Abstract
Background: One of the major migration routes for birds going between Europe and Asia is the Black Sea-Mediterranean route that converges on the Volga Delta, continuing into the area of the Caspian Sea. Cercarial dermatitis is a disorder in humans caused by schistosome trematodes that use aquatic birds and snails as hosts and is prevalent in areas of aquaculture in Northern Iran. Before the disorder can be addressed, it is necessary to determine the etiological agents and their host species. This study aimed to document whether domestic mallards are reservoir hosts and if so, to characterize the species of schistosomes. Previous work has shown that domestic mallards are reservoir hosts for a nasal schistosome. Results: In 32 of 45 domestic mallards (Anas platyrhynchos domesticus) (71.1%), the schistosome Trichobilharzia franki, previously reported only from Europe, was found in visceral veins. Morphological and molecular phylogenetic analysis confirmed the species designation. These findings extend the range of T. franki from Europe to Eurasia. Conclusion: The occurrence of cercarial dermatitis in Iran is high in areas of aquaculture. Previous studies in the area have shown that domestic mallards are reservoir hosts of T. regenti, a nasal schistosome and T. franki, as shown in this study. The genetic results support the conclusion that populations of T. franki from Iran are not differentiated from populations in Europe. Therefore, the schistosomes are distributed with their migratory duck hosts, maintaining the gene flow across populations with compatible snail hosts in Iran.
Keywords: Trichobilharzia franki, Iran, Anas platyrhynchos domesticus, Cercarial dermatitis
Abstract
Contexte : L’une des principales voies de migration des oiseaux à destination et en provenance de l’Europe et de l’Asie est la route mer Noire-Méditerranée qui converge vers le delta de la Volga et se poursuit dans la région de la mer Caspienne. La dermatite cercarienne est une affection causée chez l’homme par des trématodes Schistosomatidae utilisant des oiseaux aquatiques et des mollusques comme hôtes, qui est répandue dans les zones d’aquaculture du nord de l’Iran. Avant de pouvoir lutter contre cette parasitose, il est nécessaire de connaître les agents étiologiques et leurs espèces hôtes. Ce travail vise à documenter si les canards domestiques sont des hôtes réservoirs et si oui, à caractériser les espèces de schistosomes. Des travaux antérieurs ont montré que les canards domestiques sont des hôtes réservoirs pour un schistosome nasal. Résultats : Chez 32 de 45 canards domestiques (Anas platyrhynchos domesticus) (71,1 %), le schistosome Trichobilharzia franki, précédemment signalé uniquement en Europe, a été trouvé dans les veines viscérales. L’identification de l’espèce a été vérifiée par une analyse morphologique et phylogénétique moléculaire. Cela étend l’aire de répartition de l’Europe à l’Eurasie. Conclusion : La dermatite cercarienne est répandue dans les zones d’aquaculture en Iran. Des travaux antérieurs dans la région ont montré que les canards domestiques sont des hôtes réservoirs de T. regenti, un schistosome nasal et de T. franki, comme le montre cette étude. Les résultats génétiques soutiennent que les populations de T. franki d’Iran ne sont pas différenciées des populations d’Europe. Par conséquent, les schistosomes se dispersent avec leur hôte canard lors de la migration, maintenant un flux génétique entre les populations avec des mollusques hôtes compatibles en Iran.
Introduction
One of the major migration routes for birds going between Europe and Asia is the Black Sea-Mediterranean route that converges on the Volga Delta, continuing into the area of the Caspian Sea. Birds along this route migrate twice a year, nest, or stay for the winter. Therefore, the surrounding areas are visited year-round by migratory birds, particularly waterfowl. Aquaculture is a common occupation in many areas (covering five countries) around the Caspian Sea, but this type of farming is often associated with parasitic diseases [18, 47]. The water that is used for plants, ducks, fish, or crustaceans and other invertebrates is often inhabited by aquatic gastropods that can host a myriad of trematodes. Both domestic and wild mammals and birds use the water, as do humans, creating many opportunities for life cycles of several species of parasites to establish. One of these life cycles can result in a disorder called cercarial dermatitis (CD) or swimmer’s itch [41], caused by digenetic trematodes in the family Schistosomatidae. These worms have a two-host life cycle where adult worms live in a mammalian or avian host, and the intermediate host is an aquatic gastropod. The emerging free-swimming larval stages (cercariae) from the gastropod penetrate the skin of humans causing an allergic reaction that can last up to a week [43]. In an aquaculture environment, this involves the gastropods that naturally establish in water and domestic ducks, and sometimes migratory birds. Discovering the species of schistosome and their host diversity along a migratory route is a foundational step to initiating targeted control programs for CD. It is more manageable to control one duck species in the life cycle than all duck species, so more specific knowledge facilitates control or mitigation of the disease.
It is only within the last decade that there has been a concerted effort to study the epidemiology of CD in regions of the Middle East, as cases, particularly in rice fields, are gaining more attention [9, 26, 27, 30, 31, 35, 39, 51–53, 71]. Much of the work on CD in this area has been conducted in Iran, documenting the neglected status of the disease and narrowing down the critical hosts and worm species for transmission. A summary of research in Iran thus far shows that there are at least three common species of Trichobilharzia Skrjabin and Zakharov, 1920 that have been found in ducks and snails in Northern Iran [6, 26, 31, 51, 53, 71]. Recently, at least one species has been found in the mesenteric veins (9, 30) and a second species in the nasal tissue of its duck hosts [6, 26], particularly Spatula clypeata (Linnaeus, 1758) and Anas platyrhynchos (Linnaeus, 1758). Avian schistosomes have been recovered from the snail hosts Radix gedrosiana (Annandale & Prashad, 1919) and Radix auricularia (Linnaeus, 1758), and two species of Trichobilharzia from the mesenteric veins of their duck hosts [71]. The molecular identity of the avian schistosome from R. gedrosiana has not yet been confirmed [9, 27, 71].
Recent studies have uncovered a more detailed pattern of relationships among species of Trichobilharzia across a broader geographic range that encompasses the avian host migration routes [6, 25, 26, 37, 38, 61]. These patterns capture genetic diversity in the worms that reflect the long distances their bird hosts move (e.g. [25]). Additionally, finding these schistosomes in domestic or resident birds indicates that the snail host species is available, or at least a susceptible snail host (often a congener). The work herein provides more data demonstrating the significant impact of host mobility and ecology on the distribution and diversity of avian schistosomes [25]. This work aims to continue the survey and documentation of schistosomes in Iran, particularly in the rice-growing areas in the north. Additionally, the role of domestic mallards Anas platyrhynchos domesticus as reservoir hosts is further examined.
Materials and methods
Study area
This study was performed in Guilan Province of northern Iran, situated at the Western shores of the Caspian Sea (38° 28′ 58″ N, 50° 35′ 59″ E). This province consists of coastal plains, foothills and forested mountainous areas with a humid subtropical climate and the heaviest rainfall in the country. Mean rainfall in Guilan Province is about 1500 mm. The coastal plains along the Caspian Sea and near the foothills are mainly used for rice paddies, the same as that of Mazandaran Province, located to the east of Guilan Province, where cases of CD have also been reported. Guilan Province includes 2380 km2 of rice fields and produces about 40% of the rice products in the country. After rice harvesting from early August to late September, the rice paddies, especially those located near the farmer’s houses, become areas for livestock grazing and domestic duck breeding. The domestic mallard constitutes an important part of the diet of the indigenous population and is sold weekly at local markets. Late fall also coincides with the start of the rainy season in Guilan Province and the rice fields receive large amounts of water. These paddies then continue as suitable environments to sustain the snail intermediate hosts and maintain contact between the ducks and snails for transmission. There are also many water collections and small streams around Guilan villages, all of which have snails and sometimes ducks which widens the areas of transmission [6].
Parasite collection
Domestic ducks Anas platyrhynchos domesticus were purchased directly from villagers’ houses surrounded by rice fields where the ducks were feeding from December 2017 through October 2018 (Fig. 1), the same ducks that were collected in [6]. Locality data were determined by GPS (Table 1). The ducks were transferred to the parasitology laboratory at the Guilan University of Medical Sciences and decapitated to examine for presence of visceral schistosomes [4]. Warm saline (40–45 °C) was injected into the liver via the hepatic portal vein as well as into different parts of liver tissue (for frozen birds tap water was used). Then, the liver was cut into small pieces in saline and transferred to a series of different mesh size laboratory sieves arranged from the largest to the smallest. The liver was then slowly crushed by hand on the upper sieve, while being washed using a trigger sprayer containing warm saline. This was done for each sieve size. The bottom sieve (106 μm) was slightly tilted and the remnants of the liver washings were collected by a plastic pipette from the lower side of the sieve, and the same process was performed for 53 μm and 25 μm sieves. For microscopic examination, a small part of the collected materials was then transferred into a glass dish with clean saline solution to dilute the material and obtain a thin layer for examination under a dissection microscope for intact adults, fragments or eggs [4]. Some of the intact worms, fragments and eggs were transferred to the microtubes containing saline for rapid morphological studies (egg and adult measurements, their micrographs), and some transferred to the microtubes containing 90% alcohol for molecular studies. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted. This study was approved by the Ethics Committee of the Guilan University of Medical Sciences (IR.GUMS.REC.1398.109).
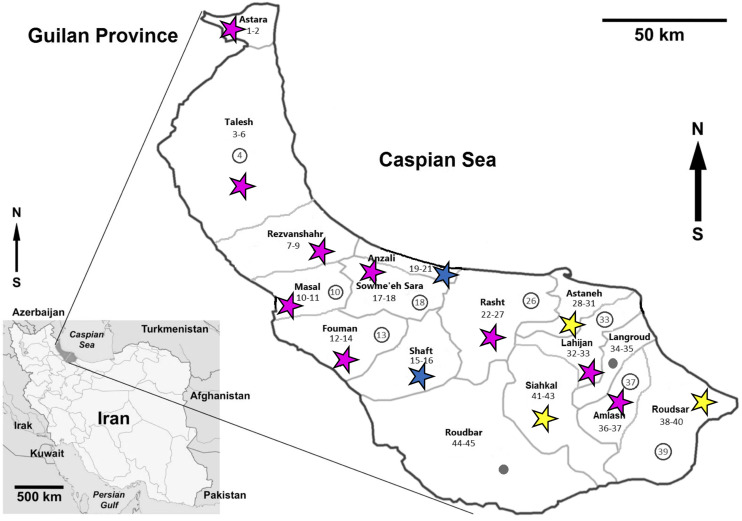
Figure 1.
Map of Iran with Guilan province highlighted, showing collecting localities. Pink stars = provinces with positive ducks for both T. franki and T. regenti, and in some cases co-infections; Yellow stars = provinces with positive ducks for only T. franki; Blue stars = provinces with positive ducks for only T. regenti; Circles without numbers = areas with negative ducks; and Circles with numbers = localities from which worms were used for sequencing.
Table 1.
