Abstract
Viral capsids are often regarded as inert structural units, but in actuality they display fascinating dynamics during different stages of their life cycle. With the advent of single-particle approaches and high-resolution techniques, it is now possible to scrutinize viral dynamics during and after their assembly and during the subsequent development pathway into infectious viruses. In this Review, the focus is on the dynamical properties of viruses, the different physical virology techniques that are being used to study them, and the physical concepts that have been developed to describe viral dynamics.
Subject terms: Biological physics, Self-assembly, Virology, Nanoscale biophysics, Supramolecular assembly
Viruses display fascinating dynamics during their life cycle. Only recently has it become possible to probe viral dynamics at the single-particle level. This Review discusses dynamical properties of viruses and recent developments in physical virology approaches to probe such dynamics.
Key points
Viruses are highly dynamic structures.
Physics approaches are well suited to study the mechanisms behind the viral life cycle.
New single-particle techniques provide unprecedented insight into viral assembly. These techniques include resistive-pulse sensing, high-speed atomic force microscopy, interferometric scattering, charge-detection mass spectrometry and optical tweezers.
Assembly studies of both empty shells and genome-filled shells provide insights into the molecular mechanism and pathways of viral self-assembly.
Maturation is a fascinating process in which large structural changes occur in viruses.
Closed-shell dynamics includes soft modes and conformational changes.
Introduction
Humans and other animals, as well as plants, are plagued by infections caused by viruses. These are parasites that cannot reproduce by themselves and that are incapable of metabolic activity. Instead, after viruses infect cells, they alter cellular molecular machinery so that it produces new viruses, which are then released into the environment. This sequence of events, the viral life cycle, is schematically discussed in Box 1. The aim of the traditional field of structural virology is to achieve a better understanding of the spatial organization of the proteins inside viral particles, an understanding that should increase our ability to develop more effective therapies.
Since the beginnings of structural virology as a field of study, physics has made important contributions. For several decades now, electron microscopy and X-ray diffraction imaging of crystals of viruses have provided high-resolution reconstructions of viral capsids with icosahedral symmetry1. The crystallographic analysis method developed by Donald Caspar and Aaron Klug in the 1960s (ref.2) provided us with a systematic classification of icosahedral capsids in terms of the so-called T-numbers (triangulation numbers), a method that has stood the test of time3. More recently, modern cryo-electron imaging methods, including tomography4,5 and asymmetric reconstructions6,7, have made it possible to reconstruct not only regular capsids, but also asymmetric protein distributions and the enclosed genome by using images of individual viruses.
These important low-temperature imaging methods are limited to the study of static viral structures. However, evidence accumulated towards the end of the twentieth century that viruses and viral capsids are in fact dynamical structures that should be viewed as active ‘nanomachines’. When a virus assembles, a highly dynamical and disorganized state of individual capsid proteins in solution progressively turns into a more ordered, collective multi-protein state of partially and eventually fully closed shells, the viral capsid. Yet these closed capsids are still quite dynamic. Studying the dynamics of viruses called for a new toolbox, in terms of both new probes and new methods of analysis and numerical modelling. We first review some of the dynamic methods that are being applied to study the assembly of empty viral capsids, then focus on the role of genome molecules (RNA/DNA) during assembly, followed by a discussion of studies of the steady-state dynamics of assembled viruses. Box 2 summarizes several experimental techniques that have been used to study viral dynamics in the past decade. In an earlier article3, we reviewed work on the equilibrium physics of viral assembly under static conditions and their mechanical properties.
Box 1 Viral life cycle.
The viral life cycle can be roughly divided into the following steps: attachment; entry; uncoating; gene expression and replication; assembly; and release. However, depending on the virus, there are variations in these steps169–172. For instance, bacteriophages typically attach to their host cell and inject the genome into the cell while the viral capsid remains outside173. During gene expression, the viral proteins are synthesized and replication involves the reproduction of the viral genome. A variety of viruses change their structure after assembly is completed. This process, called maturation, can occur inside and/or outside the cell93,113. Certain viruses also obtain a lipid envelope upon release. Cellular components, including the nucleus present in eukaryotic cells, are left out for simplicity in the schematic. In this Review we focus on the upper part of the figure, especially looking at the dynamics of assembly and the dynamics of closed shells before and after release, including steady-state dynamics and maturation.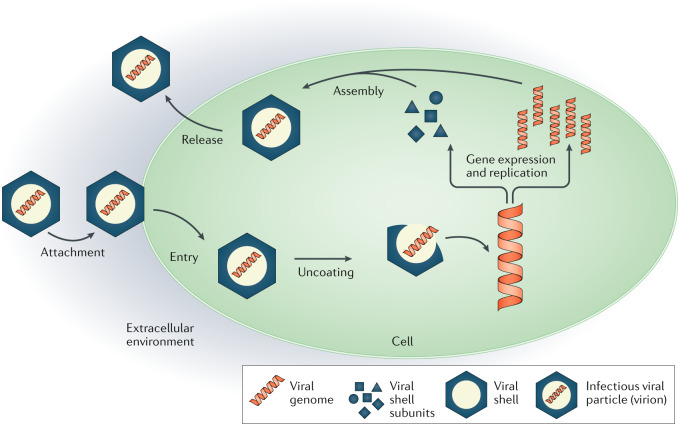
Figure adapted with permission from ref.169.
Box 2 Experimental techniques.
Besides established techniques in physical virology research such as X-ray diffraction174,175, electron microscopy176,177 and nuclear magnetic resonance178–180, over the past decade a set of single-molecule and advanced bulk techniques has been developed and optimized.
In resistive-pulse sensing48,181, single particles are shuttled over pores standing under an electrical bias (voltage V). The resistance of the system changes when a particle is present, resulting in a current change.
In small-angle X-ray scattering (SAXS)38,77, a solution containing the particles is irradiated with a monochromatic X-ray beam (k0 with radiation wavelength λ) and its scattering profile is detected. The scattered radiation k1 is registered by recording the momentum transfer s, which includes the scattering angle 2θ. By subtracting the profile from the solvent alone, the signal of the particles alone is obtained.
In atomic force microscopy (AFM), a sharp tip scans the substrate yielding a sub-nanometre-scale topographic profile of the surface-adhered particles182,183. Recent technical advances have increased the scanning velocity by several orders of magnitude, and sub-second frame rates are now possible with high-speed AFM43–46.
Optical tweezers capture dielectric, micrometre-sized beads in a focused laser beam. The set-up can be combined with fluorescence microscopy184,185. By trapping two beads and attaching a genome in between, protein–nucleic acid interactions can be studied — relevant for virus assembly.
Light-scattering studies in bulk yield insights into virus assembly20,186, and by using interferometric scattering microscopy this approach is extended to the single-particle level70,187. In single-particle studies, the destructive interference of a reference beam and scattered light yields insight on the size of the particle.
In native mass spectrometry, large non-covalent particles are ionized, and their mass can be determined188–190. Charge-detection mass spectrometry is a single-particle native mass spectrometry approach to determine the masses of individual ions by simultaneously measuring the mass to charge ratio and the charge of each ion26.
