Graphical abstract
Keywords: Reinfection, Spike protein, Long-term immunity, T-cell response, Mutation
Abstract
Coronavirus disease 19 (COVID-19) continues to challenge most scientists in the search of an effective way to either prevent infection or to avoid spreading of the disease. As result of global efforts some advances have been reached and we are more prepared today than we were at the beginning of the pandemic, however not enough to stop the transmission, and many questions remain unanswered. The possibility of reinfection of recovered individuals, the duration of the immunity, the impact of SARS-CoV-2 mutations in the spreading of the disease as well as the degree of protection that a potential vaccine could have are some of the issues under debate. A number of vaccines are under development using different platforms and clinical trials are ongoing in different countries, but even if they are licensed it will need time until reach a definite conclusion about their real safety and efficacy. Herein we discuss the different strategies used in the development of COVID-19 vaccines, the questions underlying the type of immune response they may elicit, the consequences that new mutations may have in the generation of sub-strains of SARS-CoV-2 and their impact and challenges for the efficacy of potential vaccines in a scenario postpandemic.
1. Introduction
The SARS-CoV-2 coronavirus, etiologic agent of COVID-19 has been so far responsible for more than 66,872,117 cases of infection and more than 1,536,855 deaths worldwide [1]. Great advances in the knowledge of the biology of this new coronavirus and the natural history of the disease have been reached. However, there is no effective treatment available yet and the efforts to find a drug or treatment to impair virus infection and/or to decrease the spread of the disease are still ongoing [2].
The primary target of SARS-CoV-2 infection is the lower respiratory tract causing flu-like illness with symptoms such as cough, fever, fatigue and arthralgia. However, the presentation and the course of the disease can range from asymptomatic to mild respiratory infections and pneumonia. Some infected patients develop more severe disease with acute respiratory syndrome distress (ARD) about 7–10 days after onset of symptoms, following a rapid viral replication, increased pro-inflammatory cytokine production, “cytokine storm”, as well as chemokine responses and inflammatory cell infiltrates [[3], [4], [5]]. Among other risk factors identified, age has shown to be an important factor for the development of a more severe disease. Younger individuals often are asymptomatic or present mild symptoms and thus might have a crucial role in the spread of the disease [6,7]. During viral infection both innate and adaptive immune responses play a role in the pathogenesis of SARS-CoV-2. Specific motifs present in some SARS-CoV-2 protein structure or mediators released by infected damaged cells are recognized by the conserved innate pattern recognition receptors (PRRs) and seems to be involved in the modulation of the immune response. An important feature in the pathophisiology of SARS-CoV-2 is the overproduction of early pro-inflammatory cytokines such as tumor necrosis factor (TNF), IL-6 and IL-1β which may lead to increased risk of vascular hyperpermeability, multi organ failure and eventually death if the high concentrations of cytokines is not controlled [[8], [9], [10]]. Moreover, it has been observed that adults with COVID-19 often present a decrease in both CD4+ and CD8 + T-cell subsets at the early stage of the disease what could contribute to virus replication and disease severity in some patients [11].
Innate immunity is important to inhibit viral replication and clearance, as well as to induce tissue repair and a prolonged immune response [12]. In this context, type I interferons (IFN-I, IFNα, IFNβ) play an important role in conferring antiviral activity in host cells. However, it seems that SARS-CoV-2 have evolved mechanisms to evade IFN antiviral activity [13]. Nevertheless, given the complexity of COVID-19 pathophysiology, type I interferons may have different roles at different stages of infection or at mild versus severe COVID-19 patients [14]. Despite these findings, the immunity profile in COVID-19 is not completely understood.
Even before COVID-19 was declared a pandemic on March 11th, 2020 [15]; efforts trying to develop a vaccine against SARS-CoV-2 had been initiated, when the first viral genomic sequence became available in early January. However, the recognition of the pandemic intensified and induced a rush for the development of vaccines in different countries. Thus, considering the rapid global spread of SARS-CoV-2 infection and the increasing death toll, development of an effective vaccine became priority. As result, great advances in a shorter time than expected for this research field was conquered, and several vaccine candidates are currently in phase II/III in China, UK, USA, Russia, Brazil and other countries [16].
Vaccines have played an important role in public health for decades to help prevent diseases like mumps, polio, rubella and yellow fever. Yet, we still have not understood well about their durabilities. Several questions have been raised regarding the novel vaccines being developed for SARS-CoV-2, specially concerned to the efficacy and durability. This review will highlight important structure-function relationships of key SARS-CoV-2 proteins with focus on their role in pathogenesis and ability to elicit immune responses. We also will discuss the impact of new mutations in the genome of SARS-CoV-2 and how these changes can contribute for the emergence of new sub-strains with novel fitness and transmissibility ability, which could circumvent the efficacy and durability of the vaccines under development.
2. SARS-CoV-2 genome organization, molecular structure and biological features
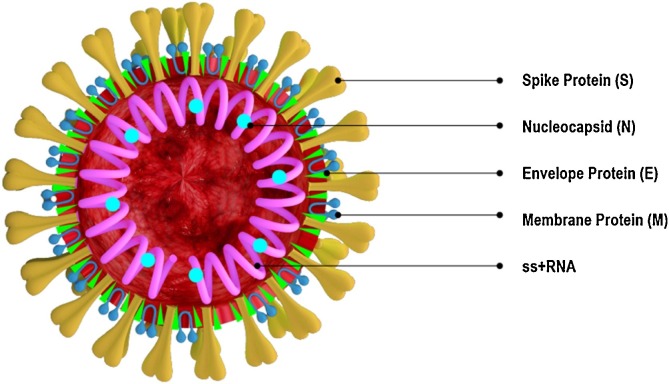
Determination of SARS-CoV-2 coronavirus structure and genome organization was pivotal to get insights on the mechanisms of viral infection and replication as well as to map immunogenic protein motifs that can elicit protective host immune responses. Moreover, detailed knowledge of the structural organization of targeted proteins has been helpful in the screening, identification and development of potential drugs and vaccines for treatment and prevention of COVID-19 [[17], [18], [19], [20]]. Genome sequence analysis demonstrated close resemblance of SARS-CoV-2 to other pathogenic coronaviruses reported earlier such as SARS-CoV and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [21,22]. SARS-CoV-2 genome is 29.9 kb in size and shares approximately 82 % nucleotide sequence identity and > 90 % protein sequence identity for essential enzymes and structural proteins with SARS-CoV and MERS-CoV coronaviruses [22,23]. In fact, SARS-CoV-2 contains four major structural proteins: spike (S), envelope (E), membrane (M) and nucleocapsid (N), which are very similar to the mentioned coronaviruses and could explain some common features of the pathogenesis mechanism (Fig. 1 ).
Fig. 1.
Structure of SARS-CoV-2 showing the main structural proteins. The virus consists of four major structural proteins: spike (S), envelope (E), membrane (M) and nucleocapsid (N). S, E and M are located in the lipid bilayer envelope while the N protein encapsulates the virus RNA genome.
SARS-CoV-2 coronavirus belongs to a group of enveloped viruses containing a positive single-stranded RNA genome [24]. They are named for their crown-like spikes present on their surface. Coronaviruses are distributed into four main sub-groupings known as alpha, beta, gamma and delta [25,26]. The alpha- and beta- coronaviruses have been receiving more attention due to their ability to cross animal-human barriers and become human pathogens [27]. Gama coronaviruses infect avian species while delta coronaviruses infect both mammals and birds. The beta-coronaviruses group, which SARS-CoV-2 belongs, can be further classified into four viral lineages named A–D [28].
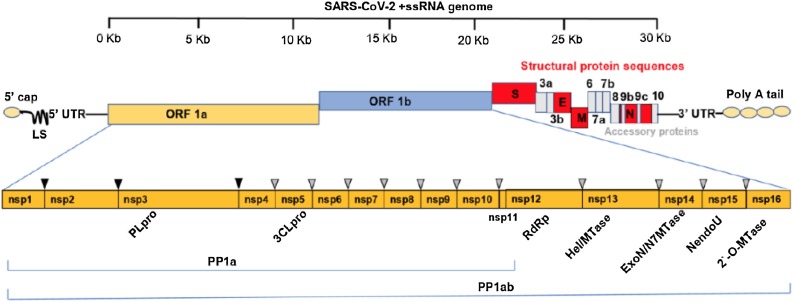
The SARS-CoV-2 genome comprises 13–15 open reading frames (ORFs) from which 12 are functional encompassing 11 protein- coding genes and 12 expressed proteins [29]. The genomic RNA has a 5’-cap and a 3’-poly(A) tail and is used for immediate translation of viral proteins once it gets inside of host cells. The ORFs are preceded by transcriptional regulatory sequences with highly structured unstranslated regions (UTRs) and are arranged sequentially following a typical 5’-3’ direction [30]. The replicase polyprotein 1a (PP1a) and polyprotein 1ab (PP1ab) are encoded by the ORFs 1a and 1b respectively. The polyprotein 1ab is the largest polyprotein and encompasses the non-structural proteins (Nsp1-16), which form the replicase complex. The 3’ end of the virus genomic RNA encodes the major S, E, M, and N structural proteins. These proteins play important role in viral entry, host cell membrane fusion and mature viral structure integrity [[31], [32], [33]]. Also, at the 3’ end are located nine ORFs coding for accessory factors interspersed among or even overlapping the structural genes [24] (Fig. 2 ). Accessory proteins are thought to play additional functions that are not required for replication, but rather are involved in the pathogenicity by modulating interferon signaling pathway [34]. Both structural and accessory proteins are translated from a set of nested subgenomic RNAs (sgRNAs) starting from a negative-sense RNA intermediates. Furthermore, the non-structural proteins (Nsps) are essential in viral pathogenesis and are involved in the modulation of early transcription regulation, gene transactivation, evasion of antiviral response, immunomodulation in addition to participate in processes related to virus replication and assembly [[35], [36], [37], [38]].
Fig. 2.
Schematic diagram of SARS-CoV-2 genomic organization. The positive single stranded 5′ capped mRNA has a leader sequence (LS), poly-A tail at 3’ end, and 5’ and 3’ UTR. The genome comprises ORF1a, ORF1b, Spike (S), ORF3a,b, Envelope (E), Membrane (M), ORF6, ORF7a, ORF7b, ORF8, Nucleocapsid (N), ORF9 and ORF10. ORF1a and ORF1b cover the 5’ two-thirds of the genome and encode two large polyproteins, pp1a and pp1ab that are cleaved by papain-like cysteine protease (PLpro) and 3C-like serine protease (3CLpro) at the cleavage sites indicated by the black and grey triangles, into 16 non-structural proteins including the mentioned proteases, the RNA dependent RNA polymerase (RdRp), Helicase (Hel/MTase) and functional domains Exonuclease (ExonN), Endonuclease (Nendo U) and 2′-O-RNA methyltransferase (2`-O-MTase). The 3’ one-third end of the genome encodes the structural and accessory proteins.
After entry into host cells, the positive ssRNA viral genome acts as messenger RNA (mRNA), which is translated by host ribosomes resulting in the translation of the 2 co-terminal and large polyproteins that are further processed by proteolysis mediated by 3C-like protease (3CLpro), also known as Main protease (Mpro), and Papain-like protease (PLpro) enzymes to generate smaller proteins [39]. These two enzymes together with the RNA-dependent RNA polymerase (RdRp) protein are the major enzymes involved in the proteolysis, replication and production of new virions [32]. The resulting processed smaller proteins forms the replication-transcription complex and the newly structural proteins synthesized are then folded and packaged together with the replicated genomic RNAs into new virions that are released to infect new cells and spread the infection. Due to the crucial role of these major enzymes together with the structural spike protein in the process of infection, survival, replication and transmission of SARS-CoV-2, they have been considered as potential targets for drug design and development of vaccines.