Districts and coordinates of collecting localities. The duck host number matches the numbers on the collection localities in Figure 1. The results for T. regenti are from Ashrafi et al., 2018 since the birds examined for that study were the same as for this one.
| District | Coordinates (latitude/longitude) | Duck No. * | T. franki | T. regenti | No. examined/infected for T. franki | |
|---|---|---|---|---|---|---|
| Astara | 38°25′11.3″ N/48°51′55.5″ E | 1 | P | P | 2/1 | |
| 38°26′08.7″ N/48°51′46.1″ E | 2 | N | N | |||
| Talesh | 37°36′46.5″ N/49°03′30.1″ E | 3 | P | P | 4/4 | |
| 37°36′46.4″ N/49°02′23.6″ E | 4* | P | P | |||
| 37°38′07.3″ N/49°02′50.7″ E | 5 | P | P | |||
| 37°49′01.0″ N/48°55′10.1″ E | 6 | P | N | |||
| Rezvanshahr | 37°33′22.1″ N/49°09′09.8″ E | 7 | N | P | 3/2 | |
| 37°33′24.5″ N/49°08′19.0″ E | 8 | P | P | |||
| 37°32′36.7″ N/49°10′00.3″ E | 9 | P | N | |||
| Masal | 37°22′28.0″ N/49°09′13.5″ E | 10* | P | P | 2/2 | |
| 37°22′17.6″ N/49°08′31.2″ E | 11 | P | N | |||
| Fouman | 37°12′55.2″ N/49°18′22.8″ E | 12 | P | P | 3/3 | |
| 37°13′01.7″ N/49°19′10.4″ E | 13* | P | P | |||
| 37°12′50.0″ N/49°18′46.1″ E | 14 | P | N | |||
| Shaft | 37°08′37.6″ N/49°23′27.9″ E | 15 | N | P | 2/0 | |
| 37°08′29.9″ N/49°22′53.1″ E | 16 | N | N | |||
| Sowme’eh Sara | 37°17′23.4″ N/49°22′51.7″ E | 17 | P | N | 2/2 | |
| 37°16′35.3″ N/49°22′20.0″ E | 18* | P | P | |||
| Anzali | 37°25′22.8″ N/49°26′11.8″ E | 19 | N | P | 3/0 | |
| 37°27′10.1″ N/49°35′49.9″ E | 20 | N | P | |||
| 37°27′32.3″ N/49°30′38.7″ E | 21 | N | N | |||
| Rasht | 37°16′46.3″ N/49°45′45.4″ E | 22 | P | N | 6/5 | |
| 37°16′16.2″ N/49°45′30.9″ E | 23 | P | P | |||
| 37°16′30.2″ N/49°45′09.0″ E | 24 | N | N | |||
| 37°10′50.5″ N/49°41′10.4″ E | 25 | P | P | |||
| 37°11′16.7″ N/49°31′44.5″ E | 26* | P | N | |||
| 37°13′04.3″ N/49°30′34.8″ E | 27 | P | N | |||
| Astaneh | 37°15′46.2″ N/49°53′30.3″ E | 28 | P | N | 4/4 | |
| 37°16′22.6″ N/49°54′30.1″ E | 29 | P | N | |||
| 37°16′22.7″ N/49°54′40.3″ E | 30 | P | N | |||
| 37°16′40.1″ N/49°55′38.3″ E | 31 | P | N | |||
| Lahijan | 37°13′06.4″ N/49°59′03.4″ E | 32 | P | N | 2/2 | |
| 37°16′03.0″ N/50°07′17.6″ E | 33* | P | P | |||
| Langroud | 37°11′11.9″ N/50°12′44.5″ E | 34 | N | N | 2/0 | |
| 37°12′12.7″ N/50°10′30.0″ E | 35 | N | N | |||
| Amlash | 37°05′40.1″ N/50°11′53.9″ E | 36 | P | N | 2/2 | |
| 37°03′37.8″ N/50°16′10.9″ E | 37* | P | P | |||
| Roudsar | 37°08′34.8″ N/50°16′47.2″ E | 38 | P | N | 3/3 | |
| 37°08′19.8″ N/50°16′37.8″ E | 39* | P | N | |||
| 37°11′29.2″ N/50°10′04.1″ E | 40 | P | N | |||
| Siahkal | 37°09′35.1″ N/49°52′58.3″ E | 41 | P | N | 3/2 | |
| 37°09′35.0″ N/49°52′57.8″ E | 42 | P | N | |||
| 37°09′51.6″ N/49°52′17.5″ E | 43 | N | N | |||
| Roudbar | 37°01′06.5″ N/49°36′46.3″ E | 44 | N | N | 2/0 | |
| 37°00′43.2″ N/49°35′55.0″ E | 45 | N | N | |||
| 45/32 | ||||||
*Numbers with an asterisk show the samples used for molecular studies; P = positive; N = negative.
Morphological and genetic analyses
For morphological studies of adults and eggs, the intact worms, large fragments and eggs were transferred onto a glass slide, covered with a coverslip and measured (Tables 2, 3) under a microscope (Olympus BX50) equipped with a digital camera (TrueChrome Metrics, China) and Nomarski Piece (U-DICT, Olympus, Japan). The length and width of the eggs were measured, and the data analyzed in SPSS v. 22 (minimum, maximum, average, and SD). The remaining eggs, intact adults and worm fragments, if any, were transferred to microtubes containing 90% alcohol for molecular studies. Some of the collected samples (full length worms, fragments and eggs) were also kept in labeled microtubes in 90% alcohol in the Department of Parasitology and Mycology of the Guilan University of Medical Sciences as a permanent museum voucher. It is critical for the evolutionary characterization of organisms to have a permanent museum voucher [33, 59, 68].
Table 2.
Measurements of fresh mounts of male worms from duck hosts Anas platyrhynchos domesticus represented as mean ± SD (min–max μm); ND = no data.
| This study |
Müller & Kimming, 1994 |
|||
|---|---|---|---|---|
| n | Mean ± SD (min – max) | n | min – max (μm) | |
| Width at esophagus (middle) | 27 | 92.4 ± 11.9 (70 – 119) | 5 | 120 – 130 |
| Width at acetabulum level | 7 | 113.4 ± 9.6 (91 – 137) | – | ND |
| Width after gynecophoric canal | 4 | 73.4 ± 9.4 (63 – 83) | – | ND |
| Width at spatulate end | 17 | 102.5 ± 16.3 (82 – 124) | – | ND |
| Oral sucker length | 29 | 63.7 ± 5.9 (50 – 76) | 5 | 51 – 77 |
| Oral sucker width | 29 | 53.1 ± 6.3 (45 – 71) | 5 | 46 – 65 |
| Acetabulum length | 18 | 58 ± 9.9 (42 – 75) | 5 | 46 – 51 |
| Acetabulum width | 18 | 69 ± 5.1 (58 – 77) | 5 | 56 – 69 |
| Acetabulum to anterior end | 16 | 499.6 ± 46.2 (407 – 562) | 5 | 458 – 530 |
| Acetabulum to gut bifurcation | 15 | 89.3 ± 14.3 (66 – 112) | 5 | 74 – 104 |
| Acetabulum to cecal reunion | 3 | 512.5 ± 63.5 (385 – 547) | – | ND |
| Acetabulum to VSE | 5 | 64.4 ± 14.7 (48 – 86) | – | ND |
| Acetabulum to gynecophoric canal | 6 | 605.7 ± 67.3 (528 – 646) | 5 | 495 – 550 |
| Vesicula seminalis externa (VSE) | 17 | 207.4 ± 37 (136 – 255) | – | ND |
| Vesicula seminalis interna (VSI) | 12 | 194.6 ± 15.5 (157 – 215) | – | ND |
| Seminal vesicle length | 4 | 381.3 ± 59.1 (293 – 419) | 5 | 265 – 315 |
| Gynecophoric canal length | 30 | 360.5 ± 54.8 (258 – 461) | 5 | 212 – 291 |
| Gynecophoric canal width | 40 | 139.5 ± 21.8 (107 – 197) | 5 | 130 – 195 |
| Gynecophoric canal to first testis | 10 | 129.1 ± 16.8 (92 – 148) | – | ND |
| Gynecophoric canal to anterior end | 14 | 1072.6 ± 119 (895 – 1290) | – | ND |
| Testis length | 56 | 69.6 ± 10.8 (49 – 96) | 5 | 95 – 106 |
| Testis width | 56 | 56.9 ± 14.8 (34 – 87) | – | ND |
| Number of testis | 11 | 47.1 ± 9.9 (35 – 65) | – | ND |
| Ceca length | 3 | 509.4 ± 53.7 (458 – 544) | 5 | 680 – 705 |
| Cecal reunion to anterior end | 8 | 1101 ± 80 (1019 – 1179) | 5 | 390 – 430 |
| Cecal bifurcation to anterior end | 8 | 451 ± 30.1 (410 – 479) | 5 | 390 – 430 |
| Body length | 3 | 3915.3 ± 75 (3830 – 3971) | 5 | 3.2 – 4.0 (mm) |
Table 3.
Measurements of fresh mounts of female worms from duck hosts Anas platyrhynchos domesticus represented as mean ± SD (min–max μm); ND = no data.