In acoustic force spectroscopy (AFS), molecules that are tethered to a surface and a bead are stretched by acoustic forces towards pressure nodes in the flow chamber191. The beads are imaged through the objective lens (OL) onto the complementary metal–oxide–semiconductor (CMOS) camera.
LED, light-emitting diode. Resistive-pulse sensing adapted with permission from ref.47. SAXS adapted with permission from ref.192. (High-speed) AFM adapted with permission from ref.193. Optical tweezers adapted with permission from ref.72. Interferometric scattering microscopy adapted with permission from ref.70. Acoustic force spectroscopy adapted with permission from ref.73.
Viral self-assembly dynamics
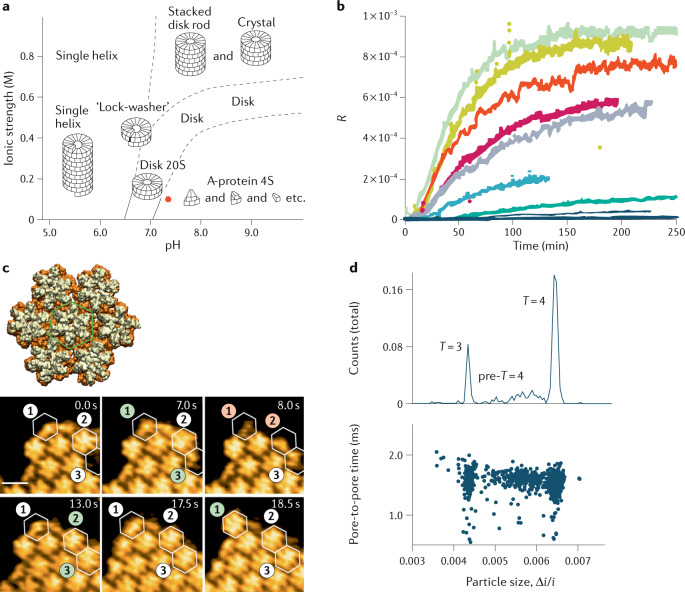
In 1955, Heinz Fraenkel-Conrat and Robley Williams discovered that solutions containing tobacco mosaic virus (TMV) capsid proteins and single-stranded (ss) RNA molecules spontaneously produced infectious viral particles (or virions)8,9, a process now known as self-assembly. Many other viruses — both with rod-like helical shapes (such as TMV) and with sphere-like icosahedral shapes10–12 — were found to self-assemble under in vitro conditions. Thermodynamic studies led to the construction of instructive phase diagrams for viral assembly, with pH and ionic strength acting as thermodynamic control parameters. For instance, for the capsid proteins of TMV without its genome molecules, increasing ionic strength and reducing pH both serve to weaken the electrostatic repulsive interactions between the amphiphilic capsid proteins, thereby allowing the competing attractive hydrophobic interactions to overcome the repulsion and drive assembly13. TMV-like capsids appear under conditions of low pH. This is not the case under physiological conditions, but adding the viral genome molecules to the solution does tilt the balance and produces infectious viruses (Fig. 1a). In comparison, the capsid proteins of the spherical cowpea chlorotic mottle virus (CCMV) form empty capsids under conditions of low pH but higher ionic strength, but form concentric multi-shells at low pH and lower ionic strength14.
Fig. 1. Assembly of empty capsids.
a | Phase diagram of tobacco mosaic virus (TMV) assembly. The red dot signifies cellular conditions. b | Light-scattering experiments yield the Rayleigh ratio R as a function of time for increasing capsid protein concentration (bottom to top) during assembly of human papillomavirus (HPV) virus-like particles. The mass average molecular weight of assembled particles can be determined from R and the concentration of the solution. c | High-speed atomic force microscopy snapshots of reversible capsid lattice assembly for human immunodeficiency virus (HIV) (lower panels). Scale bar, 10 nm. The top image shows a reconstruction of seven hexamers; the centre one is highlighted within a green hexagon. d | Resistive-pulse sensing technique to study hepatitis B virus (HBV) assembly, including complete T = 3 and T = 4 capsids and assembly intermediates (pre-T = 4). T, triangulation number. (Δi/i): normalized pulse amplitudes, with the baseline current i and the adjusted pulse amplitude Δi. Upper panel, integrated counts; lower panel, individual data points. Part a adapted with permission from ref.13. Part b adapted with permission from ref.20. Part c adapted with permission from ref.43. Part d adapted with permission from ref.47.
When empty capsids are present in such a solution, they coexist with a certain concentration of single capsid proteins (monomers) or small protein groups (oligomers). Light-scattering studies15,16 have shown that the concentrations of capsid protein monomers or oligomers and of assembled capsids obey the law of mass action (LMA) of chemical thermodynamics, a direct consequence of the application of the second law of thermodynamics to multi-component solutions in thermodynamic equilibrium. This observation suggested that viral self-assembly could be understood as an equilibrium self-assembly process, similar to the formation of micelles in surfactant-rich solutions17. Insight into the dynamics of capsid assembly was mainly achieved in the past decade, typically involving more advanced probes. We discuss some of these studies, first for the case of assembly of empty capsids and then for the case of assembly of virions.
Assembly dynamics of empty capsids
The first dynamical studies of the assembly of capsids used time-dependent light scattering and turbidity measurements to probe bulk solutions of capsid proteins18. Capsid assembly was initiated by a reduction of the pH or an increase in the salinity15,19. Through this approach, it was found that capsid assembly of human papillomavirus (HPV) typically starts after a certain lag time (Fig. 1b)20. This lag time is understood to be the formation time of a nucleation complex, followed by a ‘downhill’ protein-by-protein extension (or ‘elongation’) process that leads to completed capsids. In the limit of late times, the dependence of the capsid concentrations on the total protein concentration was consistent with the LMA of equilibrium thermodynamics.
Within the classical theory of nucleation and growth21, the nucleation complex corresponds to the critical nucleus, or transition state — that is, a maximum in the free energy of a protein cluster as a function of the number of proteins. The number of proteins or protein groups (capsomers) that constitute the nucleation complex can be determined as the slope of the log of the concentration of complete capsids as a function of the log of the concentration of free proteins measured at a given instant. For the case of HPV, such an analysis suggested that the nucleation complex is a dimer of pentamers20. Note that even hours after initiation the assembly process still is not fully complete, even though the initial lag time is in the range of minutes (Fig. 1b). An example of single-particle imaging of assembly intermediates was provided by a combined electron microscopy and atomic force microscopy (AFM) study of the assembly of the minute virus of mice (MVM)22 that provided insight into the nature of the transient assembly intermediates that interpolate between the monomer and capsid state. These experiments demonstrated the importance of single-particle approaches.
According to the equilibrium self-assembly theory of viral capsids16, capsid assembly is initiated at a critical protein aggregation concentration (the CAC). At the CAC, the critical nucleus is roughly half of a complete shell, which is much larger than the size of the experimentally observed nucleation complexes. Theoretically, the size of a nucleation complex should decrease in size for increasing levels of supersaturation21. So, according to nucleation-and-growth theory, the dynamical experiments are best understood as taking place under conditions of high levels of supersaturation — that is, far from thermodynamic equilibrium. But if that is the case, then how can the equilibrium LMA apply? Moreover, when the protein concentration is reduced after capsid formation, the capsids do not spontaneously disassemble as would happen if the system really did obey the LMA.