Considering the similarity between SARS-CoV-2 and other beta-Coronaviruses, many peptides previously identified in SARS-CoV and MERS-CoV with potential to induce strong immune response were thought to work well for SARS-CoV-2. Nonetheless, different from SARS-CoV-2 RdRp and 3CLpro proteins, which share high sequence identity with the SARS-CoV and MERS-CoV, the spike protein S is significantly different, particularly in the two regions that interact with the host cell receptor ACE2. This specific feature explains the reason why some previously developed antibodies and peptides for SARS-CoV do not work efficiently against SARS-CoV-2 [33].
3. Mechanism of infection and SARS-CoV-2-induced human immune response
The first step in the infection by SARS-CoV-2 is the binding of the virus structural protein S to the host cell through its target receptor angiotensin-converting enzyme 2 (ACE2). The glycoprotein S consists of two subunits: the subunit S1 contains a receptor-binding domain (RBD) that interacts with the ACE2 receptor while the S2 subunit mediates the fusion of viral and host cell membranes via formation of a six-helix bundle fusion core [40,41]. The serine protease TMPRSS2 also participates in this process by cleaving the S protein and allowing the fusion of its S2 subunit with cellular membrane [42]. It is believed that the formation of neutralizing antibodies targeting RDB-S1 or S2 region may block binding of protein S to ACE2 and prevent membrane fusion and entry of the virus into cells, inhibiting SARS-CoV-2 infection [43,44].
To better understand immunity towards the SARS-CoV-2 it is important to recall the basic concepts of the immune response. Innate immunity is the first line of defense against pathogens and comes from germline through both myeloid hematopoietic such as neutrophils, monocytes, and macrophages, and non-hematopoietic lymphoid such as natural killer [NK] and γδ T cells. Adaptive immunity actions mainly via T cells and B cells characterized by their somatic genetic diversification of antigen-specific responses [45]. Acquired or adaptive immunity is established at the level of the individual, either through natural infection with a pathogen or through immunization with a vaccine [46].
A variety of clinical manifestations and different outcomes have been observed in patients with COVID-19 suggesting the complexity of the interaction of this new coronavirus with human host. It has been shown that immune response against SARS-CoV-2 involves different arms of the immune system including tissue barriers, innate and adaptive response as well as modulatory molecules mediators. The infection and destruction of lung cells triggers a local immune response recruiting immune cells that release cytokines and prime adaptive T and B-cell responses. In most individuals this process is capable of resolving the infection. However, in some cases a dysfunctional immune response occurs, which triggers a cytokine storm that mediates widespread lung inflammation and leads to a severe form of COVID-19. This severe disease may lead to damage to other organs including the heart and brain [[47], [48], [49]].
After infection, the median incubation period of COVID-19 has been estimated approximately 4–5 days before symptoms onset [6,50,51]. However it has also been shown that around 97 % of symptomatic patients develop symptoms within 11.5 days and it includes fever, dry cough and less commonly, difficulty in breathing, muscle and/or joint pain, headache, dizziness, diarrhoea and nausea [[52], [53], [54], [55], [56], [57]]. Some individuals also experience temporary loss of taste (dysgeusia) and smell (anosmia) [[58], [59], [60]]. Within 5–6 days of symptoms onset, SARS-CoV-2 viral load reaches its peak [4,61]. Although majority of SARS-CoV-2-infected individuals are asymptomatic or have mild symptoms, severe COVID-19 patients can progress to acute respiratory distress syndromes (ARDs), which occurs on average around 8–9 days after symptom onset and is characterized by difficulty in breathing and decreased blood oxygen level [3,8,53]. ARDs are the cause of death in 70 % of the fatal cases of COVID-19 [8]. Studies with SARS-CoV have shown that infection reduces ACE2 expression in lung cells and this downregulation is considered to be an important factor in the COVID-19 pathophysiology [62,63]. The reason for this is related to ACE2 regulate the renin-angiotensin system (RAS) which is known to modulate blood pressure and fluid electrolyte balance. Therefore, a reduction of ACE2 function would enhance inflammation and vascular permeability in the airways [64]. Earlier studies have shown that the virus targets specially airway epithelial cells, alveolar epithelial cells, vascular endothelial cells and macrophages in the lung where ACE2 is expressed [[64], [65], [66], [67]].
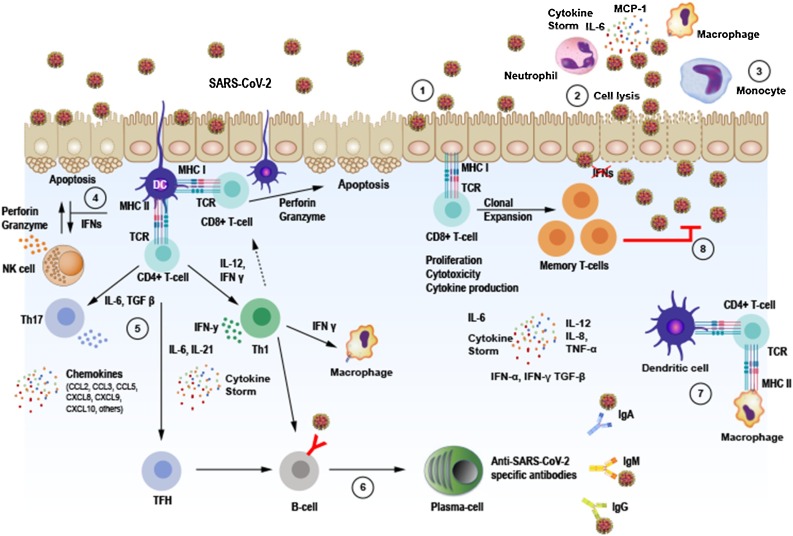
After epithelial or other type of cells are infected, the replicating SARS-CoV-2 can cause cell lysis and promote direct damage to the tissue. Infected epithelial cells may present virus antigens to CD8 + T cells, which together with natural killers (NK) cells become cytotoxic to the virus-infected epithelial cells, leading them to perforin/granzyme induced apoptosis. Dendrictic cells also can recognize antigens and present them to CD4 + T cells and induce their differentiation into memory Th1 and Th17 as well as memory T follicular helper (TFH) effector CD4 + T cells. Each cell subtype expresses different transcription factors, which regulate the function and cytokine secretion pattern of the cells and build the immune response. The TFH cells can help B-cells to divide into plasma cells (PC) and synthetize IgM, IgA and IgG anti- SARS-CoV-2 specific antibodies. The diversity of antibody production during viral infection demonstrate the ability of adaptive immune response to try to overcome the obstacles imposed by the virus. Tissue macrophages can also mediate antigen presentation to CD4 + T-cells (Fig. 3 ).
Fig. 3.
Immune response against SAR-CoV-2 infection. 1. SARS-CoV-2 infects ACE2 expressing target cells such as alveolar epitelial type 2 cells in the lungs. 2. Virus may overcome induced antiviral Interferon (IFN) responses leading to uncontrolled replication. 3. Neutrophils and monocytes/macrophages are recruited to the site of infection and may cause overproduction of pro-inflammatory cytokines such as IL-6, IL-8, IL-12,TNF-α and others, involved in the immunopathology of COVID-19 in the lungs known as “cytokine storm”. Both humoral and cellular immune responses are elicited. 4. Infected epithelial cells may present virus antigens to CD8 + T cells, which together with natural killers (NK) cells become cytotoxic to the virus-infected epithelial cells leading to apoptosis. 5. Dendrictic cells (DC) present virus antigen to CD4 + T cells and induce their differentiation into memory Th1 and Th17 as well as memory T follicular helper (TFH) effector CD4 + T cells. 6. Activated B-cells and plasma cells synthesize IgM, IgA and IgG anti- SARS-CoV-2 specific antibodies. 7. Macrophages and dendrict cells present antigens to CD4 + T cells via MHC-TCR interaction. 8. Memory T cells subset are produced and may provide immunity against reinfection with the same virus strain for a period still not well established.
Noteworthy, severe lymphopenia and eosinopenia are often observed and related to a defect in antiviral and immune regulatory immunity. T lynphocytopenia has been inversely correlated with increased peripheral pro-inflammatory cytokines in COVID-19 patients [68,69]. Furthermore, low CD8 + T cell count has been considered a predictor for high mortality risk and illness severity [70]. These observations support the hypothesis that unbalanced adaptive immune responses can potentially induce detrimental effects on acute infected patients. Also, it has been suggested that SARS-CoV-2 may infect human T-cells via a novel route that involves the CD147 receptor, known as Basigin or EMMPRIN, expressed on the surface of T-lymphocytes [71,72]. Although still controversial, the interaction between SARS-CoV-2 and T cells could interfere with immunity. CD147 plays a role in differentiation, cell proliferation, migration, inflammation and apoptosis [73]. Thus, activation of downstream CD147 signaling pathway could lead to T-cell apoptosis or contribute to severity of COVID-19, however there is no study addressing this issue and a recent study found no evidence that the proposed CD147 can act as receptor for SARS-CoV-2 [74].
SARS-CoV-2- specific CD4+ and CD8 + T cells have shown to exhibit strongest response directed to the spike protein and produce effector and Th1 cytokines, such as interferon gama (IFN-γ) and tumor necrosis factor alpha (TNF-α), in addition to Th2 and Th17 cytokines, such as interleukin (IL)-4, IL-5, IL-13 and IL-17. While Th1 cells are responsible for cell-mediated immune responses, Th2 are responsible for humoral-mediated immunity [75]. It has been shown that IL-17 can act on antigen-presenting cells (APCs) such as macrophages and dendrictic cells and induce cytokine and chemokine production [76]. SARS-CoV-2 T-cell responses have been detected not only to the spike protein S but also to M, N and other ORFs encoded proteins, although the response was more robust against spike protein, the main target of the most vaccines [77]. Thus, cytokine kinetics during COVID-19 has been crucial for understanding the fine balance between immunity and inflammation at different sites of infection.
Interestingly, reactive T cells was also detected in some health control patients not previously exposed to SARS-CoV-2, indicative of possible cross-reactivity due to past infection with coronaviruses probably from “common cold” [78]. SARS-CoV-2 seropositivity rates have been shown to vary from 2 to 73% in individuals who have probably not been exposed to the virus [79]. It is known that at least seven types of coronaviruses naturally infect humans. Besides SARS-CoV-2, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) can cause severe acute respiratory illnesses. On the other hand, four endemic genotypes, such as 229E, NL63, OC43 and HKU1, usually cause mild upper respiratory tract infections, and thus can be classified as low-pathogenic human coronaviruses [80]. Therefore, considering the genetic relationship among different coronaviruses, it is not surprising that cross-reactivity may occur and be detected in serologic assays at different degrees and in different localities. Notwhithstanding, genetically attenuated virus can induce antibodies in animal models that neutralize both SARS-CoV and SARS-CoV-2 [17]. However, when spike protein is used as antigen, human sera seems to contain only antibodies that neutralize one type of virus [81].