| n | This study |
Müller & Kimmig, 1994 |
||
|---|---|---|---|---|
| Mean ± SD (min – max) | min – max (μm) | |||
| Width at esophagus (middle) | 19 | 70.4 ± 15.9 (50 – 95) | 112 – 129 μm | |
| Width at acetabulum level | 6 | 88.7 ± 10.5 (68 – 98) | ND | |
| Width at spatulate end | 8 | 90.6 ± 15.1 (75 – 113) | ND | |
| Width before spatulate end | 3 | 56 ± 4.6 (52 – 61) | ND | |
| Oral sucker length | 16 | 56.8 ± 8.1 (40 – 68) | 57 – 64 | |
| Oral sucker width | 16 | 42.6 ± 9 (31 – 59) | 46 – 55 | |
| Acetabulum length | 12 | 42.5 ± 7 (38 – 63) | 38 – 47 | |
| Acetabulum width | 12 | 51.5 ± 7.1 (42 – 68) | 49 – 58 | |
| Acetabulum to anterior end | 10 | 390.5 ± 72.8 (312 – 494) | 455 – 545 | |
| Acetabulum to gut bifurcation | 5 | 84.4 ± 12 (75 – 98) | 62 – 70 | |
| Acetabulum to cecal reunion | 2 | 549.5 ± 98.3 (480 – 619) | ND | |
| Acetabulum to ovary | ND | 285 – 310 | ||
| Ceca length | 3 | 641 ± 114.7 (537 – 764) | 745 – 795 | |
| Cecal bifurcation to anterior end | 8 | 414.5 ± 27.6 (395 – 434) | 390 – 430 | |
| Cecal reunion to anterior end | 3 | 1119 ± 110.5 (995 – 1207) | ND | |
| Egg length in uterus | 2 | 157.5 ± 2.1 (156 – 159) | ND | |
| Egg width in uterus | 2 | 41 ± 1.4 (40 – 42) | ND | |
| Body length | 3 | 3559.7 ± 724.3 (3092 – 4394) | 4.2 – 4.6 (mm) | |
| Eggs in feces | n | Length (μm) | Width (μm) | Length/width ratio |
| Müller & Kimming (1994) | 18 | 205.8 ± 24.7 (155 – 250) | 68 ± 8.9 (52.5 – 90) | 3 |
| Skirnisson & Kolarova (2008) | 40 | 203 ± 27 (150–260) | 69 ± 6 (57–84) | 2.9 |
| This study | 99 | 193.9 ± 20.9 (154 – 250) | 62.4 ± 9.4 (45 – 85) | 3.1 |
For the genetic studies, genomic DNA was extracted from 90% ethanol-preserved worm fragments using a commercial kit (High Pure PCR Template Preparation Kit; Roche, Mannheim, Germany), according to the manufacturer’s recommended protocol. Primers BD1 (5′–GTCGTAACAAGGTTTCCGTA–3′) [12] and 4S (5′–TCTAGATGCGTTCGAARTGTCGATG–3′) [13] were used for amplification of a 1123 bp sequence of partial ITS1 nuclear rDNA. Also, Cox1_SchistoF (5′–TCTTTRGATCATAAGCG–3′) and Cox1_SchistoR (5′–TAATGCATMGGAAAAAAACA–3′) primers were employed to amplify a 1250 bp sequence of the partial mitochondrial cox1 gene [48]. PCR reaction was performed in a 30 μL reaction mixture containing 15 μL of PCR mix including 1.25 U Taq DNA polymerase, 200 μM of dNTPs and 1.5 mM MgCl2 (2 × Master Mix RED Ampliqon, Denmark), 10 pmol of each primer, and 3 μL of DNA sample. The thermal PCR profiles for the cox1 gene included an initial denaturation step at 94 °C for 2 min followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s and extension at 72 °C for 120 s, followed by a final extension step at 72 °C for 7 min. The PCR conditions of ITS1 gene amplification consisted of initial denaturation at 95 °C for 6 min, 30 cycles of 95 °C for 45 s, 55 °C for 60 s, and 70 °C for 1 min, followed by a final extension at 72 °C for 6 min. These PCR products were submitted to Bioneer Company (Korea) and sequenced in both directions using the same PCR primers.
Reconstruction of evolutionary relationships
The phylogenetic relationship of the schistosomes found in this study were reconstructed using a mitochondrial gene region of partial cytochrome oxidase 1 cox1 (695 bp) and a nuclear gene region of the internal transcribed spacer regions ITS1-5.8S-ITS2 (945 bp). Sequences were aligned by eye in Se-Al v 2.0a11 (tree.bio.ed.ac.uk). Phylogenetic analyses of the cox1 and ITS datasets were performed using Bayesian Inference in MrBayes [34] with default priors for the ITS genes (Nst = 6, rates = gamma, ngammacat = 4) and cox1 (parameters unlinked, each partition by codon had its own set of parameters; Nst = 6, rates = invgamma). The partitions by codon evolved under different rates (preset applyto = (all) ratepr = variable). Model selection was estimated using ModelTest [60]. Four chains were run simultaneously for 4 × 105 generations, the first 4000 trees discarded as burn-in. The remaining trees were used to calculate a 50% majority-rule consensus tree with posterior probabilities. Outgroups used were defined by relationships from Brant and Loker [14] and Ebbs et al. [25]. The new sequences generated by this study were deposited in GenBank (accession numbers: MF945587–MF953396; MH410291–MH410297). See Table 4 for the list of specimens, references and GenBank accession numbers.
Table 4.
Specimens used in this study.
| Avian schistosome species | Snail host | Avian host | Country of origin | Identifier | GenBank ITS | GenBank cox1 | Museum number* | Reference |
|---|---|---|---|---|---|---|---|---|
| Trichobilharzia franki | Radix auricularia | France | FORS4 | HM131184 | HM131197 | Jouet et al. [37] | ||
| Trichobilharzia franki | Radix auricularia | France | FORS3 | HM131198 | Jouet et al. [37] | |||
| Trichobilharzia franki | Radix auricularia | France | STRS2 | HM131176 | HM131202 | Jouet et al. [37] | ||
| Trichobilharzia franki | Radix auricularia | France | BERS1 | HM131199 | Jouet et al. [37] | |||
| Trichobilharzia franki | Radix auricularia | France | BERS2 | HM131182 | Jouet et al. [37] | |||
| Trichobilharzia franki | Radix auricularia | France | BERS67 | HM131200 | Jouet et al. [37] | |||
| Trichobilharzia franki | Radix auricularia | France | STRS4 | HM131178 | Jouet et al. [37] | |||
| Trichobilharzia franki | Radix auricularia | France | STRS6 | HM131180 | Jouet et al. [37] | |||
| Trichobilharzia franki | Radix auricularia | France | EAN77 | HM131183 | HM131201 | Jouet et al. [37] | ||
| Trichobilharzia franki | Radix auricularia | France | RSFO1 | AY795572 | Ferté et al., [28] | |||
| Trichobilharzia franki | Radix auricularia | Czech Republic | AF356845 | Dvorak et al. [23] | ||||
| Trichobilharzia franki | Radix auricularia | Czech Republic | FJ174530 | Brant and Loker [14] | ||||
| Trichobilharzia franki | Radix auricularia | Great Britain | HamRa6 | KJ775868 | NHMUK 2014.4.25.1 | Lawton et al. [45] | ||
| Trichobilharzia franki | Radix auricularia | Great Britain | HamRa7 | KJ775869 | NHMUK 2014.4.25.2 | Lawton et al. [45] | ||
| Trichobilharzia franki | Radix auricularia | Denmark | DK1 | KJ775869 | ZMUC-TRE-10-12 | Christiansen et al. [19] | ||
| Trichobilharzia franki | Radix auricularia | Switzerland | auri1 1100 | AJ312041 | Picard and Jousson [56] | |||
| Trichobilharzia franki | Radix auricularia | Switzerland | auri2 1100 | AJ312042 | Picard and Jousson [56] | |||
| Trichobilharzia franki | Radix auricularia | Italy | MK053632 | De Liberato et al. [20] | ||||
| Trichobilharzia franki | Radix auricularia | Italy | HM596077 | Cipriani et al. [17] | ||||
| Trichobilharzia franki | Anas p. domesticus | Iran | VT3 | MF945588 | MF945593 | Guilan University | This study | |
| Trichobilharzia franki | Anas p. domesticus | Iran | VT5 | MF945589 | MF945594 | Guilan University | This study | |
| Trichobilharzia franki | Anas p. domesticus | Iran | VT2 | MF945587 | MF945592 | Guilan University | This study | |
| Trichobilharzia franki | Anas p. domesticus | Iran | VT18 | MF945591 | MF945596 | Guilan University | This study | |
| Trichobilharzia franki | Anas p. domesticus | Iran | VT16 | MF945590 | MF945595 | Guilan University | This study | |
| Trichobilharzia franki | Anas p. domesticus | Iran | VR | MH410293 | MH410297 | Guilan University | This study | |
| Trichobilharzia franki | Anas p. domesticus | Iran | VL | MH410292 | MH410296 | Guilan University | This study | |
| Trichobilharzia franki | Anas p. domesticus | Iran | VA | MH410291 | MH410295 | Guilan University | This study | |
| Trichobilharzia sp. | Radix auricularia | Czech Republic | Ra1 | AY713969 | Rudolfova et al. [63] | |||
| Trichobilharzia sp. | Radix auricularia | Poland | Ra2 | AY713964 | Rudolfova et al. [63] | |||
| Trichobilharzia sp. | Radix auricularia | Finland | F3 | FJ609411 | Aldhoun et al. [3] | |||
| Trichobilharzia sp. | Ampullaceana balthica | Iceland | V2 | FJ469812 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. | Ampullaceana balthica | Iceland | FPC | FJ469820 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. | Ampullaceana balthica | Iceland | FPB | FJ469819 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. | Ampullaceana balthica | Iceland | 8 | FJ469816 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. | Ampullaceana balthica | Switzerland | ov2 1100 | AJ312044 | Picard and Jousson [56] | |||
| Trichobilharzia sp. | Ampullaceana balthica | Switzerland | ov1 1100 | AJ312043 | Picard and Jousson [56] | |||
| Trichobilharzia sp. | Physa marmorata | Brazil | HAP2013 | KJ855997 | KJ855996 | MSB:Para:19006 | Pinto et al. [57] | |
