cardiac function. miR-30e-3p may thus be involved in CME-induced cardiac dysfunction by regulating myocardial autophagy.30
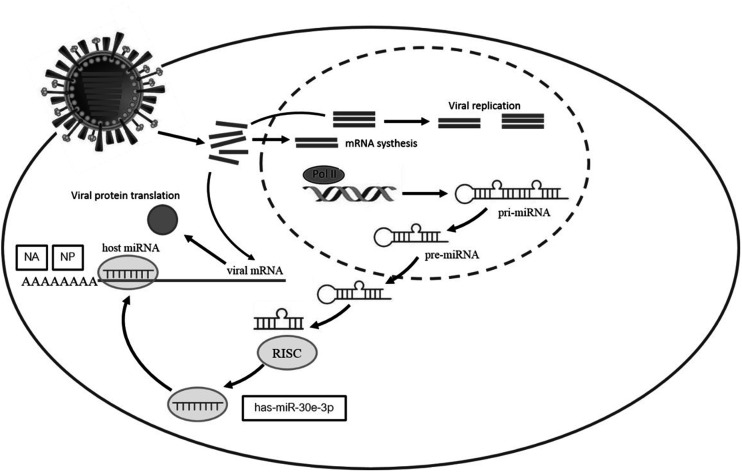
The miR-30e-3p was upregulated after Zika infection generating an inflammatory response. It was also reported to be involved in viral process by targeting B2M, YWHAE, RPL23, SUPT16H, and CBX5. Moreover, the miR-30e-3p also targets RPL23 and regulates viral life cycle and viral transcription. From our results, we found that when cells are infected with influenza B virus, some miRNAs responded to the infection by changing their expression levels. Among these miRNAs, the miR-30e-3p is one that up-regulated at the early phase of infection. In this study, this miRNA was predicted to, and we proved that it can, directly target the NA and NP genes of influenza B virus and lead to inhibition of its replication (Figure 5). This finding might be useful for a new alternative way of influenza therapeutics by using miRNA.
Figure 5.
Upregulation of hsa-miR-30e-3p directly inhibits influenza B viral NP and NA genes in influenza B virus infection.
Footnotes
Authors’ contributions: KK performed the experiment, data analysis, and drafted the manuscript. SS assisted in viral propagation. OM and PN assisted in vector construction and sequencing. TP discussed useful suggestions for data interpretations. YP provided stocks of the influenza virus. SP designed the study, revised the manuscript and coordinated the project.
Declaration OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was financially supported by the Chulalongkorn University Center of Excellence in Systems Biology; the Thailand Research Fund (TRF) (RSA6180035); Office of Higher Education Commission (NRU59-029-HR); National Science and Technology Development Agency (NSTDA) (P-17–51377); Chulalongkorn Academic Advancement into its 2nd Century Project and the Ratchadapisek Somphot Fund for Postdoctoral Fellowship, Chulalongkorn University.
ORCID iD: Sunchai Payungporn https://orcid.org/0000-0003-2668-110X
References
- 1.Ambros V. microRNAs: tiny regulators with great potential. Cell 2001; 107:823–6 [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature 2004; 431:350–5 [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–97 [DOI] [PubMed] [Google Scholar]
- 4.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 2007; 96(Suppl):R40–4 [PubMed] [Google Scholar]
- 5.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science 2005; 308:557–60 [DOI] [PubMed] [Google Scholar]
- 6.Jiang P, Zhou N, Chen X, Zhao X, Li D, Wang F, Bi L, Zhang D. Integrative analysis of differentially expressed microRNAs of pulmonary alveolar macrophages from piglets during H1N1 swine influenza a virus infection. Sci Rep 2015; 5:8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Li Z, Li Y, Liu Y, Liu J, Li X, Shen T, Duan Y, Hu M, Xu D. A computational method for predicting regulation of human microRNAs on the influenza virus genome. BMC Syst Biol 2013; 7:S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song L, Liu H, Gao S, Jiang W, Huang W. Cellular microRNAs inhibit replication of the H1N1 influenza a virus in infected cells. J Virol 2010; 84:8849–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khongnomnan K, Makkoch J, Poomipak W, Poovorawan Y, Payungporn S. Human miR-3145 inhibits influenza a viruses replication by targeting and silencing viral PB1 gene. Exp Biol Med 2015; 240:1630–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma YJ, Yang J, Fan XL, Zhao HB, Hu W, Li ZP, Yu GC, Ding XR, Wang JZ, Bo XC, Zheng XF, Zhou Z, Wang SQ. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza a virus in infected human lung epithelial cells. J Cell Mol Med 2012; 16:2539–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA. Influenza B virus in seals. Science 2000; 288:1051–3 [DOI] [PubMed] [Google Scholar]
- 12.Ran Z, Shen H, Lang Y, Kolb EA, Turan N, Zhu L, Ma J, Bawa B, Liu Q, Liu H, Quast M, Sexton G, Krammer F, Hause BM, Christopher-Hennings J, Nelson EA, Richt J, Li F, Ma W. Domestic pigs are susceptible to infection with influenza B viruses. J Virol 2015; 89:4818–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krystal M, Elliott RM, Benz EW, Jr., Young JF, Palese P. Evolution of influenza a and B viruses: conservation of structural features in the hemagglutinin genes. Proc Natl Acad Sci U S A 1982; 79:4800–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yen HL, Webster RG. Pandemic influenza as a current threat. Curr Top Microbiol Immunol 2009; 333:3–24 [DOI] [PubMed] [Google Scholar]