Not less important, genetic susceptibility to infectious disease has a close relation to the major histocompatibility complex antigen loci (HLA) [82,83]. Antigen receptors on CD4+ or CD8 + T-cells recognize the conformational structure of the antigen-binding -grove together with the associated antigen peptides. Thus, HLA molecules with increased binding specificities to SARS-CoV-2 virus peptides on the cell surface of presenting cells seems to be advantageous to individuals with determined HLA haplotype. Notwhistanding, host factors and population heterogeneity can contribute significantly to variability in cross reactivity and susceptibility to infection, specially due to the major histocompatibility complex antigen loci (MHC-HLA) [84] and possibly to polymorphisms in the gene ACE2 that encodes the receptor used for virus entry into host cells. The latter has been subject of controversial debate and need more studies to reach a conclusion [85]. MHC class I HLA-B*46:01 genotype was shown to have the fewest predicted binding peptides for SARS-CoV-2, suggesting that individuals with this allele may be particularly vulnerable to COVID-19, as they were previously shown to be for SARS. In contrast, HLAB*15:03 showed the greatest capacity to present highly conserved SARS-CoV-2 peptides that are shared among common human coronaviruses, suggesting that it could enable cross-protective T-cell-based immunity [86].
COVID-19 severe patients experience symptoms where pneumonia, acute respiratory distress syndrome and cytokine storm become part of a complex immunopathological condition [6,87]. Development of neutralizing antiviral T-cell and antibody immunity is observed in the majority of patients with self-limiting viral respiratory disease at one week post infection. Immunoglobulin (Ig)M and IgA titers against SARS-CoV-2 can be detected in median 5 days after symptom onset while IgG titers, 14 days [88]. These antibodies have been used as a measurement to predict population exposure to SARS-CoV-2 as well as to determine the cross-reactivity with other coronaviruses [89]. Regarding the duration of anti-SARS-CoV-2 antibodies in individuals that have recovered from COVID-19 there are no conclusive response. In one study, anti-SARS-CoV-2 IgG responses have been detected in most patients with either severe or mild disease, at 9 days after onset of infection and the antibody levels remained high throughout the study period which endured about 35–40 days [90]. On the other hand, titers of anti- SARS-CoV-2 IgG antibodies were found to decay in asymptomatic and early convalescent individuals after 90 days of onset of symptoms [91,92]. New studies have shown that IgA and IgM antibodies against RBD region of the spike protein were short-lived with median times to seroreversion of 71 and 49 days after symptom onset. Nevertheless, IgG antibodies decayed slowly through 90 days and these antibodies strongly correlated with anti S-neutralizing antibody titers [93]. Additionally, another study reported detection of IgA, IgM and IgG anti -SARS-CoV-2 not only in serum but also in the saliva of acute and convalescent patients and IgG could be detected for up to 3 months [94]. Interestingly, it has been reported cases of people presenting positivity for SARS-CoV-2 in molecular tests without detectable levels of protective IgG antibodies. Moreover, a recently published work analysed the antibody levels in 254 patients who had COVID-19, during a period of five months and demonstrated that SARS-CoV-2 IgG antibodies progressively decreased in outpatient and asymptomatic patients during observation up to five months post infection [95]. Together, these results indicate that acquired immunity by people with COVID-19 may not be lasting suggesting concerns about the long term efficacy of potential vaccines being developed. On the other hand, the apparent controversy on the durability of IgG could possibly be explained by variations in the methodology employed in different studies or be indicative of a more relevant role of cellular protective immune response. The most common technique used to measure anti–SARS-CoV-2 antibody titers is an enzyme-linked immunosorbent assay (ELISA). While some authors measured anti spike protein receptor-binding domain (RBD) IgG antibodies, others measured antibodies anti whole spike protein or nucleocapsid from SARS-CoV-2 [96]. As mentioned before, spike protein is present in a variety of coronaviruses and may be an important factor to be considered in terms of cross-reactivity and interpretation of kinetics of antibodies against SARS-CoV-2 infections. Also, some cases of putative reinfection have been recently reported suggesting the possibility of waning immunity and arguing in favor of a decay in antibody titers overtime. Although reinfection is currently subject of intense debate, a number of studies has been reporting cases of a second episode of positive SARS-CoV-2 infection after recovery [97]. In some cases, the patient follow up was not well conducted and the appropriate control tests were not performed, casting doubt about a true reinfection and suggesting only a persistent virus shedding [98]. A series of six cases reported in Brazil, suggest the possibility of reinfection but the results could also be due to viral reactivation [99]. On the other hand, a second episode of COVID-19 was reported in an asymptomatic patient from Hong Kong following a first symptomatic episode. SARS-CoV-2 whole genome sequencing was performed directly on respiratory specimens collected during the two episodes of COVID-19. Interestingly, viral genomes from first and second episodes were shown to belong to different clades/lineages [100]. If the virus can be reactivated after a while following recovery from a infection is an open question. Therefore, the possibility of true SARS-CoV-2 reinfection remains unclear and further studies are required.
Although it is not clear the mechanisms by which SARS-CoV-2 can subvert the body’s innate antiviral cytokine responses in some patients, it is known by studies with other coronaviruses, that multiple viral structural and non-structural proteins can antagonize interferon responses. Some identified mechanisms include preventing recognition of virus RNA, preventing downstream interferon signaling pathway or inducing host mRNA degradation and inhibiting host protein translation [[101], [102], [103]].
On the other hand, dendritic cells play an important role in immune responses by their plasticity and unique ability to induce naïve T cell activation, coordinate and regulate adaptive immune responses. They are a heterogeneous group of cells that act as the strongest antigen-presenting cells (APCs) and effectively stimulate the activation of B and T lymphocytes, thus combining innate and adaptive immunity. Notwithstanding, it has been shown that dendritic cells were significantly reduced or showed functional impairement in acute COVID-19 patients [104]. Dendritic cells are targeted through the interaction of virus S protein with dendritic cell –specific intercellular adhesion molecule 3–grabbing non-integrin (DC-SIGN) [105]. This adhesion molecule was initially discovered as an attachment factor for HIV virus and by interacting with viral glycoproteins augments infection [106]. DC-SIGN and the related protein DCSIGNR (also termed L-SIGN) have been shown to enhance infection in a variety of viruses including SARS-COV [107,108]. By targeting the C-type lectin DC-SIGN, viruses can subvert dendritic cell functions to scape immune surveillance [109].
Viral proteins such as “spike” are exposed to adaptive immune response and are the main targets of host antibodies [110]. The mechanisms underlying the generation of specific antibodies directed to a virus infection involve the production of antigenic peptides from viral proteins by the immune system B-cells. These peptides bind to the major histocompatibility complex (MHC) and then are presented at the cell surface to other cell subsets. The naïve B-cells can also become stimulated by another type of cell named helper-T-cell and become a plasma cell able to produce antibodies. The diversity of antibodies generated by different stimulated B-cells have different binding epitope and protein sequences in a specific region of the antibody which may confer different degrees of affinity to antigens. Nevertheless, viruses in a broad sense, have developed mechanisms to escape immune responses through rapid evolution of antibody-targeting epitopes, steric shielding of epitopes by glycan post-translational modifications, immune decoys such as soluble antigens that share viral spike epitopes, and immunosuppression to evade host recognition upon cellular entry [110]. Surface proteins outside of the virus structure are generally selected for antigens aiming therapeutic use, so that antibodies generated from a vaccine-primed B-cell can bind to the virus for neutralization. However, strain-specific antigens introduced as vaccines may have its use limited if variability somehow alter the antigen protein structure that may act as a mechanism of scape from the immune recognition [111].
Some of the stimulated B and T lymphocytes may become memory cells and persist for months or years in the body, allowing the mounting of faster and stronger responses in case of a new virus infection. Trained immunity is a term used for immunological memory and is thought to affect and prevent spread of virus infection. This property of lymphocytes is the basis of vaccine efficacy against specific infections [112].
Development of immunological memory and persistence of virus recognition memory responses are key for the long term-protection from SARS-CoV-2 reinfection and the durability of potentially successful vaccines. The humoral and cellular memory responses was analyzed in 15 COVID-19 recovered individuals who presented mild symptoms. Sustained neutralizing IgG antibodies and memory B-cells as well as SARS-CoV-2 specific memory T-cell were detectable up to 3 months and more than 3 months post symptom onset respectively [113,114]. The molecular mechanisms underlying long term reprogramming of immune cells are epigenetic in nature. To reach the memory status, the previously challenged cell needs to access regions of the genome that contain the target sequences and the regulatory elements of the genes involved in these processes. This process is regulated through durable epigenetic modifications, which allow unfolding of the chromatin and accessibility of transcription factors to the promoter and enhancer regions of the involved immune-related genes [115].
4. SARS-CoV-2 target proteins and strategies for vaccine development
Structural proteins that are exposed at the viruses surface are more likely targets for a vaccination approach. These include the envelope spike protein S, the small envelope protein E, the matrix protein M and the nucleocapsid protein N, although the latter is unexposed at the surface. The spike S protein is a glycoprotein composed of 1273 amino acids with three subunits S1, S2 e S2’ and is the major component of the SARS-CoV-2 envelope [116]. It is essential for host receptor binding and virus entry, and among the other viral proteins is the main focus of vaccine development. The three subunits of S protein act differently during the process of binding to the host cell receptor ACE2 and undergo conformational changes induced upon its entry into the endosomes of the host cell [33]. S1and S2 subunits form functional prefusion trimer after proteolitic cleavage. The RBD region of the S1 domain undergoes a hinge like conformational movement, which is an important determinant of host cell receptor binding [117]. Interestingly, SARS-CoV-2 S protein has a 4 amino acid insertion (PRRA) in the S1/S2 cleavage site which is different from SARS-CoV. This insertion results in a polybasic RRAAR furin-like cleavage motif that enhances infection in lung cells [26,118]. This demonstrate that the RBD region in the S protein is the most variable part of the SARS-CoV-2 and has implications with the virus pathogenesis. Potential targets for a putative SARS-CoV-2 vaccine were identified based on previous immunological studies performed with the beta coronavirus SARS-CoV. In one study, a set of B- and T-cells epitopes derived from the spike and nucleocapsid proteins that map identically to SARS-CoV-2 proteins were identified. The screening of these epitopes took into consideration only one hundred twenty SARS-CoV-2 genomic sequences available at the beginning of the COVID-19 pandemic (February 2020), and no mutation were observed in these epitopes at that time. A population coverage analysis of the associated MHC alleles was performed and based on that, it was proposed an estimated set of epitopes that could provide broad coverage [119]. It seems that the entire RBD region remains conserved in SARS-CoV-2 isolates, and some rare non-synonymous mutations in the S protein have been described V483A, L455I, F456 V and G476S [120]. However, as new genomic sequences are known novel mutations may be revealed that could impact the pathogenicity of SARS-CoV-2.
As of December 7th 2020 there was more than 245,000 genomic sequences available from SARS-CoV-2 isolated from different countries at different times since December 2019, according to the Global Initiative on Sharing All Influenza Data [121]. Therefore, it would be important to keep the analysis of SARS-CoV-2 genomes to identify and determine the potential biological effects of novel mutations in the epitopes identified preliminarly.
5. Brief history of vaccines, challenges and types of COVID-19 vaccines under development
The known vaccines currently available for different diseases such as Haemophilus influenzae type b, measles, mumps, polio, rubella among others, prevent millions of illnesses worldwide and immunization currently prevents 2–3 millions deaths every year, according to the World Health Organization [122]. Most vaccines have been created by adaptation of living organisms to growth conditions that attenuate their virulence, by preparation of suspensions of killed microrganisms or concentration of proteins or polysaccharides purified from pathogens [123,124]. However, modern technologies have provided new tools with which vaccines can be developed. These technologies have opened new possibilities to explore the cellular immune responses in addition to antibody responses essential for the success of almost all vaccines [125,126].