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | F2IS | HM131186 | Jouet et al. [37] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | FSIS | HM131190 | Jouet et al. [37] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | F5ISB | HM131189 | Jouet et al. [37] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | ICR1 | HM131191 | Jouet et al. [37] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | ls19 | FJ469808 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | ls25 | FJ469809 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | 11 | FJ469814 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | 14 | FJ469811 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | H | FJ469810 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | M2 | FJ46982 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Iceland | ls23 | FJ469805 | Aldhoun et al. [2] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | France | DOUC1 | HM131205 | Jouet et al. [36, 37] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | France | EAN57 | HM131194 | HM131204 | Jouet et al. [37] | ||
| Trichobilharzia sp. Rb | Ampullaceana balthica | France | EAN79 | HM131196 | Jouet et al. [37] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | France | EAN30 | HM131192 | Jouet et al. [36, 37] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | France | EAN31 | HM131203 | Jouet et al. [36, 37] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Norway | TFPTAK4 | KY513273 | HCIP D-735–D-750 | Soldanova et al. [67] | ||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Norway | TFPTAK1 | KY513270 | HCIP D-735–D-750 | Soldanova et al. [67] | ||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Norway | TFPTAK3 | KY513272 | HCIP D-735–D-750 | Soldanova et al. [67] | ||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Norway | TFPTAK2 | KY513271 | HCIP D-735–D-750 | Soldanova et al. [67] | ||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Switzerland | ov4 1100 | AJ312046 | Picard and Jousson [56] | |||
| Trichobilharzia sp. Rb | Ampullaceana balthica | Switzerland | ov3 1100 | AJ312045 | Picard and Jousson [56] | |||
| Trichobilharzia sp. Rb | Lymnaea stagnalis | Czech Republic | LS1 | AY713973 | Rudolfova et al. [63] | |||
| Trichobilharzia sp. Rb | Radix auricularia | Poland | Ra3 | AY713966 | Rudolfova et al. [63] | |||
| Trichobilharzia sp. A | Mareca americana | USA | W213 | FJ174570 | FJ174526 | MSB:Para:18646 | Brant and Loker [14] | |
| Trichobilharzia sp. A | Mareca americana | USA | W192 | FJ174572 | FJ174471 | MSB:Para:18609 | Brant and Loker [14] | |
| Trichobilharzia sp. A | Mareca americana | USA | W182 | FJ174573 | FJ174525 | MSB:Para:18574 | Brant and Loker [14] | |
| Trichobilharzia sp. A | Mareca americana | USA | W149 | FJ174524 | MSB:Para:18585 | Brant and Loker [14] | ||
| Trichobilharzia sp. B | Mareca americana | USA | W210 | KP788772 | MSB:Para:18643 | Ebbs et al. [25] | ||
| Trichobilharzia sp. B | Mareca americana | USA | W205 | KP788770 | FJ174528 | MSB:Para:18638 | Ebbs et al. [25] | |
| Trichobilharzia sp. C | Lophodytes cucullatus | USA | W173 | FJ174529 | MSB:Para:18562 | Brant and Loker [14] | ||
| Trichobilharzia sp. C | Aix sponsa | USA | W174 | KJ855996 | MSB:Para:18563 | Ebbs et al. [25] | ||
| Trichobilharzia physellae | Physa parkeri | USA | W234 | FJ174520 | MSB:Para:18656 | Brant and Loker [14] | ||
| Trichobilharzia physellae | Aythya affinis | USA | W171 | FJ174564 | MSB:Para:18565 | Brant and Loker [14] | ||
| Trichobilharzia physellae | Aythya affinis | USA | W212 | FJ174563 | MSB:Para:18645 | Brant and Loker [14] | ||
| Trichobilharzia physellae | Aythya affinis | USA | W193 | FJ174518 | MSB:Para:18610 | Brant and Loker [14] | ||
| Trichobilharzia physellae | Aythya collaris | USA | W194 | FJ174566 | FJ174517 | MSB:Para:18611 | Brant and Loker [14] | |
| Trichobilharzia physellae | Bucephala albeola | USA | W255 | FJ174561 | FJ174514 | MSB:Para:19159 | Brant and Loker [14] | |
| Trichobilharzia physellae | Clangula hyemalis | USA | W211 | FJ174516 | MSB:Para:18644 | Brant and Loker [14] | ||
| Trichobilharzia querquedulae | Physa gyrina | USA | W413 | HM125959 | MSB:Para:186 | Brant et al. [15] | ||
| Trichobilharzia querquedulae | Spatula discors | USA | W156 | FJ174554 | FJ174502 | MSB:Para:18590 | Brant and Loker [14] | |
| Trichobilharzia querquedulae | Spatula discors | USA | E45 | FJ174555 | FJ174510 | MSB:Para:24778 | Brant and Loker [14] | |
| Trichobilharzia querquedulae | Spatula cyanoptera | USA | W180 | FJ174505 | MSB:Para:18573 | Brant and Loker [14] | ||
| Trichobilharzia querquedulae | Spatula clypeata | USA | W135 | FJ174557 | FJ174497 | MSB:Para:183 | Brant and Loker [14] | |
| Trichobilharzia querquedulae | Spatula clypeata | USA | W203 | FJ174552 | FJ174508 | MSB:Para:18636 | Brant and Loker [14] | |
| Trichobilharzia querquedulae | Spatula clypeata | Canada | W345 | FJ174509 | MSB:Para:18626 | Brant and Loker [14] | ||
| Trichobilharzia querquedulae | Spatula rhynchotis | New Zealand | TshovNZ | KP788760 | KU057183 | MSB:Para:20794 | Ebbs et al. [25] | |
| Trichobilharzia querquedulae | Spatula rhynchotis | New Zealand | W703 | KU057181 | MSB:Para:20792 | Ebbs et al. [25] | ||
| Trichobilharzia querquedulae | Spatula smithii | South Africa | W650 | KP788765 | MSB:Para:18990 | Ebbs et al. [25] | ||
| Trichobilharzia querquedulae | Spatula smithii | South Africa | W664 | KU057180 | MSB:Para:19000 | Ebbs et al. [25] | ||
| Trichobilharzia querquedulae | Spatula platalea | Argentina | W833 | KU057184 | MSB:Para:23180 | Ebbs et al. [25] | ||
| Trichobilharzia regenti | Ampullaceana balthica | France | EAN9 | HM439499 | Jouet et al. [36, 38] | |||
| Trichobilharzia regenti | Mergus merganser | France | HAR1 | HM439501 | Jouet et al. [38] | |||
| Trichobilharzia regenti | Cygnus olor | France | CYA18 | HM439500 | Jouet et al. [38] | |||
| Trichobilharzia regenti | Anas platyrhynchos | France | BERS58 | HM439502 | Jouet et al. [38] | |||
| Trichobilharzia regenti | Anas platyrhynchos | Iceland | AC122 | HM439503 | Jouet et al. [38] | |||
| Trichobilharzia regenti | Anas platyrhynchos | Iceland | AC125 | HM439504 | Jouet et al. [38] | |||
| Trichobilharzia cf. regenti | Spatula clypeata | Iran | T39 | KR108325 | Fakhar et al. [26] | |||
| Trichobilharzia cf. regenti | Spatula clypeata | France | JIT11 | HM439505 | Jouet et al. [38] |
Results
Morphological identification and molecular characterization
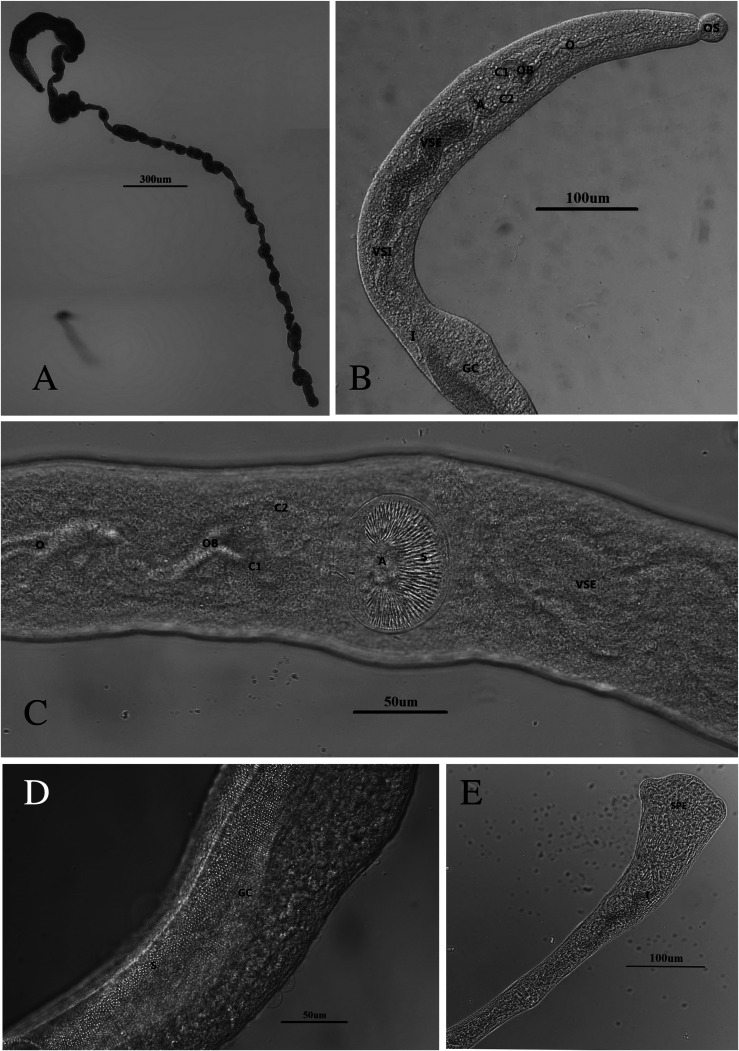
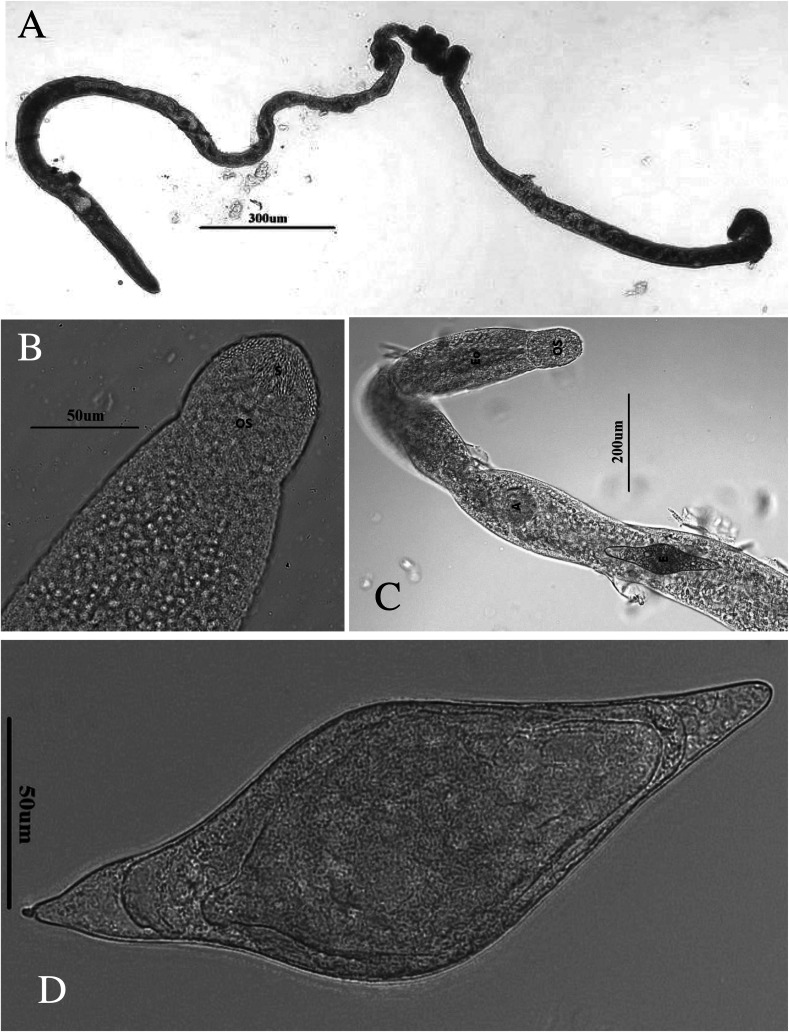
From our collection of 45 ducks from 45 localities in 16 districts, Trichobilharzia franki Müller and Kimmig, 1994 was found at 32 sites in 12 districts; worms were found in the liver of 32/45 ducks, with 71.1% prevalence (Table 1). Because this study continues the efforts of Ashrafi et al. [6], some of these ducks were infected with both T. franki and the neuropathic nasal species, T. regenti. There were 32/45 infected with T. franki, 17/45 infected with T. regenti, and 13/45 co-infected with both species (Table 1; [6]). These worms aligned morphologically with those of the original description by Müller and Kimmig (1994) of T. franki derived by infections of domestic dwarf mallards with cercariae from wild collected Radix auricularia [54]. However, some of the measurements herein were smaller. One explanation might be because Müller and Kimmig [54] put the host tissue in a trichinelloscope, which flattens the tissue to expose the live worms; their measurements might be larger with this type of preparation method. The authors even state that the measurements should not be regarded as absolute values [54]. In addition to the measurements (Tables 2 and 3) there were other features in common with the original description. Müller and Kimmig (1994) [54] found worms mostly in the veins of the liver, but in some cases, they found worms in the gut mucosa. If worms were found in the mesenteric blood vessels, they were localized near the outer wall of the intestine and were irregular in density from the duodenum to the cloaca. In the female and male worms, the oral sucker and acetabulum are spined (Figs. 2B, 2C and 3B) and in the males the gynecophoric canal is spined (Fig. 2D), but no body spines were observed. Other similarities: cecal reunion was observed between the posterior seminal vesicle and anterior gynecophoric canal and the tail is wide, spatulate and tri-lobed (Fig. 2E). Eggs in both studies are spindle-shaped with a straight longitudinal axis with one end rounded and the other end slightly less rounded, but ending in a small spine (Fig. 3D). The uterus contained only one egg at a time (Fig. 3C), and the rounded end was pointed consistently anteriad. Males and females were similarly sized in length, as was also found in Müller and Kimmig [54]. We have morphological adult comparisons only for the original description. Any specimens included in the gene trees were from fragments of adults, eggs, or cercariae, and thus no morphological descriptions are available. Jouet et al. [37] included a description of the T. franki cercariae from R. auricularia and those from Ampullaceana balthica (Linnaeus, 1758); the latter is larger than T. franki.