- 15.Krystal M, Young JF, Palese P, Wilson IA, Skehel JJ, Wiley DC. Sequential mutations in hemagglutinins of influenza B virus isolates: definition of antigenic domains. Proc Natl Acad Sci U S A 1983; 80:4527–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tambyah PA, Sepramaniam S, Mohamed Ali J, Chai SC, Swaminathan P, Armugam A, Jeyaseelan K. microRNAs in circulation are altered in response to influenza a virus infection in humans. PLoS One 2013; 8:e76811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makkoch J, Poomipak W, Saengchoowong S, Khongnomnan K, Praianantathavorn K, Jinato T, Poovorawan Y, Payungporn S. Human microRNAs profiling in response to influenza a viruses (subtypes pH1N1, H3N2, and H5N1). Exp Biol Med 2016; 241:409–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khongnomnan K, Poomipak W, Praianantathavorn K, Saengchoowong S, Pisitkun T, Poovorawan Y, Payungporn S. Human MicroRNAs expression profiles in influenza B Virus-Infected cells based on illumina MiSeq platform. MicroRNA 2018; 7:204–14 [DOI] [PubMed] [Google Scholar]
- 19.Mei Q, Li X, Meng Y, Wu Z, Guo M, Zhao Y, Fu X, Han W. A facile and specific assay for quantifying microRNA by an optimized RT-qPCR approach. PLoS One 2012; 7:e46890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suwannakarn K, Payungporn S, Chieochansin T, Samransamruajkit R, Amonsin A, Songserm T, Chaisingh A, Chamnanpood P, Chutinimitkul S, Theamboonlers A, Poovorawan Y. Typing (a/B) and subtyping (H1/H3/H5) of influenza a viruses by multiplex real-time RT-PCR assays. J Virol Methods 2008; 152:25–31 [DOI] [PubMed] [Google Scholar]
- 21.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol 2005; 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA 2004; 10:1507–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang L, Lin C, Song L, Wu J, Chen B, Ying Z, Fang L, Yan X, He M, Li J, Li M. MicroRNA-30e* promotes human glioma cell invasiveness in an orthotopic xenotransplantation model by disrupting the NF-kappaB/IkappaBalpha negative feedback loop. J Clin Invest 2012; 122:33–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan KS, Choi H, Jiang X, Yin L, Seet JE, Patzel V, Engelward BP, Chow VT. Micro-RNAs in regenerating lungs: an integrative systems biology analysis of murine influenza pneumonia. BMC Genom 2014; 15:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chartoumpekis DV, Zaravinos A, Ziros PG, Iskrenova RP, Psyrogiannis AI, Kyriazopoulou VE, Habeos IG. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS One 2012; 7:e34872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vacchi-Suzzi C, Hahne F, Scheubel P, Marcellin M, Dubost V, Westphal M, Boeglen C, Buchmann-Moller S, Cheung MS, Cordier A, De Benedetto C, Deurinck M, Frei M, Moulin P, Oakeley E, Grenet O, Grevot A, Stull R, Theil D, Moggs JG, Marrer E, Couttet P. Heart structure-specific transcriptomic atlas reveals conserved microRNA-mRNA interactions. PLoS One 2013; 8:e52442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saunders AM, Swayne LC, Schwartz JR, Moritz MW. Mediastinal mass following esophageal surgery. N J Med 1991; 88:571–2 [PubMed] [Google Scholar]
- 28.Lee H, Park CS, Deftereos G, Morihara J, Stern JE, Hawes SE, Swisher E, Kiviat NB, Feng Q. MicroRNA expression in ovarian carcinoma and its correlation with clinicopathological features. World J Surg Oncol 2012; 10:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugihara H, Ishimoto T, Watanabe M, Sawayama H, Iwatsuki M, Baba Y, Komohara Y, Takeya M, Baba H. Identification of miR-30e* regulation of Bmi1 expression mediated by tumor-associated macrophages in gastrointestinal cancer. PLoS One 2013; 8:e81839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang XT, Wu XD, Lu YX, Sun YH, Zhu HH, Liang JB, He WK, Zeng ZY, Li L. Potential involvement of MiR-30e-3p in myocardial injury induced by coronary microembolization via autophagy activation. Cell Physiol Biochem 2017; 44:1995–2004 [DOI] [PubMed] [Google Scholar]