Some insight in this strange interplay between equilibrium and non-equilibrium properties can be obtained from a simple model for the kinetics of capsid assembly23 in which the capsid is treated as a dodecahedron composed of 12 pentamers with sticky edges. The capsids assemble in a solution of pentamers. At each assembly step, a pentamer is added to a partial capsid on a location that minimizes the energy of the partial capsid. This leads to a minimum energy assembly pathway E(n) where the index n runs from 1 to 12. Mathematically, the assembly kinetics is similar to that of a one-dimensional (1D) random walk across a non-uniform energy landscape E(n). The cluster size distribution function C(n,t) is the probability that a pentamer is part of a pentamer cluster of size n. This function obeys the master equation24
with the probability current J(n,t) given by
The first term describes the growth of a cluster by the addition of a pentamer from solution with k+(n) the on-rate. The second term describes the loss of a pentamer from a cluster with k−(n) the off-rate. The on and off rates are related to the energy landscape E(n) by the detailed balance condition:
where c0 is a constant of the order of the inverse volume of a pentamer and β = 1/kBT is the inverse temperature. If one assumes that assembly is diffusion-limited, then the on-rate is a constant k+ that is independent of n and proportional to the pentamer diffusion coefficient and the pentamer size. The detailed balance condition then provides an expression for k−(n). The boundary condition for the solution of the master equation at n = 1 is J(1,t) = ΓC(1,t)2 − k−(2)C(2,t) with Γ the nucleation rate. If thermal disassembly of a completed capsid is forbidden, then the second boundary condition is that J(12,t) = 0, known as an absorber boundary condition. The initial condition is that C(1,0) = ϕ, with ϕ the total pentamer concentration, while C(n,0) = 0 for n > 1.
Through mathematical and numerical analysis, it can be shown that this model qualitatively reproduces the capsid assembly kinetics shown in Fig. 1b23,24. The dynamics has the character of a shock front of assembly intermediates that propagates in configuration space from the two-pentamer nucleation complex to the assembled capsid. The arrival time of the shock front at n = 12 corresponds to the lag time measured experimentally. It also can be demonstrated that for later times the ratio C(12,t)/C(1,t) resembles a version of the LMA, the ‘quasi-LMA’. Finally, for high nucleation rates Γ, at late times a state is produced with a large number of incomplete capsids, consistent with experimental reports.
The nature of the nucleation complex is of central importance for viral assembly, and it has been studied by different experimental methods. Native ion-mobility mass spectrometry experiments indicate that nucleation complexes of the norovirus and the hepatitis B virus (HBV) possess a five-fold symmetry axis25. Charge-detection mass spectrometry (CDMS) at the level of individual particles has been used to study the extension stage of the capsid growth, past the level of the critical nucleus. It was found that during the growth process the number of HBV proteins per particle exceeds the number of proteins in the capsid of the native T = 4 HBV virion26. In other words, late intermediates can possess a larger mass than the final closed capsid. The process of final annealing of these late intermediates with the shell shrinking to the final structure takes much longer than the initial assembly reaction. A similar case of an intermediate state with excess size is encountered for the assembly of CCMV27. One implication of these findings is that viral assembly is not just a simple protein-by-protein addition process but also involves large-scale reorganization processes of the capsid, a theme we will return to.
Kinetic trapping plays a prominent role in capsid assembly. Mutations of capsid proteins can generate kinetic traps either by suppressing assembly altogether or by producing aberrant capsid structures28,29. Kinetic trapping has also been observed in CDMS experiments on HBV assembly30. When assembly occurs in the presence of higher salt concentrations, electrostatic repulsion is weakened and thus assembly rates are increased. Such assembly rates lead to kinetically trapped intermediates. Kinetic trapping and formation of malformed capsids has been reproduced by numerical simulations of capsid assembly31–33. Kinetic trapping becomes progressively more prominent when the size of the capsid increases.
There is a useful analogy between kinetic trapping during viral assembly and kinetic trapping during protein folding. The total number of ways in which a polypeptide molecule can be folded together is far too large for all of them to be sampled by random thermal fluctuations, at least on reasonable timescales (Levinthal’s paradox)34. Similarly, there is an enormous number of ways that protein shells of various shapes and sizes can be assembled from protein monomers or oligomers. For the case of protein folding, natural selection is believed to produce kinetic assembly pathways that allow folding on reasonable timescales by avoiding kinetic traps35,36. This suggests that the capsid assembly process also may have evolved kinetic assembly pathways that avoid deep kinetic traps. Mutations that alter the assembly pathway can cause deep kinetic traps to reappear. Kinetic assembly pathways have been identified in HBV, by a combination of time-dependent small-angle X-ray scattering (SAXS) and umbrella Monte Carlo simulation with maximum entropy optimization37. Importantly, these kinetic assembly pathways appear only over narrow intervals of parameter space. Assuming that the native capsid is the lowest free-energy state, thermal annealing provides an escape route out of shallow kinetic traps through thermal activation, but thermal annealing is effective only for relatively weak protein–protein interactions. This suggests that protein–protein interaction energies should be in the range of no more than a few kBT (where T is temperature here) during initial assembly. Apart from HBV, the assembly of bacteriophage P22 procapsids and empty capsids of norovirus and CCMV has been studied by SAXS38–40, with relatively large on-path assembly intermediates reported. For norovirus, the intermediate assembly structure is a double pentamer of dimers, connected by one dimer39, whereas for CCMV, half-formed capsids with a lifetime of several seconds have been found40.
The human immunodeficiency virus (HIV) capsid consists of ~1,000–1,500 copies of the viral capsid protein41, and the self-assembly kinetics of a 2D lattice of this capsid protein has been examined by AFM42. Assembly starts at specific sites on the surface, followed by the growth of the lattice and fusion of the different patches, eventually producing full coverage of the surface. By increasing both the spatial and temporal resolution, it is possible to identify the nature of the nucleation complex and to follow assembly in real time43. The necessary increase in resolution has been achieved by using high-speed AFM (HS-AFM)44–46. The nucleation complex of the growing lattice was clearly identified as a capsid protein hexamer. Furthermore, it was shown how monomers, dimers and trimers of the capsid protein attach to, and detach from, the growing lattice (Fig. 1c). This direct visualization of the 2D assembly process revealed that the self-assembly of a lattice of capsid proteins takes place via multiple stochastic pathways.