The first vaccine ever developed has been attributed to Edward Jenner in 1798, who used material from cowpox pustules to prevent smallpox. However, the origin of the first vaccine happened long before, as a consequence of the occurrence of infectious disease in humans. There is evidence that the Chinese have employed smallpox inoculation or variolation, as such use of smallpox was named, as early as 1000 CE [127]. With the medical and technological advances over the next 200 years from the first vaccine, smallpox was considered eradicated in 1980, making vaccinia one of the most successful vaccine to date. However, it is important to note that significant side effects in the first recipients and serious, sometimes fatal, effects were observed in a proportion of individuals. This was particularly due to the lack of quality control at the initial phases of this vaccine [124]. Following the smallpox vaccine, in 1885 Louis Pasteur’s rabies vaccine was the next to demonstrate a great impact on human disease. Pasteur introduced the concept of attenuation by exposing bacterium to adverse growth conditions. Since then, a number of successful vaccines have been developed, although amid of controversies, like the two poliovirus vaccines which employed both inactivated and live virus. The former developed by Jonas Salk [128] and the latter by Albert Sabin [129]. In the 1960s three attenuated-virus vaccines were developed: one against measles virus, other against mumps virus and another against rubella virus. In the 1970s, varicella zoster vaccine was developed based on the principles of virus attenuation by passage in guinea pig cells. The 1980s were marked by the emerging of two important strategies for vaccine development: the conjugation of bacterial capsular polysaccharides to proteins and the recombinant DNA Technology, also known as genetic engineering. The first strategy was important in the initial development of Haemophilus influenzae type b vaccine and later in the development of conjugated vaccines by coupling diphtheria toxoid to the H. influenzae type b capsule and diphtheria or tetanus toxoids conjugated to meningococci and pneumococci, which were very efficacious to almost eliminate the diseases caused by meningococci and pneumococci in countries that used these vaccines. The second strategy led to the development of a vaccine against hepatitis B virus, the first to use genetic engineering approach, and then other vaccines were developed like the vaccine against human papillomaviruses, lyme disease, rotaviruses and yellow fever. Thus, the advent of the recombinant DNA technology opened a new era for the development of vaccines [124].
Rational development of vaccines began to be used in the mid-20th century, when immunology advanced to a point of distinguish protection mediated by antibodies and that mediated by lymphocytes and when passage in cell culture permitted the selection of attenuated mutants [130]. Later, other tools and approaches have been used to develop successful vaccines, such as protection studies in animals, by inference from immune responses with demonstrated protection against repeated natural infection [131], and from the use of passive immunization (antibodies) against specific antigens to verify if those antigens should be included in vaccines. However, along the history of vaccine development became clear that the success of a potential vaccine candidate depends on understanding of which type of immunological response is protective. While some vaccines induce a protective humoral response others depend on cellular immunity as it became clear in the case of M. bovis bacille Calmette–Guérin and varicella zoster virus vaccines for instance [132,133]. As new pathogenic virus emerge, more knowledge we need on the mechanism of pathogenicity and how the immune system is affected to use appropriate tools to develop effective vaccines. This has been important if we consider diseases for which natural immunity is absent or inadequate such as HIV/AIDS and recently, the new coronavirus SARS-CoV-2 for which efforts to develop a vaccine has resulted in hundreds of putative candidates at unprecedented speed.
Despite all the advances in the field of vaccinology concerns about vaccine safety still persist for some types of vaccines and include the fear that attenuated or inactivated virus vaccine can replicate into human cells and spread in the population or recombine resulting in new pathogenic strains [134].
6. Available platforms for the development of vaccines
Along the history of vaccine development several strategies were used based on the knowledge of the specific pathogens. Inactivated and live attenuated vaccines were the two platforms used in the beginning. With the emergence of breakthrough technologies new platforms were designed.
Inactivated vaccines use chemicals or physical methods or a combination of the two to inactivate the virus. Some of the agents used include ascorbic acid, ethylenimine derivatives, psorlens hydrogen peroxide, gamma irradiation, UV treatment, heat, formaldehyde and β-Propiolactone. However, only the two latter have been widely used for inactivation of licensed human viral vaccines in the last decades [135]. The live attenuated vaccine requires a genetic manipulation to create weak versions of the virus that limit the replication process, cause no disease but it is able to induce immune response similar to the ones observed in natural infections. Although live attenuated vaccines have been associated with genetic instability and residual virulence [136], several strategies are used to deal with these issues including recombination, deletion mutants, codon deoptimization, recombination and control of replication fidelity [123]. Different approaches can be used to achieve virus attenuation such as growing the virus under unfavorable conditions like suboptimal temperature and different host cells or genetically, by deleting genes involved in counteracting innate immune recognition or by codon deoptimization [137,138].
Protein based vaccines consist of specific imunogenic proteins purified from the virus or virus-infected cells. It can also consist of recombinant proteins or supramolecular structures known as virus-like particles (VLPs). VLPs may contain copies of one or more viral proteins that assemble into nanoparticles of 10−200 nm [139]. They resemble viruses but are replication-deficient since they do not carry viral genetic material. For this reason they are considered safer than whole-virus based vaccines [140,141].
Vectored vaccines are based on a carrier virus such as an adeno or pox virus which has been modified to carry a gene from a virus of interest. When this vaccine is given to a recipient, the gene will be expressed and protective immune responses will be generated. The key aspect of the vectored vaccines is the gene they carry. In some cases the relevant gene to be used in a vaccine is obvious like the haemagglutinin of measles or the G protein of rabies virus, however in practice, more genes may be required to elicit a protection than a immune response to a single antigen. Although vectored vaccines have been subject of development by many biotech companies and generated licensed gene therapy products and veterinary vaccines, no licensed human vaccines has been granted so far [124]. Some factors may contribute for this result including completeness, duration and protective efficacy of the immune response generated, commercial concerns related to large scale production to meet global demand and availability of other easier options [124]. Among vectored vaccines there are two categories: Non-Replication vectors and replication vectors. While non-replication vector vaccines enter cells and produce the vaccine antigen but are unable to replicate, the replication vector vaccines infect cells and besides the production of pathogens specific antigen they also are able to replicate and produce infectious viral vectors that will then infect new host cells and express more antigens able to stimulate a immune response.
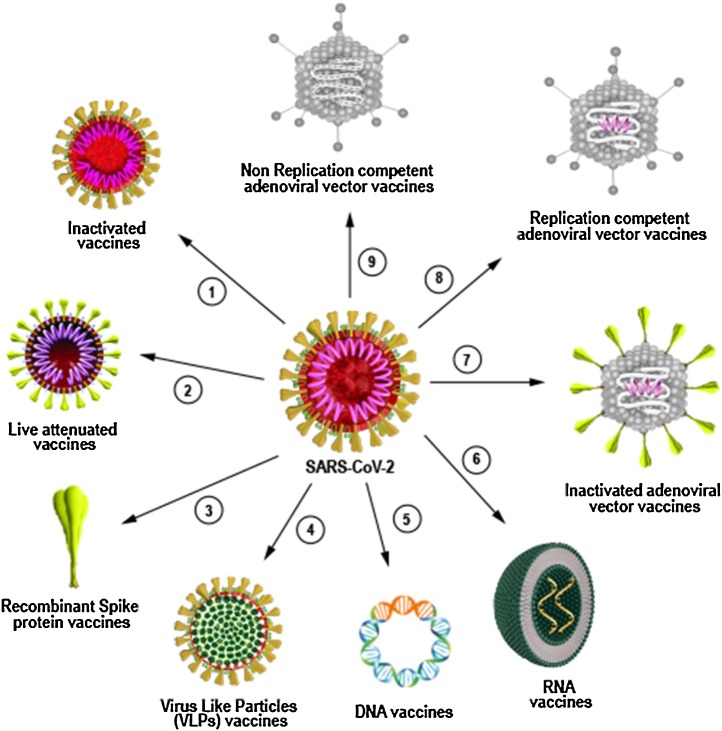
Finally, nucleic acid -based vaccines uses DNA or RNA as strategy to express imunogenic viral proteins. DNA vaccines are based on plasmid DNA to carry pathogen genes and can be produced in large scale. RNA vaccines work on the same principle as DNA based vaccines except it does not need to be translocated to the nucleus to be transcribed into RNA to be expressed. They can be based on mRNA or self-replicating RNA. However, considering the fact that mRNAs are not very stable they are synthesized with modified nucleosides to prevent degradation and a carrier molecule, such as lipid nanoparticles, is necessary to enable entry of the RNA into cells [142] (Fig. 4 ).
Fig. 4.
Different platforms used for development of COVID-19 vaccine. Classical and next generation platforms are currently being used to develop vaccines against SARS-CoV-2. Inactivated (1) and live attenuated vaccines (2) are classical platforms that uses the whole SARS-CoV-2 virus as vaccine. Since the viruses used to produce the vaccine do not replicate either by chemical inactivation or genetic modification, the vaccines require adjuvants to induce optimal immune response. Recombinant protein platform (3) uses a broad range of technologies to prepare viral proteins such as SARS-CoV-2 Spike, as imunogen to induce immune response. Viral-like particles platform (VLPs) (4) are example of new technology platform and contain copies of one or more viral proteins that assemble into nanoparticles of 10-200 nm and are used as vaccine. DNA (5) and RNA (6) vaccines are also considered new generation platforms and consist of nucleic acids containing part of virus genetic material that can be delivered by electroporation intradermally or by lipid nanoparticles respectively. Inactivated adenoviral vectors (7), replication competent adenoviral vectors (8) and non replication competent adenoviral vectors (9) are known as viral-based vectors vaccine and use genetic modified adenovirus as vector to carry SARS-CoV-2 imunogenic protein coding sequences to induce immune response.
6.1. COVID-19 vaccines under development
Pontential antivirals and other treatment alternatives under development are important to treat infected individuals and to decrease disease burden during current COVID-19 pandemic. However, only effective vaccines will be able to prevent and control the disease as expected. Several vaccines against SARS-CoV-2 are currently in clinical trials at different phases, some of them reaching the final steps to be licensed in different countries (Table 1 ). By 22nd september 2020, the World Health Organization (WHO) estimated that 38 candidate vaccines were in clinical evaluation and 149 in pre-clinical evaluation [16]. These vaccines have been developed using different platforms including non-replicating or replicating viral vectors, live attenuated virus, inactivated virus, RNA, DNA, recombinant protein subunits and virus-like particles (VLPs) vaccines.
Table 1.