Figure 2.
Images of an adult male worm in saline. a) full length male; b) anterior end OS = oral sucker, O = esophagus, OB = esophagus bifurcation, C1/C2 = cecum1 and cecum 2, A = acetabulum, VSE = external seminal vesicle, VSI = internal seminal vesicle, I = intestine, and GC = gynecophoric canal; c) spines on the acetabulum, codes the same as defined in (b); d) fine spines in the gynecophoric canal; e) posterior end of worm showing spatulate tail.
Figure 3.
Images of an adult female worm in saline. a) full length female; b) oral sucker showing fine spines; c) anterior end showing single egg in the uterus, OS = oral sucker, OE = esophagus, A = acetabulum, E = egg; d) egg from liver washings.
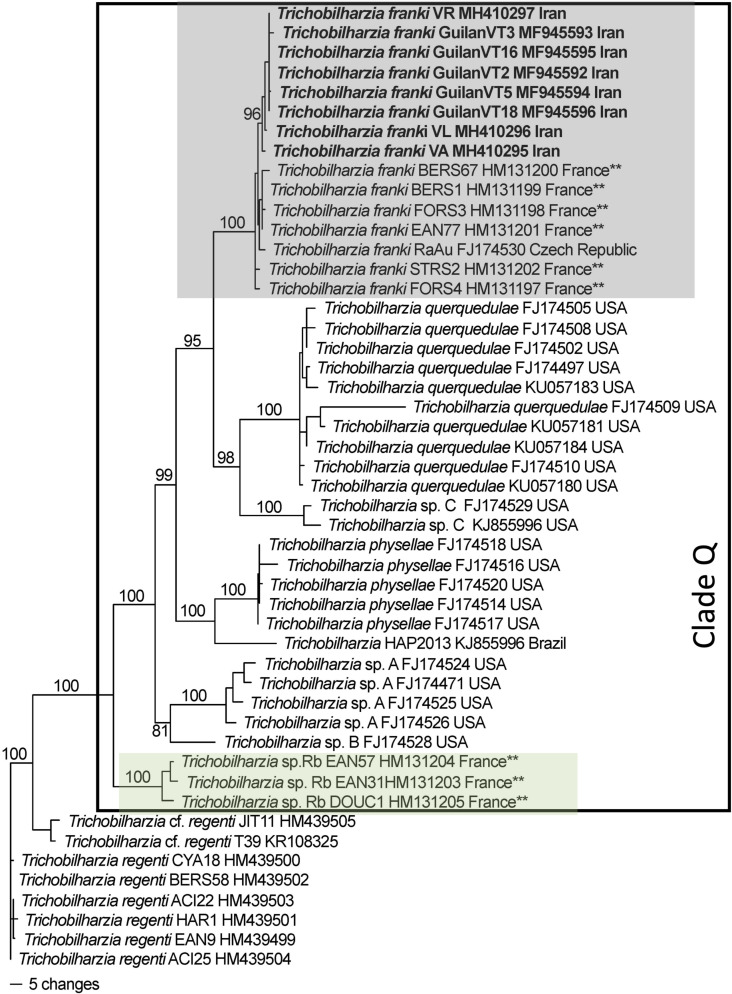
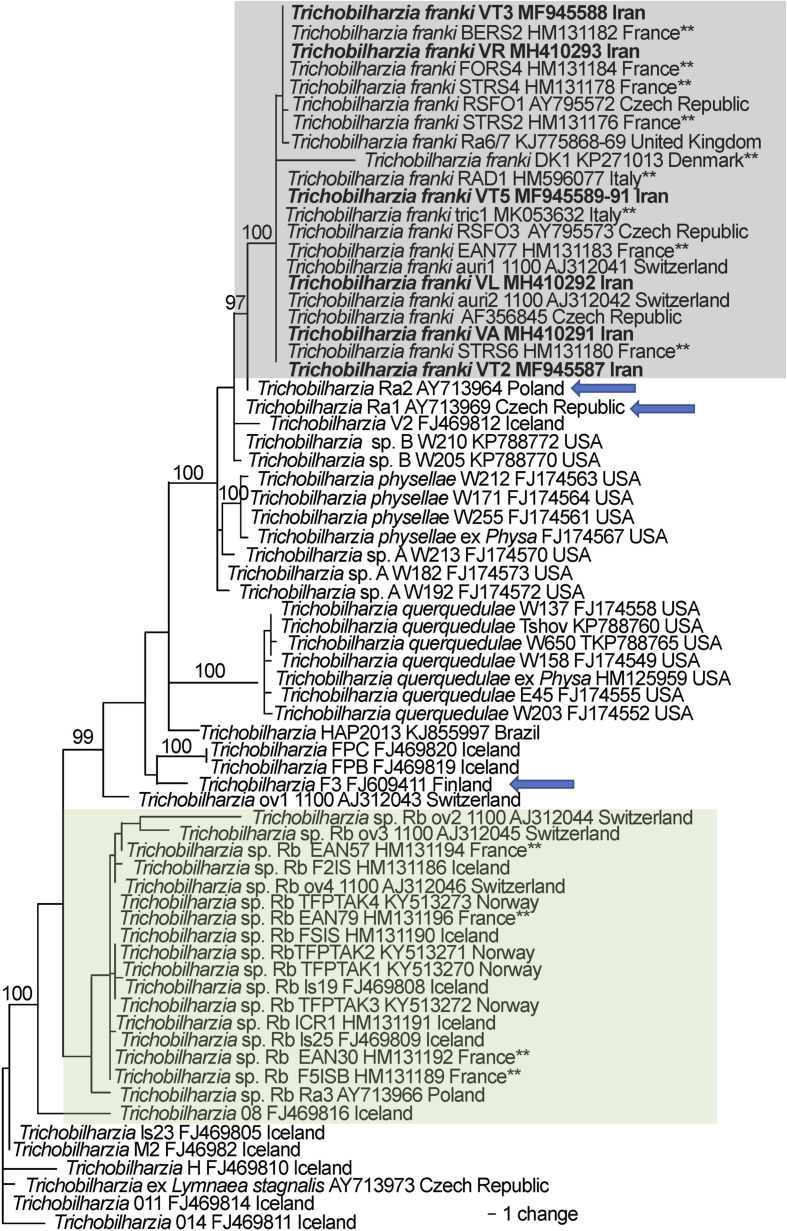
The phylogenetic analysis of both the cox1 (Fig. 4) and ITS1-5.8S-ITS2 (Fig. 5) datasets placed the samples from this study within specimens described as T. franki from Radix auricularia snail intermediate hosts. Some of the previous studies that submitted sequences to GenBank labeled as T. franki were not monophyletic and most of those sequences belong to an undescribed species Trichobilharzia franki haplotype “peregra” (sensu [37] and were referred to as Trichobilharzia sp. Rb from A. balthica snail intermediate hosts. Furthermore, there were many haplotypes labeled T. franki that did not group with any previously defined clade. The clades of T. franki and Trichobilharzia sp. Rb were also found by Soldanova et al. [67]. Using uncorrected p-distances as a measure of genetic diversity and as a proxy for species differentiation, T. franki specimens from Iran were not very divergent from the available specimens of T. franki from GenBank. The average within species diversity was 0.1% for ITS and 0.7% for cox1, which is consistent with other species of Trichobilharzia (Table 5).
Figure 4.
Phylogenetic tree based on Bayesian analysis of cox1 with nodal support indicated on the branches by posterior probabilities. The outer box defines taxa in Clade Q sensu Brant and Loker [14]. The gray boxed clade includes individuals of Trichobilharzia franki with the samples from this study in bold, all other samples are from R. auricularia snails. The green box highlights the “peregra” group (sensu [37]); these species are often confused for T. franki but mostly come from A. balthica. The double asterisk indicates that the snail host in the study was also characterized genetically. Blue arrows indicate schistosomes from R. auricularia but did not group within the clade for T. franki. Taxa are listed with their corresponding GenBank accession number, followed by the country of collections (see Table 4).
Figure 5.
Phylogenetic tree based on Bayesian analysis of ITS1-5.8S-ITS2 with nodal support indicated on the branches by posterior probabilities. The gray boxed clade includes individuals of Trichobilharzia franki, all samples from snails are from Radix auricularia and the samples from this study are in bold. Blue arrows point to specimens that were reported from putative R. auricularia outside of the T. franki clade. The green box highlights the “peregra” group (sensu [37]); these species are often confused for T. franki but mostly come from A. balthica. The double asterisk indicates that the snail host in the study was also characterized genetically. Taxa are listed with their corresponding GenBank accession number followed by the country of collections (see Table 4).
Table 5.