Other new probes have also been used to study assembly. For example, single-particle resistive-pulse sensing was used to study how HBV assembly depends on ionic strength47, and complete capsids and assembly intermediates have been identified (Fig. 1d). Subsequently, this technique was applied to test the effect of antivirals that specifically target the capsid assembly of HBV48. For weak protein–protein interactions, the heteroaryldihydropyrimidine (HAP) antiviral agents induce the formation of aberrant capsid structures. However, strong protein interactions cannot be overcome by the HAPs, in which case the native T = 4 icosahedral capsid is formed. Such results show that understanding (and potentially interfering in) kinetic assembly pathways can have implications for the development of antiviral drugs. In particular, as there is a considerable structural similarity in the architecture of the capsid proteins between most icosahedral viruses (the ‘Swiss roll’ motif, also called the ‘jelly roll’ motif49), such interference could be a promising general ‘nanomedicine’ strategy for different viral families.
Capsid assembly around the viral genome
The assembly of a virion — the fully infectious viral particle containing the viral genome molecule(s) — is a more delicate enterprise than the assembly of an empty capsid. In the infected cell, the viral genome molecules are surrounded by host genome material, and only the virus genome should be packaged in the viral shell. The associated search process to locate the viral DNA or RNA is often carried out by the capsid proteins, in order to specifically package this viral genome. The assembly of most double-stranded (ds) DNA and RNA viruses involves the insertion of viral genome molecules into pre-assembled, empty procapsids. This insertion is an active process driven by a powerful rotary molecular motor that derives its energy from ATP (adenosine triphosphate) hydrolysis50,51. The viral DNA molecules are marked by an enzyme (terminase) that becomes a component of the packaging motor. Well-studied examples of this process are provided by the dsDNA viruses that infect bacteria (bacteriophages) and by some animal viruses such as the herpesvirus and adenoviruses50,51. The assembled virion is characterized by large osmotic pressures (in the range of 10 atm) exerted by the highly compacted genome on the capsid wall52. This pressure probably plays an important role in the life cycle of the virus by assisting the release of genome molecules into host cells. Both dsDNA insertion and release have been successfully replicated under laboratory conditions. Packaging studies of bacteriophage DNA have been done with optical tweezers51,53–55 whereas genome release studies of archaeal, prokaryotic and eukaryotic viruses used bulk and other single-molecule approaches52,56–58.
Apart from active packaging, the recruitment of genome molecules can also occur as a passive co-assembly process, driven by a form of chemical affinity between genome molecules and capsid proteins. In this process, there is no build-up of large osmotic pressures. It is the prevalent assembly process of viruses with ss genome molecules as well as for some dsDNA viruses such as simian virus 40 (SV40). Co-assembly was first studied in TMV, leading to the suggestion that ssRNA genome molecules can act as assembly templates. Whereas TMV capsid proteins can form oligomers when the pH and salinity level is approximately at a physiological level, electrostatic repulsion is — under such conditions — too strong to allow formation of empty capsids. However, the association of negatively charged RNA nucleotides with the positively charged amino acid residues that line the interior of disk-like protein oligomers weakens the electrostatic repulsion and tips the free energy balance towards assembly. Stacking the TMV protein disks on top of each other produces a helical capsid with the RNA molecule incorporated in the capsid interior (Fig. 2a). The length of the RNA molecule acts as a caliper that determines the length of the virus. For icosahedral viruses, genome length also can alter particle size, as exemplified by later studies on SV4059 and CCMV60. In both cases, two different capsid sizes were observed, depending on the length of the encapsidated genome. These examples show that the RNA genome cargo both aids viral self-assembly and influences the outcome of assembly.
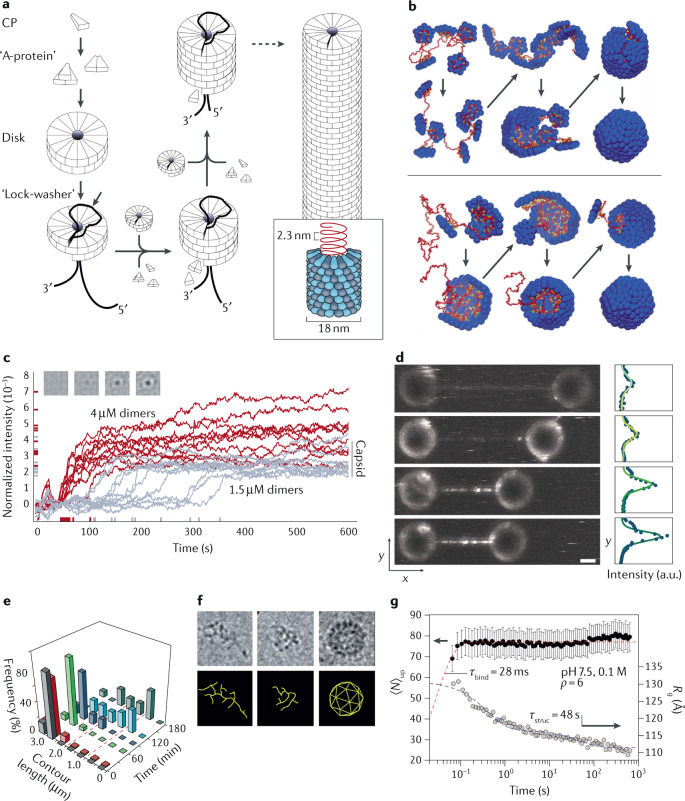
Fig. 2. Assembly around a genome.
a | Tobacco mosaic virus (TMV) capsid proteins (CP) assemble into A-proteins and subsequently disks. By insertion of the RNA (black thread in main image; red in inset) into a protein disk, a conformational change to a helical ‘lock-washer’ configuration occurs. The virion grows by the sequential addition of protein disks and A-proteins. b | Simulated assembly around a genome (red) following an en masse (upper) or nucleation-and-growth pathway where the genome acts as an ‘antenna’ (lower). c | Assembly traces of individual MS2 bacteriophage particles for different capsid protein concentrations, recorded by interferometric scattering microscopy. Inset: images of a single assembling particle. d | Fluorescence optical tweezers measurements of DNA packaging by synthetic capsid proteins. Progressive virus-like particle (VLP) assembly decreases the DNA end-to-end distance and pulls the beads together (left panel). Scale bar 2.5 µm. The fluorescence intensity profiles reflect the increase in bound polypeptides (right panel). e | Acoustic force spectroscopy data reveals the decrease in DNA contour length during assembly of simian virus 40 (SV40) VLPs. f | Electron micrograph (top) and corresponding schematic (bottom) of cowpea chlorotic mottle virus (CCMV) assembly around brome mosaic virus (BMV) genome. The images show (left to right): RNA without protein; decrease in size of the complex after adding capsid protein to the RNA; capsid formation after a reduction in pH. g | Time evolution of mean number of subunits 〈N〉up (black circles) and radius of gyration Rg (grey circles) after mixing of CCMV capsid proteins and genome at a mass ratio ρ of 6:1. The dashed lines are decay functions for the binding time τbind (red) and the structural relaxation time τstruc (blue). Error bars indicate standard error of the mean. Part a adapted with permission from ref.167 and T. Splettstößer. Part b adapted with permission from ref.67. Part c adapted with permission from ref.70. Part d adapted with permission from ref.72. Part e adapted with permission from ref.73. Part f adapted with permission from ref.69 and ref.168. Part g adapted with permission from ref.77.