Main vaccines against SARS-CoV-2 under Phase II/III clinical trial.
| Name | Developer | Platform | Target | Status |
|---|---|---|---|---|
| AZD1222 (ChAdOx1nCoV19) | University of Oxford & AstraZeneca | Adenovirus (non-replicating) | Spike protein | Phase III |
| Ad5-nCoV | CanSino Biologics | Adenovirus (non-replicating) | Spike protein | Phase II |
| Ad26-SARS-Cov-2 | Johnson & Johnson - Jannssen | Adenovirus (non-replicating) | Spike protein | Phase II/II |
| Sputinik V | Gamaleya Research Institute of Epidemiology and Microbiology | Heterologous Adenovirus -Ad5 and Ad26 (non-replicating) | Spike protein | Phase IIIa |
| INO-4800 | Inovio Pharmaceuticals | DNA | Spike protein | Phase I/II |
| Unnamed | Osaka University/ AnGes/ Takara Bio | DNA | Spike protein | Phase I/II |
| Unnamed | Cadila Healthcare Limited | DNA | undisclosed | Phase I/II |
| GX-19 | Genexine Consortium | DNA | Spike protein | Phase I/II |
| BNT162 | BioNTech and Pfizer | RNA | 3CLpro, NSP5, Mpro, other | Phase IIIb |
| mRNA-1273 | Moderna and NIAID | RNA | Spike protein | Phase III |
| Unnamed | Curevac | RNA | Spike protein | Phase I/II |
| ARCT-021 | Arcturus/Duke-NUS | RNA (Lipid nanoparticle) | Spike protein (prefusion) | Phase I/II |
| Unnamed | Wuhan Institute of Biological Products and Sinopharm | Inactivated virus | Whole virus | Phase I/II |
| BBIBPCorV | Beijing Institute of Biological Products and Sinopharm | Inactivated virus, plus adjuvant | Whole virus | Phase I/II |
| CovidVax | Institute of Medical Biology and Chinese Academy of Medical Sciences | Inactivated virus | Whole virus | Phase I/II |
| CoronaVac (PiCoVacc) | Sinopharm/Sinovac Biotech | Inactivated virus, plus adjuvant | Whole virus, (Spike RBD main immunogen) | Phase III |
| Unnamed | Research Institute for Biological Safety Problems, Rep of Kazakhstan | Inactivated virus, plus adjuvant | Whole virus | Phase I/II |
| Covaxin | Bharat Biotech | Inactivated virus | Whole virus | Phase I/II |
| EpiVacCorona | Vector Institute - novosibirsk | Protein subunit | Synthetic peptide antigens | Phase I/II |
| SCB-2019 | Clover Biopharmaceuticals | Protein subunit | Spike trimer | Phase I/II |
| NVX-CoV2373 | Novavax | Protein subunit | Spike protein (prefusion) | Phase I/II |
| Unnamed | Anhui Zhifei Longcom Biopharmaceutical/Institute of Microbiology, Chinese Academy of Sciences | Protein subunit | Spike protein (RBD dimer) | Phase I/II |
| Unnamed | Kentucky Bioprocessing, Inc | Protein subunit | Spike protein (RBD) | Phase I/II |
| Unnamed | Sanofi Pasteur/GSK | Protein subunit (baculovirus production) | Protein subunit | Phase I/II |
Vaccination campaign started in Russia on December 5th, 2020.
Vaccination programme started in UK on December 8th, 2020.
Source: ClinicalTrials.gov.
Thus more than 180 vaccines against SARS-CoV-2 using different platforms are currently under development or already being tested in humans. Previous work on vaccines developed for MERS-CoV and SARS-CoV have shown that both humoral and cellular immune responses are important to induce protection against infection [143,144]. In this context, some vaccines based on recombinant viral vectors have demonstrated this ability to induce both responses and built protective immunity after one or two doses [145,146]. However, vaccination experimentation against SARS-CoV-2 in non-human primates have shown that neutralizing antibodies, but not T-cell response correlates with protection [147]. Furthermore, it has also been shown that in these animal models, infection with SARS-CoV-2 is able to protect against a re-infection [148].
SARS-CoV-2 inactivated vaccines have been produced by growing the virus in cell culture, such as Vero cells, and performing chemical inactivation [149,150]. Examples of this type of vaccine include the vaccine named coronavac, developed by the Chinese company Sinovac Biotech Ltd., and the vaccines developed by the companies Bharat Biotech in India and by the Research Institute for Biological Safety Problems in Kazakhstan. Inactivated vaccines are generally administered via intramuscular and requires an adjuvant to induce immune response. This type of vaccine elicits immune response directed to different virus proteins and not only S-protein since the whole virus is being used to challenge the immune system. A pilot-scale production of an inactivated vaccine candidate, named BBIBP-CorV, was reported that is able to induce high levels of neutralizing antibodies titers in mice, rats, guinea pigs, rabbits, and nonhuman primates. This vaccine was given at two-dose immunization regimem and provided efficient protection against SARS-CoV-2 intratracheal challenge in rhesus macaques [150].
A heterologous COVID-19 vaccine consisting of a recombinant adenovirus type 26 (rAd26) vector and a recombinant adenovirus type 5 (rAd5) vector was recently developed in Russia. In this vaccine both vectors carry the SARS-CoV-2 Spike protein gene. In two open, non-randomised phase 1/2 studies (clinical trials NCT04436471 and NCT04437875) it was demonstrated that two formulations (frozen and lyophilised) of the vaccines presented good safety profile and induced strong humoral and cellular immune response in the 76 enrolled participants [151]. This vaccine named Sputnik V, has just completed phase III clinical trial and was approved for vaccination in Russia beginning December 5th 2020.
A randomised, double-blind, placebo-controlled, phase two trial was performed to assess the immunogenicity and safety of one candidate non-replicating adenovirus type-5 (Ad5)-vectored COVID-19 [152]. 508 participants 18 years or older eligible to participate were randomly assigned to receive the vaccine (1 × 10¹¹ viral particles n = 253; 5 × 10¹⁰ viral particles n = 129) or placebo (n = 126). It was shown that both doses of the vaccine induced significant neutralising antibody responses to live SARS-CoV-2 after a single immunisation. No serious adverse reactions were documented but solicited adverse reactions were reported in 73 % and severe adverse reactions in 9 % of the participants [152].
The university of Oxford and AstraZeneca developed a recombinant vaccine (AZD1222), formely known as ChAdOx1 nCoV-19, by using a non-replicating chimpanzee adenovirus to deliver a SARS-CoV-2 spike protein to induce an immune response. Preclinical studies indicated that this vaccine is able to induce rapid immune responses mediated by type -1 and type-2 T helper cells against SARS-CoV-2 in mice and rhesus macaques [153]. A phase 1/2, single-blind, randomised controlled trial with 1077 enrolled volunteers demonstrated acceptable safety profile and induction of both humoral and cellular immune responses [154]. Currently this vaccine entered a multi-site phase III clinical trial in the United States and is being evaluated in Phase 2/3 trials in the U.K. and Brazil and in a Phase 1/2 trial in South Africa. The Janssen Pharmaceutical Companies of Johnson & Johnson developed the recombinant adenoviral vector vaccine JNJ-78436725 also known as Ad.26.COV2.S. Preclinical studies demonstrated that the vaccine induced robust neutralizing antibody responses and provided complete or near-complete protection in bronchoalveolar lavage and nasal swabs after SARS-CoV-2 challenge [155]. This vaccine is currently at phase III clinical trial.
NVX-CoV2373 is a recombinant nanoparticle vaccine consisting of trimeric full-length SARS-CoV-2 spike glycoproteins, with the polybasic cleavage site deleted and the two stabilizing proline mutations, and Matrix-M1 adjuvant. Phase 1–2 clinical trial was performed with 131 healthy adult participants 18–59 years of age where two-dose regimens of 5 μg and 25 μg of rSARS-CoV-2 plus the Matrix-M1 adjuvant were used. The results demonstrated absence or mild reactogenicity, which was more common in participants who received the Matrix-M1 as adjuvant but no severe adverse events. This vaccine was able to elicit immune responses that exceeded levels in COVID-19 convalescent serum and the Matrix-M1 adjuvant induced CD4 + T-cell responses that were biased toward a Th1 phenotype [156].
Many advanced vaccine candidates have been using emerging technology platforms. The mRNA-1273 developed by Moderna is an example of nucleotide based vaccines. Similar to tradicional live-virus vaccines, it delivers a genetic sequence into a host cell and co-opt host machinery to express antigens of interest. However, the mRNA template is transported by synthetic lipid nanoparticle instead of weakened SARS-CoV-2. In this vaccine the spike protein is the target used to elicit a immune response. Another nucleic acid based vaccine named BNT162 mRNA was developed by Pfizer and BioNTech. It passed phase III clinical trial enrolling up to 44,000 participants globally and it was approved on December 2nd for emergency use by the Medicines & Healthcare Products Regulatory Agency (MHRA) in the United Kingdon, paving the way for mass vaccination. This vaccine became the first one to be granted emergency use by a health regulatory agency.
A next generation vaccine strategy was reported where antigen optimization and nanoparticle display were combined to create a self-assembling protein nanoparticles (SApNPs) to use receptor binding domain (RBD) from SARS-CoV-2 spike protein as antigen. The nanoparticles induced neutralizing antibody response as well as T-cell immunity in murine animal model [157].
It has been proposed that the bacillus Calmette-Guérin (BCG) vaccine against tuberculosis could reduce the severity of COVID-19. This hypothesis was based on epidemiological studies suggesting a negative association between BCG vaccination policy and the prevalence and mortality of COVID-19 [158,159]. The supposed protection effect is thougth to be mediated by the general long-term boosting of innate immune mechanisms [160]. Interestingly, the hypothesis that BCG vaccine may protect against unrelated infectious agents and specially respiratory tract infections, particularly in children, was strengthened by a study in Guinea-Bissau showing that BCG reduced the incidence of respiratory syncytial virus infection in Indonesia [161,162]. Studies trying to understand the cellular and molecular mechanisms of this effect has revealed some interesting results. BCG vaccination in healthy volunteers leaded to enhanced production of pro-inflammatory cytokines including IL-1β, tumour necrosis factor (TNF) and IL-6, following ex-vivo stimulation of monocytes with unrelated pathogens. Additionally, transcriptional, epigenetic and metabolic reprograming were observed in myeloid cells of vaccinated individuals [163,164]. BCG vaccine is a live attenuated vaccine that was developed at the beginning of the 20th century against tuberculosis. While there are data suggesting a protective effect of BCG vaccine, there are no definitive proof of causality. A phase III clinical trial double-blind, randomized placebo-controlled is currently ongoing to compare the efficacy of BCG vaccination to that of placebo in reducing severity of COVID-19 (NCT04534803). Similar study is being performed in Brazil with Healthcare workers in a research collaboration between Oswaldo Cruz Foundation (Fiocruz) and the Murdoch Children’s Research Institute, in Australia.
Despite all the challenges imposed by COVID-19 pandemic as demonstrated above we are being able to at least develop promising vaccines candidate at record speed. Based on the preliminary results obtained so far for different vaccines there is reason to believe that successful vaccines will be soon released for mass vaccination and decrease the mortality observed around the world. Nevertheless, even if COVID-19 vaccines are developed, continuing follow up must occur to assess how long the protection will last and how the dynamics of transmission will change overtime. Vaccine-induced immunity wanes overtime and the protection level differs with each disease. As it happens with influenza, mumps, pertussis, and yellow fever, it is important to realize that immune responses against all these diseases, may disappear at a faster rate than appreciated, and this fact call the attention for the timing of booster shots recommended by health officials [165]. For instance, it has been demonstrated by assessing transmission dynamic of measles in Japan, that despite verification of measles elimination, multiple generations of transmission have occurred following importation events in 2016. Therefore it was suggested that despite vaccination, because importation events may continue, supplementary vaccination of adults should be considered [166].
Additionally, the question about how long a vaccine-induced protection can last was subject of another study aiming to investigate 10 years immune response and long term safety profile of a vaccine. It was evaluated the effect of human papillomavirus HPV-16/18 AS04-adjuvanted vaccine in females aged between 15 and 55 years at first vaccination. After 10 years, seropositivity rates for anti-HPV-16 remained high (≥96.3 %) in all age groups while seropositive for anti-HPV-18 was 99.2 % for aged group 15−25-years old compared to 93.7 % and 83.8 % for 26−45-years old and 45−55-years old group, respectively [167]. In this case, vaccine elicited sustained immunogenicity suggesting long term protection against HPV, however, this is not always true for all vaccines.
Thus, it is important to keep the surveillance since we still do not have the whole picture of COVID-19 natural history and the role of mutations in the emergence of new SARS-COV-2 sub-strains with novel properties as discussed below.
7. The impact of virus mutations for the transmissibility, pathogenesis and efficacy of potential vaccines
Genetic variability of viruses due to mutations is of considerable medical and biological relevance as it has great impact not only in the prevention and diagnosis of infectious disease but also for potential therapy perspectives. Mutations can be considered as the ground for evolution since they can provide the necessary variation upon which the natural selection can act and generate diversity [168]. Nevertheless, not always mutations are beneficial to the organism, in fact most of them are not and may have as consequence organisms leaving fewer descendants overtime. In contrast, the mutations that are beneficial may provide enough diversity to survival and better fitness of an organism facing an changing environment.