Average pairwise uncorrected p-distances among taxa in the phylogenetic trees.
| ITS1 (%) | cox1 (%) | |
|---|---|---|
| Intraspecific variation | ||
| Trichobilharzia franki | 0.1 | 0.7 |
| Trichobilharzia sp. Rb | 0.3 | 0.6 |
| Trichobilharzia querquedulae | 0.2 | 0.3% – 1.6% 2.4% |
| Trichobilharzia physellae | 0.4 | 0.7 |
| Interspecific variation | ||
| T. franki – Trichobilharzia sp. Rb | 2.7 | 11.5 |
| T. querquedulae – T. franki | 2.6 | 9.2 |
| T. querquedulae – T. physellae | 1.9 | 10.2 |
| T. physellae – T. franki | 1.2 | 10.6 |
| T. physellae – Trichobilharzia sp. Rb | 2.5 | 11.5 |
| T. querquedulae – Trichobilharzia sp. Rb | 3.7 | 11.1 |
Discussion
Our results show that T. franki, a species of avian schistosome that occurs in the visceral veins of its anatid hosts, can be found in domestic ducks in Iran. Duck breeding is a routine activity in almost all rural areas and towns in the flatlands and foothills of Guilan Province, as well as other provinces such as Mazandaran [6, 9, 26]. In these areas, Radix spp. and Physa sp. snail are also found since they also do very well in these modified habitats. It can be assumed that with 71% prevalence in ducks found in this study, T. franki is a common species maintained over time and space by the ubiquity of the intermediate host in the same modified aquaculture habitats as well as the wide distribution and use of domestic mallards, an ideal reservoir host. In addition, the ducks examined here were the same ducks examined in Ashrafi et al. [6] for the neuropathogenic species T. regenti. An interesting question to consider is whether the diversity of Trichobilharzia in domestic mallards over time would reflect the diversity of schistosome species found in migratory birds that also cycle through the commonly available snail intermediate hosts? Or given the prevalence of T. franki and T. regenti, are they well established enough in a domestic life cycle that finding the rarer species from migratory birds would be difficult, or competition in either or the snail or duck hosts? Mallards are listed to host at least 10 named species (7 Trichobilharzia spp.) of schistosomes. Some adult schistosome species are only known from experimental infections of domestic mallards using cercariae from captured wild snails, such as T. franki. Distinguishing between wild mallards and resident mallards is not often defined in most papers. It is known that mallards can host schistosomes, and certainly they have made excellent experimental hosts, but it is difficult to ascertain the distribution and diversity of schistosomes in wild mallards, or if the diversity reflects what can be found in the co-occurring anseriforms.
Until now, T. franki had been confirmed genetically only in Europe to include mostly northern countries (France, Great Britain, Denmark, Switzerland, Czech Republic, Austria, western Russia) [3, 16, 17, 19, 20, 23, 28, 32, 36–38, 42, 45, 61, 62] and now the geographic range is extended to include Europe and Western Asia (Iran). This is the first study to find intact adults of T. franki and characterize them since the original description. Previous reports were based mostly on cercariae, and some on adult fragments or eggs. Trichobilharzia franki was first described from southwestern Germany by Müller and Kimmig (1994) from wild Radix auricularia snail hosts, then cycled experimentally through domestic mallards to obtain the adult worms. Since 1994, there have been few confirmed reports of adults other than small fragments or eggs, and very little effort has been made to sequence more than the nuclear ITS, particularly ITS2 (which is not a useful marker for congeners) and little effort to voucher the specimens or snail hosts. There are very few mitochondrial sequences available for T. franki, most of which have come from cercariae in France (one from the Czech Republic and Austria), making it difficult to characterize genetic diversity across time, space and hosts, with few exceptions [37, 61]. This study has expanded geographic sampling and suggests that T. franki populations are not isolated, at least spanning from France to Iran. The results herein also suggest that if Radix auricularia (or closely related permissive species of Radix) and ducks use the same water body, then it is likely to find T. franki along the migratory route of these birds. Some of these ducks will migrate to northern Africa and if there is R. auricularia or perhaps another permissive snail, then likely this schistosome can extend all along the migration route. Migration was also suggested as the cause of haplotype sharing in T. franki from the UK and Austria [45, 61].
The question remains, how do you designate a species as T. franki in the absence of adult worm morphology and genetic identification of the snail host? Certainly, by genetic comparisons, other named species can mostly be eliminated as the gene trees show them grouping to the exclusion of others with strong support. However, as of this writing, GenBank does have sequences vouchered as T. franki, but they are the ITS2 region which is not informative for species discrimination from Vietnam (MT892757, MT895500, MT919390–MT919394), and Russia and Belarus [49, 62]. For the above sequences, the authors stated that the cercariae were recovered from putative R. auricularia but there is no genetic confirmation of this identification or even a morphological justification. The tree presented in Figure 3 of reference [62] highlights their samples with other putative samples of T. franki. However, there is no monophyletic clade of T. franki and their clade includes other species of Trichobilharzia making it impossible to assign a species based only on their gene tree. Unfortunately, the tree presented in Lopatkin et al. [49] did not include samples from GenBank for comparison and even though they collected cox1 data, it was not vouchered in any publicly available sequence repository. At least for the snails from Russia and Belarus, likely the snail identifications are correct since it is expected that the snail can be found in these geographic areas. The status of the GenBank record from Vietnam is unclear and the sequences currently are not featured in a publication
The natural definitive hosts of T. franki are not well known. There are a few short 28S sequences (552 bp) available in GenBank from duck hosts (FJ793813–FJ793818, FJ793820–FJ793822), and of these, the ones that form a clade with T. franki, are from Anas platyrhynchos and one Cygnus olor (Gmelin, 1789). The other duck hosts reported are Anas crecca (Linnaeus, 1758), whose schistosomes did not group with any clade, and Aythya ferina (Linnaeus, 1758), whose schistosomes grouped with the Trichobilharzia franki “peregra” group (sensu [37] and see [66]; Trichobilharzia sp. Rb herein), a clade that is not closely related to T. franki (Figs. 4 and 5, Table 5). However, eggs were not found in Ay. fuligula or A. crecca hosts; therefore it difficult to know whether worms would produce offspring or if these are dead-end hosts [37]. The occurrence of T. franki is likely facilitated by the widespread intermediate snail host, species of Radix (likely R. auricularia) plus the long distance migration of the anatid hosts. While the snail host in Iran is not yet known, different species of Lymnaeidae, Galba shiraziensis (Küster, 1862 [44]), Stagnicola palustris, Radix auricularia and Radix sp. have been reported in Guilan Province and are potential hosts [5, 7] for T. franki. There is some evidence that R. auricularia (= L. gedrosiana) is an intermediate host of Trichobilharzia spp. in Iran [9, 30, 31] but more studies are needed to confirm this hypothesis.
The systematics and taxonomy of the identity and distribution of R. auricularia are not well understood. Recent studies using more variable gene regions (cox1) have shown that this species may include a complex of clades [46, 70]; however, no samples from the Middle East region were included. Furthermore, it has been suggested that R. auricularia might be an invasive snail and/or more widespread than previously thought, but this proposition has not yet been tested [8, 40, 46]. It appears that R. auricularia likely plays a major role in transmission of avian schistosomes in the country, but the species has not yet been verified and unidentified species of Radix have yet to be characterized. Furthermore, Aksenova et al. [1] suggest that Radix euphratica may be more widespread in the area and Vinarski et al. [69] suggest R. gedrosiana should be synonymized under R. euphratica. The first sequences to be described as T. franki are from R. auricularia from Switzerland [56], but there is no mention in the paper about what they based their species identification on, other than that the original description from R. auricularia hosts [6]. This assumption of host specificity might be reasonable given that the molecular results have shown over time that cercariae from R. auricularia most often group with haplotypes named T. franki (see some clades from [3, 63] and Figs. 4 and 5 herein), but T. franki had not been sampled widely with genetic confirmation of their snail hosts. At least in the ITS tree, not all samples from R. auricularia form a clade (see blue arrows in Fig. 5) and many of them from Ampullaceana balthica (= Radix peregra, = R. ovata) form a clade [1, 37]). Brant and Loker [14] suggested that T. franki might be found in North America, but the results for both cox1 (Fig. 3; FJ174528) and ITS (Fig. 4; KP788772, KP788770) show a position outside T. franki. The ITS tree shows a grouping with a haplotype from R. auricularia from Poland (Fig. 5; Table 4; AY713969).
Much of the genetic diversity of Trichobilharzia lineages in Clade Q (sensu [14]; Fig. 4) that includes T. franki from GenBank sequences is represented by ITS sequences, and many of these samples do not form clades (Fig. 5). This might suggest that more diversity is yet to be discovered. Processes that may contribute to our understanding of diversity in Trichobilharzia that emerges from sequencing surveys include the following: (A) Incomplete lineage sorting – the ITS tree is based only on a nuclear region, which may have a slower mutation rate relative to a faster evolving gene, thus ancestral polymorphisms are retained, or the ancestral population was large and thus takes more time. Also, it could be that speciation within at least Clade Q has been relatively recent and nuclear copies do not match mitochondrial gene trees or species trees. However, in general, most sequences fall into taxa that group according to species (or lineages if only cercariae) and there is little or no evidence of widespread mito-nuclear discordance. (B) Hybridization – within avian schistosomes, hybridization has not been studied. However, mito-nuclear discordance is not reliable evidence of hybridization (see [58]) but a more variable gene certainly helps in diversity characterizations and species delineations. It is impossible to obtain mitochondrial or genomic data to explore hybridization or any other question with the individual worms available currently in GenBank, because there are no museum vouchers for re-evaluation. The few specimens that were vouchered are not available for destructive sampling and thus it is strongly recommended that vials of adult and cercariae (and hosts, particularly gastropods) are deposited so that we have a record of the past and material available for new investigations. (C) Host-induced variation – though poorly understood, host-induced variation can contribute to morphological variability, but it is not known how much it would affect genetic diversity in schistosomes, and most studies have used morphology, not genetics in this context, with adult worms (see [10, 65]). (D) Ecological speciation is plausible given that the offspring of the worms in the migratory hosts are being distributed along the route, at each locality the miracidia might be exposed to snails that are normally compatible but might also be exposed to novel putatively susceptible snails. For parasites, this is akin to host switching events. For a high-quality review see [64]. (E) Poor host taxonomy. One of the consequences of genetic characterizations is that it has provided a yardstick to define in more detail lineage diversity, which may not be reflected in morphological diversity. Invertebrates in particular have fallen in this category as they can often have little variation to compare and some of the observed variation is subject to change based on a myriad of abiotic and biotic influences (such as parasitism, temperature, water chemistry, etc.) rather than phylogeny. Gastropods in particular have been shown to be more species-rich than previously considered based on morphology [1, 29]. If every genetic report of Trichobilharzia spp. was accompanied by genetic assessment of the snail host, then we could understand more about host-parasite relationship specificity. It could be that there are more species of Radix transmitting these species of Trichobilharzia than is reported based solely on morphology (e.g. [21, 22, 46]).
Until there is more effort to include multiple and variable gene regions for schistosomes (or any organism) it is not possible to understand the phylogeography or epidemiology of disease-causing helminths. It has been shown repeatedly that variable mitochondrial genes are ideal for assessing cryptic diversity compared to nuclear genes. When only ITS2 is used without ITS1, it is not possible to find enough variation within congeners, particularly if they are closely related. However, a study should not rely only on a single gene (and if it does so, it should be variable and useful for the future), as the diversity revealed is gene diversity, not always directly reflecting species, which must be tested for congruency [50, 55]. The specimens available in GenBank that had variable cox1 sequences available represent mostly Western Europe (Table 1). Yet, given the geographic distance between these specimens and Northern Iran, there was very little genetic differentiation in either cox1 or ITS (Table 5). This result should not be surprising if the long-distance migration of the wild hosts and the suitability of domestic mallards as reservoir hosts are considered in transmission dynamics (also see [4]). Furthermore, Ebbs et al. [25] showed that the intraspecific genetic diversity in Trichobilharzia querquedulae was within average range for species schistosomes, even though the comparison included individual worms from across 5 continents.