The proper assembly of a TMV particle requires that viral RNA molecules are distinguished from host RNA molecules, such as messenger RNA (mRNA) molecules. Packaging signals are short, evolutionarily conserved RNA sequences with specific affinity for the capsid proteins of a virus. The initiation of virion assembly starts when capsid proteins bind to the packaging signal(s) of a viral RNA molecule. The subsequent growth process (also known as elongation) may involve additional packaging signals, as in the case for the well-studied MS2 bacteriophage virus, but it can also be driven entirely by the generic electrostatic affinity of the negatively charged ssRNA molecules for positively charged amino acid residues of capsid proteins. An example of the latter case is TMV, which has only a single packaging signal of 20 nucleotides.
The combination of specific and nonspecific protein–genome affinity with the hydrophobic affinity between capsid proteins drives the co-assembly process. Box 3 summarizes common assembly models. Below, we discuss generic aspects of the co-assembly process and then virus-specific aspects.
Box 3 Assembly around the genome.
We describe four possible strategies for capsid–genome co-assembly71. For the nucleation–elongation model (figure panel a), the genome molecule is assumed to be condensed prior to assembly, for example by polyvalent counterions194,195. Affinity of positively charged residues of the capsid proteins (CPs) for the negatively charged surface of the condensate drives capsid assembly.
For the other three cases, the genome molecule is assumed to be extended prior to assembly. For the micellar condensation model (figure panel b), generic electrostatic affinity of capsid proteins for the RNA produces a nucleoprotein condensate. An ordering transition transforms ‘en masse’ the disordered condensate into an ordered virion17,61. In the RNA antenna model (figure panel c), the capsid assembles protein-by-protein, like an empty capsid, with the genome molecule adhering to positively charged residues along the interior68. The part of the genome molecule that protrudes from the partially formed shell can act as an ‘antenna’ that assists the capture of capsid protein monomers, which can diffuse along the genome molecule to the capsid. This scenario is expected for the case of strong, non-specific protein–RNA interaction. In assembly mediated by packaging signals (PSs) (figure panel d), the capsid also grows protein-by-protein, but with the capsid proteins having a specific affinity for packaging sequences of the genome molecule196. Because assembly is strongly influenced by ionic strength and pH, certain viruses seem to be able to follow different assembly pathways depending on the environmental conditions, revealing the flexibility in viral dynamics71.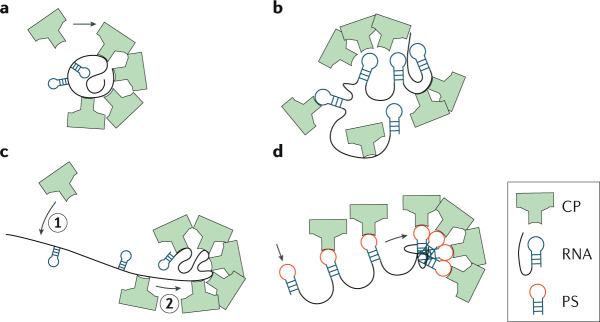
Figure adapted with permission from ref.71.
Non-specific assembly
The capsid proteins of CCMV assemble around non-genomic ssRNA molecules without viral packaging signals. Furthermore, CCMV capsid proteins even assemble around negatively charged polyelectrolytes61,62, forming virus-like particles (VLPs). CCMV is thus a suitable ‘laboratory’ in which to study how non-specific interactions can drive viral assembly. It is possible to carry out ‘packaging competition’ experiments on CCMV to see whether different types of RNA molecules are packaged more readily than others27. As might be expected from packing considerations and considerations based on the electrostatics of capacitive charging and charge neutralization, there is an optimal size for the packaging of RNA molecules in a capsid of a given size. For the T = 3 wild-type CCMV particles, this optimal size is, for instance, ~3,200 nucleotides. Unexpectedly, the ‘alien’ RNA genome of brome mosaic virus (BMV) was packaged by CCMV capsid proteins with threefold higher efficiency than the RNA molecules of CCMV itself, even though the two RNA molecules have virtually identical lengths. This result suggested that the secondary structure of an ssRNA molecule influences in some manner the non-specific packaging efficiency. This conjecture was nicely confirmed by packaging competition experiments carried out on polyU and polyA RNA chains of the same length but with no secondary structure63. The negative charges of polyU and polyA closely compensate the positive charges of the capsid protein tails, in contrast with the wild-type CCMV which shows pronounced electrostatic overcharging64,65. The secondary structure of virus genomes is much more branched than that of randomized variants66. The degree of branching can be expressed quantitatively through the maximum separation between two nucleotides of the secondary structure66, known as the maximum ladder distance.
Obtaining information about the detailed sequence of events of virus assembly is an experimental challenge. Before turning to the methods that are being used, we first discuss the outcomes of numerical simulations of the assembly dynamics of simplified coarse-grained models, which act as an aid to interpreting the experiments. For instance, simulations of assembly of a dodecahedral capsid composed of 12 pentagonal pentamers67 that have an affinity for a linear genome molecule exhibit different assembly sequences depending on the capsid–genome interactions (Fig. 2b). The ‘antenna’ scenario is an orderly assembly sequence that resembles the nucleation-and-growth scenario of empty capsids. The genome molecule is attracted to the interior of the partially formed capsid. The part of the genome that is not yet packaged acts as an ‘antenna’ for the diffusive influx of capsomers from infinity (Fig. 2b, lower panel, and Box 3)68. By contrast, in the ‘en masse’ scenario, a disordered protein/genome condensate forms before the assembly of the particle. In effect, the condensate provides a local ‘milieu’ with high protein concentration that aids assembly. This scenario appears for increasing attraction strength between the capsid proteins and the genome (Fig. 2b, upper panel, and Box 3). Theoretical work on RNA-directed assembly of small viruses indicates that such a condensate can even be a feature of the equilibrium assembly phase diagram69.
Although bulk studies have difficulty distinguishing between these different assembly scenarios, a number of promising single-particle probes have been developed. One approach is based on interferometric scatter microscopy, in which interference between light scattered from a particle and reflection from a nearby surface provides information about the size of that particle. The size evolution then can be followed with high temporal resolution. This technique has been used to study the assembly of individual phage MS2 particles around surface-tethered ssRNA molecules (Fig. 2c)70. The tethering surface provides the reflected wave required for the technique, and the scattering intensities give the growth curves of individual particles. For a capsid protein dimer concentration of 1.5 µM, most scattering intensities approach a value consistent with that of a full capsid. In addition, a few reached significantly larger values. Recall that assembly intermediate states with an excess number of capsid proteins are also encountered for the assembly of empty HBV and CCMV capsids26. The lag time between assembly initiation and capsid growth has a strikingly stochastic character, which suggests that assembly initiation requires thermal activation over a free-energy barrier that is substantially larger than the thermal energy. Note that once assembly starts, it continues to completion with little disassembly. Conversely, for 4 µM concentrations, the lag time has little stochasticity, indicating a lower activation barrier. Most assemblies have excess protein material in this case. These results support the nucleation-and-growth picture with an activation energy barrier that decreases with increasing levels of supersaturation. Although no specific packaging signals were identified by these experiments, based on literature and the observed assembly pathway an assembly sequence in which packaging signals mediate assembly (Box 3) was proposed70,71.