RNA virus in general have a higher mutation rate compared to their hosts and these high rates are correlated with enhanced virulence and evolvability, traits that are considered beneficial for virus [169,170]. Mutagenesis in the viral genome depends on the enzymes involved in the process of nucleic acid replication and is greatly influenced by few or no proofreading or repair mechanisms. It is known that in most viruses, RNA polymerase lacks proofreading activity, with few exceptions that include coronaviruses such as SARS-CoV-2. In addition to mutations, genomic recombination is a common event during the replication process in coronaviruses and may have an important role in the generation of diversity [171]. It has been demonstrated that coronaviruses recombination is associated with increased spread, severe disease and vaccine failure at least in livestock coronavirus epidemics [172,173]. Recombination is driven by the non structural protein nsp14 3’-to-5’ exoribonuclease (Nsp14-ExoN). In vitro genetic inactivation of Nsp14-ExoN has shown to significantly decrease the frequency and altered patterns of recombination in both infected cells and released virions [171]. Thus, the high rate of mutations observed in RNA virus due to the lack or deficient proofreading activity, place them facing an apparent vital paradox: high mutation rate leads to a genetic heterogeneity that help them to adapt and overcome environmental challenges such as host switch, antiviral treatment and immune responses, but on the other hand, the accumulation of excessive deleterious mutations can result in errors that can lead the extinction of the viral species [169,174].
It seems that if an organism is subjected to variable environments where it is less fit, an increased mutation rate would be favored allowing the organism a greater chance to have a beneficial mutation that could improve its fitness [170]. Thus the environment may exert a selective pressure on the mechanisms involved in adjusting mutation rate by controlling the replication process of large genomes organisms including coronaviruses like SARS-CoV-2 which among other RNA virus contain larger genomes and present 3′-5′ ExoN activity differently from shorter genome virus where this activity lacks [175]. The mutation rate estimated based on the global diversity of SARS-CoV-2 was of ∼6 × 10−4 nucleotides/genome/year, which is in the range to the rates calculated for other RNA viruses [176].
Interestingly, a common feature of viruses is that increased transmissibility is accompanied by decreased virulence and this characteristics can be observed for SARS-CoV-2. This is suggested by the finding that infected patients from early stage of the pandemic in Wuhan province presented more severe or critical disease compared to patients from a later stage where the transmission started to increase or to infections occurred in other regions like Zhejiang province at a later stage. Genetic analysis of SARS-CoV-2 strains isolated at later stages of pandemic demonstrated that mutations near a furin cleavage site located at the surface of the spike protein could affect its structure and the binding of the receptor ACE2 resulting in mild symptoms in infected patients [177].
As the virus evolves overtime different lineages are formed and can be grouped as clades. By December 7th, 2020 there was more than 245,000 SARS-CoV-2 genomic sequences available in the Global Initiative on Sharing All Influenza Data database - GISAID [121]. These sequences were obtained from viruses isolated in different countries at different times since december 2019. At the beginning of the pandemic two types of viruses, classified as L type and S type, were identified based on differences in two single nucleotide polymorphisms located in ORF1ab and ORF8 [178]. Further analysis with more genomes categorized the virus into three types named A, B, and C regarding amino acid changes and corresponding to outbreaks [179]. Later on, more clades were revealed as new genome sequences were included in the database such as V, G, GR, GH, T and O. Currently, these nomenclatures depend on the database used. While Nextstrain [180,181] and GISAID [182] databases provide a broad categorization based on globally circulating SARS-CoV-2 diversity, cov.lineages.org database name lineages based on the outbreaks [183].
Despite different used nomenclatures, whole genomic sequences have been crucial to determine broad geographical and temporal trends in the distribution of SARS-CoV-2 genetic clades. This approach may be useful for epidemiological surveillance, investigation of transmission dynamics and introduction of novel genetic variants, the understanding of the impact of response measures on the virus population; the assessment of the impact of mutations on the performance of antiviral drugs and the modelling the antigenic properties of the virus to assess the risk of escape from a potential vaccine [184].
A phylogenetic network analysis of SARS-CoV-2 genomes from across the world revealed that they are under evolutionary selection in the human host and sometime with parallel evolution events as indicated by the emergence of same mutation in two different human host [179]. A comparison of 10,022 SARS-CoV-2 genomes from 68 countries, recovered from data repository, with a reference genome sequenced in December 2019, revealed a total 65,776 variants with 5775 distinct variants. From these 2969 were missense mutations; 1965 were synonymous mutations, 484 mutations in the non-coding regions, 142 were non-coding deletions, 100 in-frame deletions, 66 non-coding insertions, 36 stop codon variants, 11 frameshift deletions and two in-frame insertions [185]. Khailany et al. [186] performed mutation analysis on ninety five SARS-CoV-2 complete genome sequences and found a total of one hundred fifty six variations from which one hundred sixteen were unique mutations. Among these mutations, the most frequent were the 8782 C > T in ORF1ab gene, the 28,144 T > C in ORF8 gene and the 29,095 C > T in the N gene. The genome sequences investigated in this study were from SARS-CoV-2 isolated at different times and from different locations. The comparative analysis of genomic signatures demonstrated a strong association between the time of sample collection, the location and accumulation of genetic diversity. Despite the diversity observed, at the current stage of COVID-19 this variability is considered a moderate genetic diversity with an estimated average pairwise difference of 9.6 SNPs between any two genomes, which supports the hypothesis of a relatively recent common ancestor for SARS-CoV-2 [176].
One mutation in particular has been calling the attention of several researchers, the D614 G mutation which leads to a replacement of the amino acid glycin in the spike protein S. It is believed that this mutation interferes with the binding affinity of SARS-CoV-2 to its human cell receptor ACE2. This mutation is suspected to be related to increased transmissibility of the strains that carry it. The shift from the original D614 form to the G614 variant has been consistently observed in different geographical regions worldwide and has become the most prevalent form of SARS-CoV-2, suggesting a fitness advantage of the variant [187]. A study performed in Houston, Texas, USA showed that after genome sequencing of 5085 SARS-CoV-2 strains recovered from two different waves of infection, virtually all strains in the second wave presented the mutation D614 G. Moreover, patients infected with this strain presented significantly higher virus loads in the nasopharynx on initial diagnosis, although no significant evidence was found regarding the relationship between virus genotype and altered virulence [188]. Moreover, it was recently demonstrated that this variant exhibits more efficient infection, replication, and competitive fitness in human airway epithelial cells, but maintains similar morphology and in vitro neutralization properties, compared with the ancestral wild-type SARS-CoV-2 virus [189]. The results obtained so far related to the D614 G mutation, suggests that when the population immunity reaches a level high enough, it is possíble that SARS-CoV-2 may find a way to overcome this immune response. If this holds true, we could be in the same situation with the flu regarding the durability of protection of potential vaccines.
Noteworthy, it was recently reported a new variant of SARS-CoV-2, featuring a mutation A222 V in the spike protein, circulating in Europe. Although this mutation is not located at the RBD region, the frequency of variants bearing this mutation is increasing within Europe and it is demonstrating an age group bias towards young adults [190]. Moreover, another mutation (Y453 F) has been repeatedly found in mink farms in Denmark, Italy, Spain, the Netherlands, Sweden and the United States. This mutation has been identified in more than 300 SARS-CoV-2 genome sequences isolated from humans across the globe. The finding of virus bearing this mutation in minks led Denmark authorities to order the culling of millions of minks over the concern of potential transmission to humans [191]. Further studies are necessary to evaluate the potential risk of this mutation.
Despite the confirmation of the circulation of different SARS-CoV-2 sub-strains worldwide, it is still necessary to better understand the implications of these variabilities for the severity and spread of the SARS-CoV-2. In addition, since the possibility of reinfection with SARS-CoV-2 is not well understood, it is also important to determine at which degree mutation could contribute to reinfection or to a second wave of transmission as already observed in some countries. Most important, even if a vaccine is developed we will still need to understand if and how these new virus versions could affect the efficacy of a vaccine in use and the endurance of the protection.
Since mutations may lead to escape from immune recognition, virus sub-strains with different mutations should be taken into account during development of vaccines. Thus, the identification and detailed characterization of the effects of a particular mutation for the pathogenesis of SAR-CoV-2 is crucial. It has been shown that by using epitope information along with variants of virus, the 23403A > G variant (D614 G at protein level) in spike protein B-cell epitope is frequently observed in European countries but in contrast it is rare in China [185]. Interestingly, a detailed mutation map was performed in the spike protein and it was identified mutations that could prevent binding of ten human antibodies and possibly favor escape of virus from immune response [192]. Also, reinfection and relapse of COVID-19 have been reported in some countries [193]. Secondary infection cases with SARS-CoV-2 have been reported in Hong Kong [100], the Netherlands and Belgium [194], Ecuador [195], Brazil [99] and the United States [196]. Important to notice the second infection was more severe than the first infection at least for the cases reported in Ecuador and in the United States. Genomic analysis were performed in some of these reported cases and showed significant differences between each variant associated with each time of infection. Therefore, although still under debate, these observations suggest that the difference found in the two SARS-CoV-2 specimens was greater than could be expected for a short term in vivo evolution. Also, there is still not a definitive answer about the degree which the immune response mounted after a first infection could be protective against a subsequent infection.
One of the hallmarks of a vaccine is the induction of long-term protective immunity against the pathogen. However, one of the problems that can affect the efficacy of the vaccines is related to the possibility of the induced immunity wane overtime and be less effective against more virulent strains. Sub-strains can emerge due to genetic mutations conferring functional differences associated to infectivity, tissue tropism and even transmissibility. Considering a scenario where most of population in the world is still susceptible to the SARS-CoV-2, immunity may not be a major factor that could impact the evolution of the virus. However, as the immunity reaches a wide range of the population by natural infection or vaccination, it is possible that this pressure select immune-evading mutations helping SARS-CoV-2 become a more established and common infection, although less severe.
Finally, it is clear that understanding the molecular evolution of SARS-CoV-2 is pivotal since the consequences of some mutations may have implications for the formulation of vaccines and even if immunity can be reached through infections or a vaccine, the selective pressure could induce emergence of strains able to evade the immune response requiring a continuous surveillance even post pandemic.
8. Concluding remarks
Generally vaccines are approved and come to market years before it is really known how long protection endures. On the other hand, interruption of protection can go unnoticed because a vaccine may largely eliminate transmission and this way a reemergence of infection may be rare. The truth is that the reason for the long lasting immunity induced by some vaccines but not others is not completely understood, however we know that it depends of the immunologic memory. Some COVID-19 vaccines were recently approved before they have accomplished all the needed clinical trials and politicization is playing a negative influence on this process. Vaccine safety is crucial and any indication that demonstrate lack of safety may increase antivaccination movements, which could jeopardize the desired effect of reaching herd immunity. Currently we know much more about COVID-19 than we knew at the beginning of the pandemic and this has helped somehow decrease the rate of deaths compared to the number of cases. Even if COVID-19 may be brought under control in the near future, unexpected outbreaks and development of SARS-CoV-2 resistant to treatments or even vaccines are highly possible due to new mutations.
Declaration of Competing Interest
I hereby declare that I have no conflict of interest.
Acknowledgements
We would like to dedicate this work to every health professional who has been working in the front line against COVID-19 especially those who have lost their life during the pandemic. We also would like to express our sincere gratitude to the researchers worldwide whose hard work is enabling a better understanding of the disease and giving hope to the development of an effective vaccine.