The occurrence of CD in Iran is high in areas of aquaculture. In addition to wild duck hosts, previous work in the area has shown that domestic mallards are reservoir hosts of T. regenti, a nasal schistosome [6], and as well for T. franki, shown in this study and thus maintain high prevalence of CD. The genetic results support the finding that populations of T. franki from Iran are not differentiated from populations in Europe. Therefore, the schistosomes are dispersed with their migratory duck host, maintaining the gene flow across populations with compatible snail hosts in Iran. It is not surprising that species of Trichobilharzia are thought to be one of the common etiological agents of CD; several of these species use snail hosts that are widespread and/or invasive (e.g. [24]) and prefer or at least are easily established in modified aquatic habitats used by domestic animals and humans. Added to this, their definitive hosts travel long distances, further facilitating transmission from one continent to another.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This study was performed with the collaboration of the Guilan University of Medical Sciences, Iran and the University of New Mexico, USA. The authors would like to thank Mrs. Behnaz Rahmati and Mr. Alireza Noroosta for their kind help in performing this study. The authors thank two anonymous reviewers for suggestions that improved the content of the manuscript.
Cite this article as: Ashrafi K, Sharifdini M, Darjani A & Brant SV. 2021. Migratory routes, domesticated birds and cercarial dermatitis: the distribution of Trichobilharzia franki in Northern Iran. Parasite 28, 4.
References
- 1.Aksenova OV, Bolotov IN, Gofarov MY, Kondakov AV, Vinarski MV, Bespalaya YV, Kolosova YS, Palatov DM, Sokolova SE, Spitsyn VM, Tomilova AA, Travina OV, Vikhrev IV. 2018. Species richness, molecular taxonomy and biogeography of the radicine pond snails (Gastropoda: Lymnaeidae) in the Old World. Scientific Reports, 8, 11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldhoun J, Kolarova L, Skirnisson K, Horak P. 2009a. Bird schistosome diversity in Iceland: molecular evidence. Journal of Helminthology, 83, 173–180. [DOI] [PubMed] [Google Scholar]
- 3.Aldhoun J, Faltynkova A, Karvonen A, Horak P. 2009b. Schistosomes in the North: A unique finding from a prosobranch snail using molecular tools. Parasitology International, 58, 314–317. [DOI] [PubMed] [Google Scholar]
- 4.Ashrafi K, Brant SV. 2020. An efficient method for collecting the full-length adults, fragments, and eggs of Trichobilharzia spp. from the liver of definitive hosts. Parasitology Research, 119, 1167–1172. [DOI] [PubMed] [Google Scholar]
- 5.Ashrafi K, Mas-Coma S. 2014. Fasciola gigantica transmission in the zoonotic fascioliasis endemic lowlands of Guilan, Iran: experimental assessment. Veterinary Parasitology, 205, 96–106. [DOI] [PubMed] [Google Scholar]
- 6.Ashrafi K, Nouroosta A, Sharifdini M, Mahmoudi MR, Rahmati B, Brant SV. 2018. Genetic diversity of an avian nasal schistosome causing cercarial dermatitis in the Black Sea-Mediterranean migratory route. Parasitology Research, 117, 3821–3833. [DOI] [PubMed] [Google Scholar]
- 7.Ashrafi K, Valero MA, Peixoto RV, Artigas P, Panova M, Mas-Coma S. 2015. Distribution of Fasciola hepatica and F. gigantica in the endemic area of Guilan, Iran: Relationships between zonal overlap and phenotypic traits. Infections, Genetics and Evolution, 31, 95–109. [DOI] [PubMed] [Google Scholar]
- 8.Aksenova O, Vinarski M, Bolotov I, Kondakov A, Besplalaya Y, Tomilova A, Palster I, Gofarov M. 2017. Two Radix spp. (Gastropoda: Lymnaeidae) endemic to thermal springs around Lake Baikal represent ecotypes of the widespread Radix auricularia. Journal of Zoological Systematics and Evolutionary Research, 55, 298–309. [Google Scholar]
- 9.Athari A, Gohardehi S, Rostami-Jalilian M. 2006. Determination of definitive and intermediate hosts of cercarial dermatitis producing agents in northern Iran. Archives of Iranian Medicine, 9, 11–15. [PubMed] [Google Scholar]
- 10.Bayssade-Dufour C, Jouet D, Rudolfova J, Horak P, Ferté H. 2006. Seasonal morphological variations in bird schistosomes. Parasite, 13, 205–214. [DOI] [PubMed] [Google Scholar]
- 11.Blair D, Islam KS. 1983. The life cycle and morphology of Trichobilharzia australis n. sp. from the nasal blood vessels of the black duck in Australia with a review of the genus Trichobilharzia. Systematic Parasitology, 5, 89–117. [Google Scholar]
- 12.Bowles J, McManus DP. 1993. Rapid discrimination of Echinococcus species and strains using a PCR-based RFLP method. Molecular and Biochemical Parasitology, 57, 231–239. [DOI] [PubMed] [Google Scholar]
- 13.Bowles J, Blair D, McManus DP. 1995. A molecular phylogeny of the human schistosomes. Molecular Phylogenetics and Evolution, 4, 103–109. [DOI] [PubMed] [Google Scholar]
- 14.Brant SV, Loker ES. 2009. Molecular systematics of the avian schistosome genus Trichobilharzia (Trematoda: Schistosomatidae) in North America. Journal of Parasitology, 95, 941–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brant SV, Bochte CA, Loker ES. 2011. New intermediate host records for the avian schistosomes Dendritobilharzia pulverulenta, Gigantobilharzia huonensis, and Trichobilharzia querquedulae from North America. Journal of Parasitology, 97, 946–949. [DOI] [PubMed] [Google Scholar]
- 16.Caron Y, Cabaraux A, Marechal F, Losson B. 2017. Swimmer’s itch in Belgium: first recorded outbreaks, molecular identification of the parasite species and intermediate hosts. Vector-Borne and Zoonotic Diseases, 17, 3. [DOI] [PubMed] [Google Scholar]
- 17.Cipriani P, Mattiucci S, Paoletti M, Scialanca F, Nascetti G. 2011. Molecular evidence of Trichobilharzia franki Müller and Kimmig, 1994 (Digenea: Schistosomatidae) in Radix auricularia from Central Italy. Parasitology Research, 109, 935–940. [DOI] [PubMed] [Google Scholar]
- 18.Clausen JH, Madsen H, Van PT, Dalsgaard A, Murrell KD. 2015. Integrated parasite management: path to sustainable control of fishborne trematodes in aquaculture. Trends in Parasitology, 31, 8–15. [DOI] [PubMed] [Google Scholar]
- 19.Christiansen AO, Olsen A, Buchmann K, Kania PW, Nejsum P, Vennervald BJ. 2016. Molecular diversity of avian schistosomes in Danish freshwater snails. Parasitology Research, 115, 1027–1037. [DOI] [PubMed] [Google Scholar]
- 20.De Liberato C, Berrilli F, Bossu T, Magliano A, Montalbano Di Filippo M, Di Cave D. 2019. Outbreak of swimmer’s itch in Central Italy: description, causative agent and preventive measures. Zoonoses and Public Health, 66, 377–381. [DOI] [PubMed] [Google Scholar]
- 21.Devkota R, Brant SV, Loker ES. 2015. The Schistosoma indicum species group in Nepal: presence of a new lineage of schistosome and use of the Indoplanorbis exustus species comples of snail hosts. International Journal for Parasitology, 45, 857–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devkota R, Brant SV, Loker ES. 2016. A genetically distinct Schistosoma from Radix luteola from Nepal related to Schistosoma turkestanicum: a phylogenetic study of schistosome and snail host. Acta Tropica, 164, 45–53. [DOI] [PubMed] [Google Scholar]
- 23.Dvorák J, Vanácová S, Hampl V, Flegr J, Horák P. 2002. Comparison of European Trichobilharzia species based on ITS1 and ITS2 sequences. Parasitology, 124, 307–313. [DOI] [PubMed] [Google Scholar]
- 24.Ebbs ET, Loker ES, Brant SV. 2018. Phylogeography and genetics of the globally invasive snail Physa acuta Draparnaud 1805, and its potential to serve as an intermediate host to larval digenetic trematodes. BMC Evolutionary Biology, 18, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebbs ET, Loker ES, Davis NE, Flores V, Veleizan A, Brant SV. 2016. Schistosomes with wings: how host phylogeny and ecology shape the global distribution of Trichobilharzia querquedulae (Schistosomatidae). International Journal for Parasitology, 46, 669–677. [DOI] [PubMed] [Google Scholar]
- 26.Fakhar M, Ghobaditara M, Brant SV, Karamian M, Gohardehi S, Bastani R. 2016. Phylogenetic analysis of nasal avian schistosomes (Trichobilharzia) from aquatic birds in Mazandaran Province, northern Iran. Parasitology International, 65, 151–158. [DOI] [PubMed] [Google Scholar]
- 27.Farahnak A, Essalat M. 2003. A study on cercarial dermatitis in Khuzestan province, southwestern Iran. BMC Public Health, 3, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferté H, Depaquit J, Carre S, Villena I, Léger N. 2005. The presence of Trichobilharzia szidati in Lymnaea stagnalis and T. franki in Radix auricularia in northeastern France: molecular evidence. Parasitology Research, 95, 150–154. [DOI] [PubMed] [Google Scholar]
- 29.Gauffre-Autelin P, von Rintelen T, Stelbrink B, Albrecht C. 2017. Recent range expansion of an intermediate host for animal schistosome parasites in the Indo-Australian Archipelago: phylogeography of the freshwater gastropod Indoplanorbis exustus in South and Southeast Asia. Parasites & Vectors, 10, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghobaditara M, Fakhar M, Sharif M. 2015. An overview on the present situation of cercarial dermatitis: a neglected zoonotic disease in Iran and the World. Journal of Mazandaran University of Medical Sciences, 24, 446–460. [Google Scholar]
- 31.Gohardehi S, Fakhar M, Madjidaei M. 2012. Avian schistosomes and human cercarial dermatitis in a wildlife refuge in Mazandaran Province, northern Iran. Zoonoses and Public Health, 60, 442–447. [DOI] [PubMed] [Google Scholar]
- 32.Gulyas K, Soldanova M, Orosova M, Oros M. 2020. Confirmation of the presence of zoonotic Trichobilharzia franki following a human cercarial dermatitis outbreak in recreational water in Slovakia. Parasitology Research, 119, 2531–2537. [DOI] [PubMed] [Google Scholar]