In a different single-particle approach, a dsDNA strand was stretched between two beads in a double optical tweezers set-up72 and then exposed to a solution of fluorescently labelled synthetic polypeptide strands, which played the role of model capsid proteins. Different assembly stages of the resulting VLP were visualized in real time with millisecond temporal resolution (Fig. 2d). Smaller oligomers moved diffusively along the DNA strand during the initial stages of assembly, as expected in the antenna picture, but pentameric or larger oligomers were stably attached to the DNA. Fitting the observations to a kinetic version of Langmuir adsorption theory led to an estimation of the effective binding free energy of pentamers to the DNA of ~25kBT. To examine the elongation stage following nucleation, acoustic force spectroscopy (AFS) was used. The results indicated formation of a helical assembly with a repeat period of 30 nm, consistent with the observed packaging ratio. The stretched DNA strand functioned in this experiment as an antenna for catching polypeptides (Box 3). The same approach has been used to examine the assembly of VLPs of SV40 around dsDNA73. SV40 has a T = 7 icosahedral structure composed of pentameric capsomers, and packages both viral and heterologous dsDNA74,75. In addition, SV40 assembles around ss genomes, for which previously a two-step process was observed by SAXS76. A combination of optical tweezers, AFS and AFM was used to reveal a multi-step assembly mechanism around dsDNA. As in the previous example, the stretched DNA strand functioned as an antenna catching diffusing proteins, followed by formation of DNA-associated protein clusters. The interaction of capsid proteins with each other and with the DNA was sufficient to overcome the tension of the DNA strand, resulting in genome packaging, as revealed by a decrease in DNA contour length during assembly (Fig. 2e). For the future, such experiments open the prospect of quantitative determination of the assembly kinetics by measuring the time dependence of the forces exerted on two optical traps.
In these two examples, capsid assembly was initiated at a specific instant of time by a rapid change in solution conditions. An alternative method is to change solution conditions sufficiently slowly that the system remains near thermodynamic equilibrium during the assembly process. For the case of CCMV (Fig. 2f), the capsid proteins have positively charged, disordered tail groups with generic electrostatic affinity for the negatively charged RNA. By using BMV genome molecules in a solution with CCMV capsid proteins, all effects due to CCMV-specific packaging signals can be avoided69. The branched viral RNA molecules, which are highly charged and swollen when free in solution, reduce in size when CCMV capsid proteins are added to the solution (Fig. 2f, second panel). At high pH — that is, for weakened affinity between capsid proteins — disordered nucleoprotein condensates are formed, consistent with self-assembly theory69 and simulations of assembly67. The disordered condensates transform into ordered VLPs after reduction of the pH.
The protein-to-RNA concentration ratio is a critical parameter for the assembly ‘phase diagram’ of CCMV. This ratio must be carefully distinguished from the dependence of the assembly kinetics of MS2 on the absolute capsid protein concentration that was discussed earlier. The optimal mixing ratio is defined as the minimum value of the protein-to-RNA concentration ratio for which all of the RNA molecules are packaged. One would expect the optimal mixing ratio to equal the stochiometric protein-to-RNA ratio of CCMV virions, but in fact it was found to be considerably larger, corresponding to excess capsid proteins and reduced electrostatic overcharging. The final assembly step requires shedding of these excess proteins, as discussed above. The scenario of an excess of genome-bound proteins in the first phase of assembly fits well with the en masse pathway (Box 3) that SAXS revealed for the initial steps in CCMV assembly around its genome (Fig. 2g)77. Interestingly, these latter experiments also showed that final closure of the shell seems to occur through an activated process from a disordered condensate to an ordered closed particle.
Packaging signals
As discussed above, the assembly of ssRNA viruses is initiated and guided by genomic ssRNA molecules that have both a specific and a non-specific affinity for the capsid proteins. An important example of ssRNA viruses is HIV, which has a single packaging signal at the 5′ end of the viral RNA molecules, known as the Ψ sequence. It seems to act only on the nucleation step, not on the subsequent growth78,79 (Fig. 3A). This RNA sequence adopts an intricate structure with several high-affinity binding sites for the Gag proteinnucleocapsid domain80,81. If the Ψ sequence is removed, then cellular RNA material is packaged, driven by generic electrostatic affinity between RNA and the positively charged nucleocapsid domain of the Gag capsid protein82. The assembly thermodynamics of these VLPs is essentially the same as that of HIV virions, indicating that genome selection during HIV assembly is a purely kinetic process.
Fig. 3. Packaging signals.
A | HIV packaging efficiency of RNA containing the packaging signal Ψ, compared with RNA without Ψ (control). B | Asymmetric structure of the genome of bacteriophage MS2. C | Schematic of MS2 assembly: A-protein (AP) attaches to RNA and to capsid protein dimers (CP2) (panel Ca); additional CP2 binds and capsid formation starts (panel Cb); CP2 is not only recruited by existing RNA stem-loops (SLs) but also triggers SL formation (panel Cc); further condensation of the RNA and closed-shell formation (panel Cd); formation of a stable virion (panel Ce). D | Hamiltonian path analysis. Reconstruction of RNA inside MS2 (panel Da); the RNA shell as polyhedral cage (panel Db); 3D view of the Hamiltonian path (panel Dc); planar representation of the same Hamiltonian path with quasi-equivalent MS2 capsid subunits (panel Dd). Part A adapted with permission from ref.79. Part B adapted with permission from ref.86. Part C adapted with permission from ref.86. Part D adapted with permission from ref.88.
Dynamical observations of assembly are challenging, but it may be possible to gather information about the assembly history of a virion from the structure of the encapsidated genome. Cryo-electron microscopy (cryo-EM) combined with icosahedral averaging allows for high-resolution structural studies of the genome density inside icosahedral capsids83,84. However, a consequence of the averaging approach is that only those parts of the genome density that possess icosahedral symmetry can be imaged. In most cases, only a small fraction of the genome of icosahedral ssRNA viruses displays icosahedral symmetry. An interesting exception is the family of the Nodaviridae, in which a notable amount of the ssRNA genome obeys icosahedral symmetry in the form of double-stranded sequences lining the capsid edges85. However, it has also become possible to visualize the genomes of individual viruses by asymmetric reconstruction, which has been performed for BMV7 and MS2 (Fig. 3B)86.