References
- 1.2021. European Centre for Disease Prevention and Control. COVID-19 Situation Dashboard.https://www.ecdc.europa.eu/en/covid-19/situation-updates (Accessed December 7, 2020) [Google Scholar]
- 2.Dos Santos W.G. Natural history of COVID-19 and current knowledge on treatment therapeutic options. Biomed. Pharmacother. 2020;129 doi: 10.1016/j.biopha.2020.110493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou L. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J. Transl. Med. 2020;18:206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan W.J. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kronbichler A. Asymptomatic patients as a source of COVID-19 infections: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;98:180–186. doi: 10.1016/j.ijid.2020.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B. Preprint at medRxiv; 2020. Clinical Characteristics of 82 Death Cases With COVID-19. [DOI] [Google Scholar]
- 9.Tang Y. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meduri G.U., Kohler G., Headley S., Tolley E., Stentz F., Postlethwaite A. Inflammatory cytokines in the BAL of patients with ARDS. Persistent elevation over time predicts poor outcome. Chest. 1995;108:1303–1314. doi: 10.1378/chest.108.5.1303. [DOI] [PubMed] [Google Scholar]
- 11.Zhou F. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadjadj J. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J.S., Shin E. The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol. 2020;20:585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.2021. World Health Organization.https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-mission-briefing-on-covid-19---12-march-2020 (Accessed September 24, 2020) [Google Scholar]
- 16.WHO . 2020. DRAFT Landscape of COVID-19 Candidate Vaccines – 22 September 2020.https://www.who.int/who-documents-detail/draf-landscape-of-covid-19-candidate-vaccines (Accessed 23 September 23, 2020) [Google Scholar]
- 17.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y.W., Yiu C.B., Wong K.Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CLpro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res. 2020;9:129. doi: 10.12688/f1000research.22457.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon D.E. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan M.I. Editorial. Recent advances in the structure-based drug design and discovery. Curr. Top. Med. Chem. 2016;16:899–900. doi: 10.2174/1568026616999150918145640. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Z. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020;21:224. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu R. Genomic characterisation and epidemiology of 2019 novel coronavirus:implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naqvi A.A.T. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta. Mol. Basis. Dis. 2020;1866(10):165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu F. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou P. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman C.M., Frieman M.B. Coronaviruses: important emerging human pathogens. J. Virol. 2014;88:5209–5212. doi: 10.1128/JVI.03488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel C.J. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020;17:131. doi: 10.1186/s12985-020-01402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R.A. Structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9(5):1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham R.L. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133:88–100. doi: 10.1016/j.virusres.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang J. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrapp D. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV 429 and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue B. Structural disorder in viral proteins. Chem. Rev. 2014;114:6880–6911. doi: 10.1021/cr4005692. [DOI] [PubMed] [Google Scholar]
- 36.Alsadi E.A.J., Jones I.M. Membrane binding proteins of coronaviruses. Future Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang C. Helicase of Type 2 porcine reproductive and respiratory syndrome virus strain hv reveals a unique structure. Viruses. 2020;12:215. doi: 10.3390/v12020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller C. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antiviral Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Othman H. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem. Biophys. Res. Commun. 2020;527:702–708. doi: 10.1016/j.bbrc.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffmann M. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang D. Immunoinformatic analysis of T- and B-cell epitopes for SARS-CoV-2 vaccine design. Vaccines. 2020;8:355. doi: 10.3390/vaccines8030355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moens L., Meyts I. Recent human genetic errors of innate immunity leading to increased susceptibility to infection. Curr. Opin. Immunol. 2020;62:79–90. doi: 10.1016/j.coi.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Randolph H.E., Barreiro L.B. Herd immunity: understanding COVID-19. Immunity. 2020;52(5):737–741. doi: 10.1016/j.immuni.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14(2):185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang L. Cardiac involvement in recovered COVID-19 patients identified by magnetic resonance imaging. JACC Cardiovasc. Imaging. 2020 doi: 10.1016/j.jcmg.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kandemirli S.G. Brain MRI findings in patients in the intensive care unit with COVID-19 infection. Radiology. 2020;297:232–235. doi: 10.1148/radiol.2020201697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pung R. Investigation of three clusters of COVID-19 in Singapore: implications for surveillance and response measures. Lancet. 2020;395:1039–1046. doi: 10.1016/S0140-6736(20)30528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lauer S.A. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/m20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan J.F. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang C. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen G. Clinical and immunologic features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/jci137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen N. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phan L.T. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N. Engl. J. Med. 2020;382:872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lechien J.R. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur. Arch. Otorhinolaryngol. 2020;277(8):2251–2261. doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakalli E., Temirbekov D., Bayri E., Alis E.E., Erdurak S.C., Bayraktaroglu M. Ear nose throat-related symptoms with a focus on loss of smell and/or taste in COVID-19 patients. Am. J. Otolaryngol. 2020;41(6):102622. doi: 10.1016/j.amjoto.2020.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hopkins C., Surda P., Kumar N. Presentation of new onset anosmia during the COVID-19 pandemic. Rhinology. 2020;58:295–298. doi: 10.4193/Rhin20.116. [DOI] [PubMed] [Google Scholar]
- 61.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/s1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuba K. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imai Y., Kuba K., Penninger J.M. The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Exp. Physiol. 2008;93:543–548. doi: 10.1113/expphysiol.2007.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hamming I. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia H.P. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu H. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Y. Preprint at bioRxiv; 2020. Single-cell RNA Expression Profiling of ACE2, the Putative Receptor of Wuhan 2019-nCov. [DOI] [Google Scholar]
- 68.Chiappelli F., Khakshooy A., Greenberg G. CoViD-19 immunopathology and immunotherapy. Bioinformation. 2020;16(3):219–222. doi: 10.6026/97320630016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng Y. Epidemiological characteristics and clinical features of 32 critical and 67 noncritical cases of COVID-19 in Chengdu. J. Clin. Virol. 2020;127:104366. doi: 10.1016/j.jcv.2020.104366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du R.H. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur. Respir. J. 2020;55(5) doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang K. Preprint at bioRxiv; 2020. SARS-CoV-2 Invades Host Cells Via a Novel Route: CD147-spike Protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ulrich H., Pillat M.M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev. Rep. 2020;16(3):434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grass G.D., Toole B.P. How, with whom and when: an overview of CD147-mediated regulatory networks influencing matrix metalloproteinase activity. Biosci. Rep. 2015;36(1):e00283. doi: 10.1042/BSR20150256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shilts J., Wright G.J. Preprint at bioRxiv; 2020. No Evidence for Basigin/CD147 As a Direct SARS-CoV-2 Spike Binding Receptor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berger A. Th1 and Th2 responses: what are they? BMJ. 2000;321:424. doi: 10.1136/bmj.321.7258.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishigame H. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30(1):108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 77.Grifoni A. Targets of t cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weiskopf D. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5(48):eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.To K.K.-W. Seroprevalence of SARS-CoV-2 in Hong Kong and in residents evacuated from Hubei province, China: a multicohort study. Lancet Microbe. 2020;1:e111–e118. doi: 10.1016/S2666-5247(20)30053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma Z., Li P., Ji Y., Ikram A., Pan Q. Cross-reactivity towards SARS-CoV-2: the potential role of low-pathogenic human coronaviruses. Lancet Microbe. 2020;1(4):e151. doi: 10.1016/S2666-5247(20)30098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ou X. Characterization of spike glycoprotein of SARSCoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blackwell J.M., Jamieson S.E., Burgner D. HLA and infectious diseases. Clin. Microbiol. Rev. 2009;22:370–385. doi: 10.1128/CMR.00048-08.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matzaraki V., Kumar V., Wijmenga C., Zhernakova A. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 2017;18:76. doi: 10.1186/s13059-017-1207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi Y. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Delanghe J.R., Speeckaert M.M., De Buyzere M.L. ACE polymorphism and COVID-19 outcome. Endocrine. 2020;70(1):13–14. doi: 10.1007/s12020-020-02454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nguyen A., David J.K., Maden S.K., Wood M.A., Weeder B.R., Nellore A., Thompson R.F. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 2020;94(13):e00510–00520. doi: 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dong X. Eleven faces of coronavirus disease 2019. Allergy. 2020;75:1699–1709. doi: 10.1111/all.14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo L. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Azkur A.K. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75(7):1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y., Zhang L., Sang L. Kinetics of viral load and antibody response in relation to COVID-19 severity. J. Clin. Invest. 2020;31(August) doi: 10.1172/JCI138759. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Long Q. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 92.Ibarrondo F.J. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild Covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iyer A.S. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020 doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Isho B. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020 doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Röltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan C.A., Najeeb J., Hunter M., Wang H., Sahoo M.K., Huang C., Yamamoto F., Manohar M., Manalac J., Otrelo-Cardoso A.R., Pham T.D., Rustagi A., Rogers A.J., Shah N.H., Blish C.A., Cochran J.R., Jardetzky T.S., Zehnder J.L., Wang T.T., Narasimhan B., Gombar S., Tibshirani R., Nadeau K.C., Kim P.S., Pinsky B.A., Boyd S.D. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020;5(54):eabe0240. doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Premkumar L. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARSCoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bonifácio L.P. Are SARS-CoV-2 reinfection and Covid-19 recurrence possible? A case report from Brazil. Rev. Soc. Bras. Med. Trop. 2020;53:e20200619. doi: 10.1590/0037-8682-0619-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Duggan N.M., Ludy S.M., Shannon B.C., Reisner A.T., Wilcox S.R. Is novel coronavirus 2019 reinfection possible? Interpreting dynamic SARS-CoV-2 test results through a case report. Am. J. Emerg. Med. 2020 doi: 10.1016/j.ajem.2020.06.079. S0735-6757(20), 30583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takeda C.F.V. Case report: recurrent clinical symptoms of COVID-19 in healthcare professionals: a series of cases from Brazil. Am. J. Trop. Med. Hyg. 2020:1–4. doi: 10.4269/ajtmh.20-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.To K.K. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020;25 doi: 10.1093/cid/ciaa1275. ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Versteeg G.A., Bredenbeek P.J., van den Worm S.H., Spaan W.J. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology. 2007;361:18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Narayanan K. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jin D.Y. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell. Mol. Immunol. 2014;11:141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou R. Acute SARS-CoV-2 infection impairs dendritic cell and t cell responses. Immunity. 2020;4 doi: 10.1016/j.immuni.2020.07.026. S1074-7613(20)30333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bai B., Hu Q., Hu H., Zhou P., Shi Z., Meng J. Virus-like particles of SARS-like coronavirus formed by membrane proteins from different origins demonstrate stimulating activity in human dendritic cells. PLoS One. 2008;3(2685) doi: 10.1371/journal.pone.0002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Geijtenbeek T.B. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 107.Marzi A. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78(21):12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang Z.Y. PH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van Kooyk Y., Geijtenbeek T.B. DC-SIGN: escape mechanism for pathogens. Nat. Rev. Immunol. 2003;3(9):697–709. doi: 10.1038/nri1182. [DOI] [PubMed] [Google Scholar]