- 33.Hoberg EP, Pilitt PA, Galbreath KE. 2009. Why museums matter: a tale of pinworms (Oxyuroidea: Heteroxynematidae) among pikas (Ochotona princeps and O. collaris) in the American west. Journal of Parasitology, 95, 490–501. [DOI] [PubMed] [Google Scholar]
- 34.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics Application Notes, 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 35.Imani-Baran A, Yakhchali M, Malekzadeh-Viayeh R, Farahnak A. 2013. Seasonal and geographical distribution of cercarial infection in Lymnaea gedrosiana (Pulmunata: Lymnaeidae) in northwest Iran. Iranian Journal of Parasitology, 8, 423–429. [PMC free article] [PubMed] [Google Scholar]
- 36.Jouet D, Ferté H, Depaquit J, Rudolfova J, Latour P, Zanella D, Kaltenbach ML, Léger N. 2008. Trichobilharzia spp. in natural conditions in Annecy Lake, France. Parasitology Research, 103, 51–58. [DOI] [PubMed] [Google Scholar]
- 37.Jouet D, Skirnisson K, Kolářová L, Ferté H. 2010a. Molecular diversity of Trichobilharzia franki in two intermediate hosts (Radix auricularia and Radix peregra): a complex of species. Infection, Genetics and Evolution, 10, 1218–1227. [DOI] [PubMed] [Google Scholar]
- 38.Jouet D, Skirnisson K, Kolářová L, Ferté H. 2010b. Final hosts and variability of Trichobilharzia regenti under natural conditions. Parasitology Research, 107, 923–930. [DOI] [PubMed] [Google Scholar]
- 39.Karamian M, Aldhoun JA, Maraghi S, Hatam G, Farhangmehr B, Sadjjadi SM. 2011. Parasitological and molecular study of the furcocercariae from Melanoides tuberculate as a probable agent of cercarial dermatitis. Parasitology Research, 108, 955–962. [DOI] [PubMed] [Google Scholar]
- 40.Kipp RM, Benson AJ, Larson J, Fusaro A. 2017. Radix auricularia. USGS Nonindigenous Aquatic Species Database: Gainesville, FL: http://nas.er.usgs.gov/queries/factsheet.aspx?SpeciesID = 1012. [Google Scholar]
- 41.Kolářová L, Horák P, Skírnisson K, Marečková H, Doenhoff M. 2013. Cercarial dermatitis, a neglected allergic disease. Clinical Reviews in Allergy and Immunology, 45, 63–74. [DOI] [PubMed] [Google Scholar]
- 42.Korsunenka AV, Chrisanfova GG, Ryskov AP, Movseessian SO, Vasilyev VA, Semyenova SK. 2010. Detection of European Trichobilharzia schistosomes (T. franki, T. szidati, and T. regenti) based on novel genome sequences. Journal of Parasitology, 96, 802–806. [DOI] [PubMed] [Google Scholar]
- 43.Kourilová P, Hogg KG, Kolářová L, Mountford AP. 2004. Cercarial dermatitis caused by bird schistosomes comprises both immediate and late phase cutaneous hypersensitivity reaction. Journal of Immunology, 172, 3766–3774. [DOI] [PubMed] [Google Scholar]
- 44.Küster HC. 1862. Die Gattungen Limnaeus, Amphipeplea, Chilina, Isidora und Physopsis, in Systematisches Conchylien-Cabinet, 2nd edn Martini C, Editor. Nürnberg: Bauer & Raspe, I.17 b: issues 180-182: 1-48, plates 1-11 (1862); issue 184: 49-77, plate 12 (1863). [Google Scholar]
- 45.Lawton SP, Lim RM, Dukes JP, Cook RT, Walker AJ, Kirk RS. 2014. Identification of a major causative agent of human cercarial dermatitis, Trichobilharzia franki (Müller and Kimmig, 1994), in southern England and its evolutionary relationships with other European populations. Parasites & Vectors, 7, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawton SP, Lim RM, Dukes JP, Kett SM, Cook RT, Walker AJ, Kirk RS. 2015. Unravelling the riddle of Radix: DNA barcoding for species identification of freshwater snail intermediate host of zoonotic digeneans and estimating their inter-population evolutionary relationships. Infection, Genetics and Evolution, 35, 63–74. [DOI] [PubMed] [Google Scholar]
- 47.Lima dos Santos CAM, Howgate P. 2011. Fishborne zoonotic parasites and aquaculture: A review. Aquaculture, 318, 253–261. [Google Scholar]
- 48.Lockyer AE, Olson PD, Ostergaard P, Rollinson D, Johnston DA, Attwood SW, Southgate VR, Horak P, Snyder SD, Le TH, Agatsuma T, McManus DP, Carmichael AC, Naem S, Littlewood DT. 2003. The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology, 126, 203–224. [DOI] [PubMed] [Google Scholar]
- 49.Lopatkin AA, Chrisanfova GG, Voronin MV, Zazornova OP, Beer SA, Semyenova SK. 2010. Polymorphism of the cox1 gene in cercariae isolates of bird schistosomes (Trematoda: Schistosomatidae) from ponds of Moscow and Moscow region. Russian Journal of Genetics, 46, 873–880. [PubMed] [Google Scholar]
- 50.Maddison WP. 1997. Gene trees in species trees. Systematic Biology, 46, 523–536. [Google Scholar]
- 51.Mahdavi SA, Farahnak A, Mobedi I, Rad MBM, Azadeh H. 2013. Survey of migratory birds (Anatidae: Anas platyrhynchos) for schistosome parasites from Mazandaran Province, northern Iran. Iranian Journal of Parasitology, 8, 333–336. [PMC free article] [PubMed] [Google Scholar]
- 52.Mahdavi SA, Farahnak A, Mousavi SJ, Mobedi I, Rezaeian M, Golmohamadi T, Azadeh H, Gohardehi S. 2013. Prevalence of schistosome induced cercarial dermatitis in north of Iran. Asian Pacific Journal of Tropical Disease, 3, 37–40. [Google Scholar]
- 53.Maleki SH, Athari A, Haghighi A, Taghipour N, Gohardehi SH, Tabaei SS. 2012. Species identification of birds nasal Trichobilharzia in Sari, north of Iran. Iranian Journal of Parasitology, 7, 82–85. [PMC free article] [PubMed] [Google Scholar]
- 54.Müller V, Kimmig P. 1994. Trichobilharzia franki n. sp.–the cause of swimmer’s dermatitis in southwest German dredged lakes. Applied Parasitology, 35, 12–31. [PubMed] [Google Scholar]
- 55.Nakhleh L. 2013. Computational approaches to species phylogeny inference and gene tree reconciliation. Trends in Ecology and Evolution, 28, 719–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Picard D, Jousson O. 2001. Genetic variability among cercariae of the Schistosomatidae (Trematoda: Digenea) causing swimmer’s itch in Europe. Parasite, 8, 237–242. [DOI] [PubMed] [Google Scholar]
- 57.Pinto HA, Brant SV, de Melo AL. 2014. Physa marmorata (Mollusca: Physidae) as a natural intermediate host of Trichobilharzia (Trematoda: Schistosomatidae), a potential causative agent of avian mercurial dermatitis in Brazil. Acta Tropica, 138, 38–43. [DOI] [PubMed] [Google Scholar]
- 58.Platt RN III, McDew-White M, Le Clech W, Chevalier FD, Allan F, Emery AM, Garba A, Hamidou AA, Ame SM, Webster JP, Rollinson D, Webster BL, Anderson TJC. 2019. Ancient hybridization and adaptive introgression of an invadolysin gene in schistosome parasites. Molecular Biology and Evolution, 36, 2127–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pleijel F, Jondelius U, Norlinder E, Nygren A, Oxelman B, Schander C, Sundberg P, Thollesson M. 2008. Phylogenies without roots? A plea for the use of vouchers in molecular phylogenetic studies. Molecular Phylogenetics and Evolution, 48, 369–371. [DOI] [PubMed] [Google Scholar]
- 60.Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics, 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 61.Reier S, Haring E, Billinger F, Blatterer H, Duda M, Corofsky C, Grasser HP, Heinisch W, Horweg C, Druckenhauser L, Szucish NU, Wanka A, Sattmann H. 2020. First confirmed record of Trichobilharzia franki Müller & Kimmig, 1994, from Radix auricularia (Linnaeus, 1758) for Austria. Parasitology Research, 119, 4135–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rizevsky SV, Cherviakovsky EM, Kurchenko VP. 2010. Molecular taxonomic identification of Schistosomatidae from Naroch Lake and Polonevichi Lake in Belarus. Biochemical Systematics and Ecology, 39, 14–21. [Google Scholar]
- 63.Rudolfova J, Hampl V, Bayssade-Dufour C, Lockyer AE, Littlewood DT, Horák P. 2005. Validity reassessment of Trichobilharzia species using Lymnaea stagnalis as the intermediate host. Parasitology Research, 95, 79–89. [DOI] [PubMed] [Google Scholar]
- 64.Schluter D. 2001. The ecology and origin of species. Trends in Ecology and Evolution, 16, 372–380. [DOI] [PubMed] [Google Scholar]
- 65.Skírnisson K, Kolářová L, Horák P, Ferté H, Jouet D. 2012. Morphological features of the nasal blood fluke Trichobilharzia regenti (Schistosomatidae, Digenea) from naturally infected hosts. Parasitology Research, 110, 1881–1892. [DOI] [PubMed] [Google Scholar]
- 66.Skrjabin KI, Zakharov NP. 1920. Zweineue Trematodengattugen aus den Blutgefassen der Volgel. Izvestnik Donskovo Veterinarnovo Instituta, 2, 1–6. [Google Scholar]
- 67.Soldanova M, Georgieva S, Rohacova J, Knudsen R, Kuhn JA, Henriksen EH, Siwertsson A, Shaw JC, Kuris AM, Amundsen PA, Scholz T, Lafferty KD, Kostadinova A. 2017. Molecular analyses reveal high species diversity of trematodes in a sub-Arctic lake. International Journal for Parasitology, 47, 327–345. [DOI] [PubMed] [Google Scholar]
- 68.Valkiunas G, Atkinson CT, Bensch S, Sehgal RNM, Ricklefs RE. 2008. Parasite misidentifications in GenBank: how to minimize their number? Trends in Parasitology, 24, 247–248. [DOI] [PubMed] [Google Scholar]
- 69.Vinarski MV, Aksenova OV, Bolotov IN. 2020. Taxonomic assessment of genetically-delineated species of radicine snails (Mollusca, Gastropoda, Lymnaeidae). Zoosystematics and Evolution, 96, 577–608. [Google Scholar]
- 70.Von Oheimb PV, Albrecht C, Riedel F, Du L, Yang J, Aldridge DC, Bößneck U, Zhang H, Wilke T. 2011. Freshwater biogeography and limnological evolution of the Tibetan Plateau – Insights from a plateau-wide distributed gastropod taxon (Radix spp.). PLoS One, 6(10), e26307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yakhchali M, Hosseinpanahi A, Malekzadeh-Viayeh R. 2016. Molecular evidence of Trichobilharzia species (Digenea: Schistosomatidae) in the snails of Lymnaea auricularia from Urmia suburb, northwest Iran. Iranian Journal of Parasitology, 11, 296–302. [PMC free article] [PubMed] [Google Scholar]