A large fraction of the MS2 genome is in the form of double-stranded sequences organized in a pattern that has little icosahedral symmetry. A number of the double-stranded sequences are stem-loops closely associated with protein dimers located along one half of the protein capsid. A natural explanation for this organization is that a specific sequence of stem-loops of the secondary structure of the genome molecule in solution associate with protein dimers during the early stages of assembly of the virion. Once half of the capsid has formed, the remainder ‘snaps together’ more rapidly, forcing additional condensation that transforms ssRNA sequences into double-stranded material86 (Fig. 3C). A study identified 80% of the MS2 genome material inside the virion6 and showed that specific stem-loops of the secondary structure of the MS2 genome are associated with specific capsid protein dimers. A geometrical method for reconstructing the MS2 assembly history has been proposed87,88 (Fig. 3D). It starts from the cryo-EM reconstruction of the RNA density, which is then represented as a cage of lines with icosahedral symmetry. A path is constructed on this cage that visits all vertices of the icosahedron, known as a Hamiltonian path89. Such a path no longer has icosahedral symmetry. Through a geometrical construction, the Hamiltonian path can be translated into an assembly sequence. This form of assembly, with a disperse set of packaging signals guiding the process90, should be contrasted with that of TMV, BMV and the retroviruses, in which packaging signals only initiate assembly. Indeed, reconstruction of the BMV genome indicates a much weaker correlation between capsid and genome structures7. It would be interesting to compare measurements of the kinetics of virion assembly with kinetic models for the assembly kinetics as discussed for the case of empty capsids, no such model is not yet available. To support the development of such a model, systematic experimental comparisons of the assembly kinetics of empty capsids with that of virions would be very useful.
Dynamics of closed shells
The initial assembly of a capsid often is only the first step in a shorter or longer process of dynamical development known as maturation. Not all viruses undergo maturation, but a wide variety of larger viruses do mature after assembly. Maturation involves conformational changes of the capsid proteins that either strengthen the capsid or prime the virion for genome release. It may have an evolutionary character, as exemplified by the sequence of conformational changes of the Escherichia coli phage HK97, or the character of a striking capsid metamorphosis, as exemplified by HIV-1. Apart from the irreversible dynamics of the maturation process, capsids in thermal equilibrium are also subject to reversible thermal shape fluctuations and soft modes.
Viral maturation
Our first example of maturation is the family of retroviruses, which, at least initially, have ssRNA genomes. Retroviruses assemble either inside the cytoplasm of an infected cell or on the cytoplasmic membrane (hereafter PM)91. In either case, they acquire a lipid envelope when they bud from the PM92. The resulting spherical immature particle is not yet infectious. This immature particle undergoes a conformational transformation induced by proteolytic cleavage of the Gag structural proteins of the capsid, resulting in the formation of a new, smaller, capsid surrounded by a spherical lipid–protein membrane93,94. The mature capsid itself may be cone-like, in the case of HIV-1, or spherical or sphero-cylindrical for other retroviruses. The mature virion is infectious. After its membrane proteins bind to host cell receptors — for instance, CD4 receptors of a T cell for HIV infection — the outer membrane fuses with the host cell membrane93,95,96. This fusion occurs either at the cell surface or in intracellular vesicles, and results in the release of the mature capsid into the cytoplasm of the cell.
Maturation is promoted and inhibited in various processes. Structural and dynamic studies using solution nuclear magnetic resonance (NMR) and solid-state NMR have shed light on proteolytic processing of Gag97 and the effect of maturation inhibitors98. Furthermore, cryo-electron tomography that included subtomogram averaging99 was used to study Gag regions that are targeted by maturation inhibitors100. Besides stimulating assembly of HIV particles, inositol phosphate — a compound that is present in all mammalian cells — also promotes maturation101. By shielding repulsive charges, it first aids structural rearrangements of Gag, and during maturation it stimulates hexamer formation.
Approaches focusing on the mechanical properties of retroviral capsids using AFM nanoindentation102–106 have further elucidated the physics of maturation. The retrovirus Moloney murine leukaemia virus undergoes a marked softening of the shell during maturation107, as expected in view of the proteolytic cleavage of the capsid proteins. A similar softening of the shell occurs for HIV-1108,109. A second marked change in the mechanical properties of the capsid takes place during infection. After entry into the new host cell, the very flexible ssRNA genome inside the capsid is transformed by the co-packaged reverse transcriptase enzyme into a much stiffer dsDNA genome. The necessary nucleotides for reverse transcription centre the capsid through dynamic, size-selective pores that are present in the centres of the hexamers110. The mechanical stress exerted by the newly formed enclosed dsDNA on the mature capsid causes it to rupture, allowing release of the genome into the cell. The ruptured capsids have been visualized both by AFM and electron microscopy111 (Fig. 4a). Under in vivo conditions, this rupture probably takes place after the mature capsid has docked on a nuclear pore complex. Softening and rupture of the capsid is a key for successful infection of the host cell by the viral particles: this reveals that there is a direct link between capsid mechanics and the infectivity of retroviruses112.
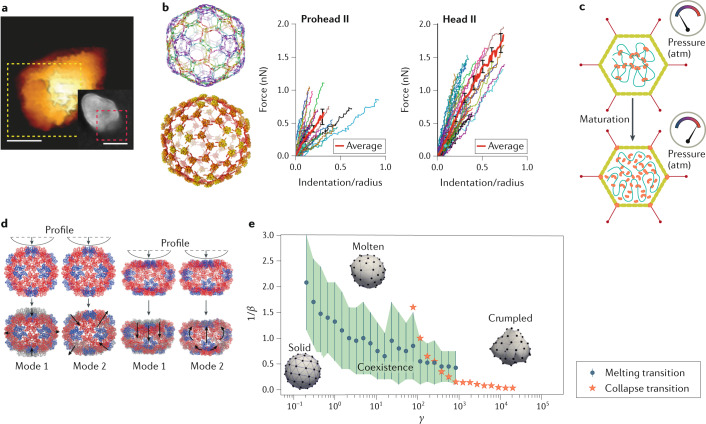
Fig. 4. Closed-shell dynamics.
a | Atomic force microscopy (AFM) image of human immunodeficiency virus (HIV) capsid with opening (rectangle) due to reverse transcription. Inset: electron microscopy image of a different particle with an opening. Scale bars, 50 nm. b | Phages HK97 (top left) and λ (bottom left) strengthen the same parts of their icosahedral shells, as visualized by these reconstructions highlighting the differences between the two particles. AFM nanoindentation curves (right panels) show the difference in mechanical response of HK97 Prohead II and Head II. Error bars indicate standard error of the mean. c | Schematic of the effect of maturation in adenovirus. d | Cowpea chlorotic mottle virus (CCMV) nanoindentation in silico, showing the first two collective excitation modes (black arrows). Pentamers are shown in blue and hexamers in red. e | Phase diagram of capsid soft modes with the dimensionless temperature β−1 as a function of the Föppl–von Kármán number γ. Vertical blue bars indicate the regions of phase coexistence of solid and molten states. Part a adapted with permission from ref.111. Part b adapted with permission from refs115,118. Part c adapted with permission from ref.123. Part d adapted with permission from ref.139. Part e adapted with permission from ref.152.