- 110.Murin C.D., Wilson I.A., Ward A.B. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat. Microbiol. 2019;4(5):734–747. doi: 10.1038/s41564-019-0392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Koyama T., Weeraratne D., Snowdon J.L., Parida L. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens. 2020;9(5):324. doi: 10.3390/pathogens9050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Netea M.G., Giamarellos-Bourboulis E.J., Domínguez-Andrés J., Curtis N., van Crevel R., van de Veerdonk F.L., Bonten M. Trained immunity: a tool for reducing susceptibility to and the severity of SARS-CoV-2 infection. Cell. 2020;181(5):969–977. doi: 10.1016/j.cell.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Alrubayyi A. Coordinated and sustained immune memory responses after mild COVID-19. Nat. Rev. Immunol. 2020;20:648. doi: 10.1038/s41577-020-00450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rodda L.B. Preprint at medRxiv; 2020. Functional SARS-CoV-2-specific Immune Memory Persists After Mild COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Netea M.G. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020;20(6):375–388. doi: 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Salvatori G. SARS-CoV-2 Spike protein: an optimal immunological target for vaccines. J. Transl. Med. 2020;18:222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hoffmann H., Kleine-Weber S., Pohlmann A. Multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.2021. Global Initiative on Sharing All Influenza Data-GISAID. Over 151,000 Viral Genomic Sequences of hCoV-19 Shared With Unprecedented Speed Via GISAID.https://www.gisaid.org/ (Accessed October 19, 2020) [Google Scholar]
- 122.World Health Organization – WHO . World Health Organization; 2021. Immunization Coverage.https://www.who.int/news-room/fact-sheets/detail/immunization-coverage (Accessed October 19, 2020) [Google Scholar]
- 123.Plotkin S.A. Vaccines: the fourth century. Clin. Vaccine Immunol. 2009;16(12):1709–1719. doi: 10.1128/CVI.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Minor P.D. Live attenuated vaccines: historical successes and current challenges. Virology. 2015;479–480:379–392. doi: 10.1016/j.virol.2015.03.032. [DOI] [PubMed] [Google Scholar]
- 125.Plotkin S.A., Plotkin S.L. The development of vaccines: how the past led to the future. Nat. Rev. Microbiol. 2011;9(12):889–893. doi: 10.1038/nrmicro2668. [DOI] [PubMed] [Google Scholar]
- 126.Rappuoli R., Black S., Lambert P.H. Vaccine discovery and translation of new vaccine technology. Lancet. 2011;378:360–368. doi: 10.1016/S0140-6736(11)60440-6. [DOI] [PubMed] [Google Scholar]
- 127.The College of Physicians of Philadelphia . 2020. The History of Vaccines.https://www.historyofvaccines.org/timeline/all (Accessed September 09, 2020) [Google Scholar]
- 128.Salk J.E. Formaldehyde treatment and safety testing of experimental poliomyelitis vaccines. Am. J. Public Health Nations Health. 1954;44:563–570. doi: 10.2105/ajph.44.5.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sabin A.B., Hennessen W.A., Winsser J. Studies on variants of poliomyelitis virus. I. Experimental segregation and properties of avirulent variants of three immunological types. J. Exp. Med. 1954;99:551–576. doi: 10.1084/jem.99.6.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Plotkin S. History of vaccination. Proc. Natl. Acad. Sci. U. S. A. 2014;111(34):12283–12287. doi: 10.1073/pnas.1400472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Plotkin S.A., Gilbert P.B., P. B Nomenclature for immune correlates of protection after vaccination. Clin. Infect. Dis. 2012;54(11):1615–1617. doi: 10.1093/cid/cis238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Miller J.A. Scientific odyssey: unravelling the secrets of the thymus. Med. J. Aust. 2005;183:582–584. doi: 10.5694/j.1326-5377.2005.tb00041.x. [DOI] [PubMed] [Google Scholar]
- 133.Oxman M.N. A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N. Engl. J. Med. 2005;352:2271–2284. doi: 10.1056/NEJMoa051016. [DOI] [PubMed] [Google Scholar]
- 134.Zepeda-Cervantes J., Vaca L. Induction of adaptive immune response by self-aggregating peptides. Expert Rev. Vaccines. 2018;17:723–738. doi: 10.1080/14760584.2018.1507742. [DOI] [PubMed] [Google Scholar]
- 135.Sanders B., Koldijk M., Schuitemaker H. Inactivated viral vaccines. In: Nunnally B., Turula V., Sitrin R., editors. Vaccine Analysis: Strategies, Principles, and Control. Springer; Berlin, Heidelberg: 2015. [DOI] [Google Scholar]
- 136.Ehrenfeld E., Modlin J., Chumakov K. Future of polio vaccines. Expert Rev. Vaccines. 2009;8:899–905. doi: 10.1586/erv.09.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Talon J. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4309–4314. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Broadbent A.J. Evaluation of the attenuation, immunogenicity, and efficacy of a live virus vaccine generated by codon-pair bias de-optimization of the 2009 pandemic H1N1 influenza virus, in ferrets. Vaccine. 2016;34:563–570. doi: 10.1016/j.vaccine.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zeltins A. Construction and characterization of virus-like particles: a review. Mol. Biotechnol. 2013;53:92–107. doi: 10.1007/s12033-012-9598-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chackerian B. Virus-like particles: flexible platforms for vaccine development. Expert Rev. Vaccines. 2007;6:381–390. doi: 10.1586/14760584.6.3.381. [DOI] [PubMed] [Google Scholar]
- 141.Crisci E., Bárcena J., Montoya M. Virus-like particles: the new frontier of vaccines for animal viral infections. Vet. Immunol. Immunopathol. 2012;148:211–225. doi: 10.1016/j.vetimm.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vogel A.B. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26(2):446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mubarak A., Alturaiki W., Hemida M.G. Middle East respiratory syndrome coronavirus (MERS-CoV): infection, immunological response, and vaccine development. J. Immunol. Res. 2019;2019 doi: 10.1155/2019/6491738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu X., Gao X. Immunological responses against SARS-coronavirus infection in humans. Cell. Mol. Immunol. 2004;1:119–122. [PubMed] [Google Scholar]
- 145.Dolzhikova I.V. Virus-vectored Ebola vaccines. Acta Naturae. 2017;9:4–11. [PMC free article] [PubMed] [Google Scholar]
- 146.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yu J. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Deng W. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gao Q. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang H. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182(3):713–721. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Logunov D.Y. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;S0140-6736(20) doi: 10.1016/S0140-6736(20)31866-3. 31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhu F.C. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396(10249):479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.van Doremalen N. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Folegatti P.M. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mercado N.B. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Keech C. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2026920. NEJMoa2026920 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.He L. Preprint at bioRxiv; 2020. Self-assembling Nanoparticles Presenting Receptor Binding Domain and Stabilized Spike As Next-generation COVID-19 Vaccines. [DOI] [Google Scholar]
- 158.Escobar L.E., Molina-Cruz A., Barillas-Mury C. BCG vaccine protection from severe coronavirus disease 2019 (COVID-19) Proc. Natl. Acad. Sci. U. S. A. 2020;117(30):17720–17726. doi: 10.1073/pnas.2008410117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gursel M., Gursel I. Is global BCG vaccination-induced trained immunity relevant to the progression of SARS-CoV-2 pandemic? Allergy. 2020;75(7):1815–1819. doi: 10.1111/all.14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.O’Neill L.A.J., Netea M.G. BCG-induced trained immunity: can it offer protection against COVID-19? Nat. Rev. Immunol. 2020;20(6):335–337. doi: 10.1038/s41577-020-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stensballe L.G. Acute lower respiratory tract infections and respiratory syncytial virus in infants in Guinea-Bissau: a beneficial effect of BCG vaccination for girls community based case-control study. Vaccine. 2005;2:1251–1257. doi: 10.1016/j.vaccine.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 162.Wardhana The efficacy of Bacillus calmette-Guerin vaccinations for the prevention of acute upper respiratory tract infection in the elderly. Acta Med. Indones. 2011;43:185–190. [PubMed] [Google Scholar]
- 163.Kleinnijenhuis J. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Moorlag S. Non-specific effects of BCG vaccine on viral infections. Clin. Microbiol. Infect. 2019;25:1473–1478. doi: 10.1016/j.cmi.2019.04.020. [DOI] [PubMed] [Google Scholar]
- 165.Cohen J. Waning immunity. Science. 2019;364(6437):224–227. doi: 10.1126/science.364.6437.224. [DOI] [PubMed] [Google Scholar]
- 166.Nishiura H., Mizumoto K., Asai Y. Assessing the transmission dynamics of measles in Japan, 2016. Epidemics. 2017;20:67–72. doi: 10.1016/j.epidem.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 167.Schwarz T.F. Ten-year immune persistence and safety of the HPV-16/18 AS04-adjuvanted vaccine in females vaccinated at 15-55 years of age. Cancer Med. 2017;6(11):2723–2731. doi: 10.1002/cam4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Baer C.F. Does mutation rate depend on itself. PLoS Biol. 2008;6(2):e52. doi: 10.1371/journal.pbio.0060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Drake J.W. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 1993;90:4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gribble J. Preprint at bioRxiv; 2020. The Coronavirus Proofreading Exoribonuclease Mediates Extensive Viral Recombination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen N. Two novel porcine epidemic diarrhea virus (PEDV) recombinants from a natural recombinant and distinct subtypes of PEDV variants. Virus Res. 2017;242:90–95. doi: 10.1016/j.virusres.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 173.Feng K.Y. Molecular characteristic and pathogenicity analysis of a virulent recombinant avain infectious bronchitis virus isolated in China. Poult. Sci. 2018;97(10):3519–3531. doi: 10.3382/ps/pey237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Eigen M. Error catastrophe and antiviral strategy. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13374–13376. doi: 10.1073/pnas.212514799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.van Dorp L. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;8 doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jin X. Virus strain from a mild COVID-19 patient in Hangzhou represents a new trend in SARS-CoV-2 evolution potentially related to Furin cleavage site. Emerg. Microbes Infect. 2020;9(1):1474–1488. doi: 10.1080/22221751.2020.1781551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tang X. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hadfield J. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics (Oxford, England) 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.2020. Nextstrain: Genomic Epidemiology of Novel Coronavirus - Global Subsampling.https://nextstrain.org/ncov Accessed: 17 October 17. [Google Scholar]
- 182.GISAID- Global Initiative on Sharing All Influenza Data . GISAID; Munich: 2020. Clade and Lineage Nomenclature Aids in Genomic Epidemiology Studies of Active hCoV-19 Viruses.https://www.gisaid.org/references/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/ 4 Jul Available from: [Google Scholar]
- 183.Rambaut A. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Alm E. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Euro Surveill. 2020;25(32) doi: 10.2807/1560-7917.ES.2020.25.32.2001410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Korber B. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Long S.W. Preprint at medRxiv; 2020. Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., III, Leist S.R., Schäfer A., Nakajima N., Takahashi K., Lee R.E., Mascenik T.M., Graham R., Edwards C.E., Tse L.V., Okuda K., Markmann A.J., Bartelt L., de Silva A., Margolis D.M., Boucher R.C., Scott H., Randell S.H., Suzuki T., Gralinski L.E., Kawaoka Y., Baric R.S. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020 doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.GISAID . 2020. New hCoV-19 Variant in Europe, Part of Natural Evolution.https://www.gisaid.org/ [Google Scholar]
- 191.GISAID . 2020. Mutations in Spike Putatively Linked to Outbreak at Danish Mink Farms.https://www.gisaid.org/ [Google Scholar]
- 192.Greaney A.J. Preprint at bioRxiv; 2020. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-binding Domain That Escape Antibody Recognition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gousseff M. Clinical recurrences of COVID-19 symptoms after recovery: Viral relapse, reinfection or inflammatory rebound? J. Infect. 2020 doi: 10.1016/j.jinf.2020.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Van Elslande J. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Prado-Vivar B. Preprint at SSRN; 2020. COVID-19 Re-infection by a Phylogenetically Distinct SARS-CoV-2 Variant, First Confirmed Event in South America. [DOI] [Google Scholar]
- 196.Tillett R.L. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]