Whereas the capsids of retroviruses soften during maturation, bacteriophages undergo a maturation transition that strengthens their capsids113. Recall that, on the one hand, the capsids of phage viruses must withstand very large osmotic pressures exerted by the enclosed dsDNA genome. On the other hand, during assembly the interactions between capsid proteins must be sufficiently weak to allow for the thermal annealing of kinetic traps. Phage viruses thus must transform their capsids from the initial, relatively unstable, procapsid into a very robust shell. As in the case of retroviruses, phage maturation involves a conformational rearrangement of the shell. For instance, the bacteriophage λ and HK97 virus initially assemble as ~55-nm-diameter empty shells that, through a sequence of stages, mature into ~65-nm-diameter genome-packed particles with a thinner shell. This maturation transition goes hand in hand with a strengthening of the capsid, as observed by AFM nanoindentation studies of phage λ114 and HK97115. For HK97, capsid maturation is composed of a sequence of coordinated, discrete steps ending with a covalent interlocking network of bonds that resembles the chainmail of medieval soldiers. The basic structural motif of the capsid protein of HK97 that governs the maturation steps is shared with λ and other bacteriophages such as P22, as well as the eukaryotic herpesviruses116. In fact, these viruses share genetic motifs117. In the initial procapsid state, the capsid protein can be represented as a compressed spring, which relaxes during the swelling and thinning of the capsid. Interestingly, although the HK97 and λ phages have different pathways to strengthen their shells (covalent crosslinking in the case of HK97 and addition of gpD protein in the case of the λ phage), the icosahedral symmetry sites on the capsid that are strengthened are the same118 (Fig. 4b). Furthermore, the increase in mechanical capsid strength during maturation (shown in Fig. 4b for HK97) is similar, even though the covalent crosslinking that occurs during HK97 maturation renders a particle that can be deformed to a larger extent than phage λ before failure occurs115. The maturation of bacteriophages P22 and T7 also leads to an increase in stability, as revealed by nanoindentation119,120.
Not every dsDNA virus undergoes the same maturation process. The maturation of the dsDNA adenovirus is markedly different from that of dsDNA bacteriophages. Maturation of adenovirus involves proteolytic cleavage of a variety of viral precursor proteins121. Comparison of immature and mature particles revealed a clear difference in terms of mechanical response and DNA decondensation. An increase in flexibility of the DNA during maturation — and hence an increase in osmotic pressure — seems to be an essential condition for effective genome release122,123. In concert with DNA decondensation, the pentameric structures of the capsid are also destabilized during maturation124,125, presumably assisting the egress of DNA (Fig. 4c). By the successive action of host-cell integrin molecules, which serve as secondary receptors during adenovirus docking, the pentameric structures become loose enough to be released into the endosome126,127. In other words, whereas dsDNA bacteriophages are strengthened during maturation, adenoviruses become more flexible during maturation, reminiscent of retrovirus maturation.
Soft modes and conformational dynamics
Structurally ordered capsids are reminiscent of tiny crystals. Although viral maturation is associated with complex biochemical processes, it is worth comparing the maturation steps discussed above with the physical phenomena that occur when the structure of a crystal changes as a function of some thermodynamic control parameter, such as temperature. An important feature of such structural phase transitions is the appearance of soft modes128. These are collective modes whose energy vanishes in the limit of long wavelengths and that can be viewed as precursors of the structural transition. Near the transition point, the stiffness of these modes softens substantially. As discussed below, very similar soft modes have been encountered for viral capsids. A Ginzburg–Landau theory that was developed to describe structural transitions and soft modes in solids can be adapted directly to structural transitions of viral shells129. Soft modes of viral capsids are generally associated with specific conformational changes of the capsid protein. However, according to thin-shell elasticity theory, any icosahedral shell should show mode softening at the buckling transition130,131. At a buckling transition, the shape of a shell with icosahedral symmetry changes from spherical to polyhedral. According to this theory, the buckling transition and the response of a shell to mechanical deformation should be determined by just a single dimensionless number, the Föppl–von Kármán (FvK) number γ, which is the ratio of the in-plane Young’s modulus of the shell and the out-of-plane bending modulus132. Soft modes associated with buckling have indeed been reported in molecular dynamics (MD) simulations of the capsid maturation of HK97133
The pioneering cryogenic X-ray and electron microscopy structural studies mentioned earlier reported that viral capsids were static crystallographic structures. However, more recent structural studies of capsids carried under room-temperature conditions found that, in fact, capsids are dynamical structures even when they are not undergoing maturation134,135. An interesting case study is the swelling transition of CCMV. The radius of a CCMV capsid increases substantially when the pH is raised from 5 to 7 (ref.136), whereas its bending stiffness, which can be probed by AFM nanoindentation132, shows a marked reduction. Such a reduction of the bending stiffness of a spherical surface indicates that the relaxation rates of the various spherical harmonic modes of the surface should be reduced137. Reduced relaxation rates, or ‘mode softening’, have indeed been observed for CCMV138. The highly dynamical nature of the CCMV capsid is illustrated at a more local level by the fact that protease enzymes are able to selectively digest the N-termini of the capsid proteins of CCMV capsids135. This is surprising because low-temperature X-ray diffraction and cryo-EM studies136 reported that these N-termini are located in the capsid interior even though the proteases are too large to centre the capsid. The implication is that capsids can be much more dynamic than indicated by low-temperature structural studies.
The effect of both in-plane and out-of-plane soft modes on the capsid thermodynamics of CCMV has been probed quantitatively through the coupling of numerical simulations to AFM nanoindentation data139 (Fig. 4d). The results of MD simulations can also be compared with studies by Raman spectroscopy140. The equilibrium dynamics of the capsid of HBV has been examined in considerable detail by MD, and the results were used to reinterpret cryo-EM imaging of HBV capsids141. MD in combination with solid-state NMR or AFM has been used to study the effect of host factor protein binding on HIV-1 assemblies, revealing that such factors could modulate capsid stability and thereby possibly infection142,143. One can also observe changes by hydrogen–deuterium exchange mass spectrometry, as for instance shown for MVM and dengue virus144,145; conformational fluctuations under equilibrium conditions of the closed MVM shells, termed ‘breathing’, have been observed144. Vibrational modes of solvated HIV-1 capsids were also studied by MD simulations, revealing the presence of breathing motions that simultaneously are directed radially as well as in the plane of the capsid surface146.
All-atom MD simulations of viruses147–149 — which have also been used to study the permeability of poliovirus capsids to water150 and the binding of ions to HIV-1 capsids146, for example — indicate that mutated capsids may collapse because of thermal fluctuations151. Relatedly, Brownian dynamics simulations of a coarse-grained representation of capsids revealed that they can melt, crumple or collapse under the action of thermal fluctuations, depending on the materials properties of the shell152 (Fig. 4e). The finite temperature phase behaviour is largely determined by the same FvK number that determines the response to mechanical deformation in the absence of thermal fluctuations. Viruses whose capsids lack structural order are known as pleiomorphic153, and some of these pleiomorphic viruses may well have capsids that are effectively in a liquid state154. A particularly interesting case is the family of archaeal viruses155. Certain archaeal viruses, whose capsids bear no resemblance to the classical rod and sphere viral geometries, can smoothly transform the Gaussian curvature of their capsids, which suggests that these capsids are in a liquid state153–157.
Conclusion
In this Review, we have discussed how recent developments in experimental and numerical techniques in the burgeoning field of physical virology have allowed a move from static descriptions of viral properties to descriptions of dynamical properties. With further refinement of the existing techniques, as well as the development of new techniques, in the coming years we expect large steps in terms of understanding the dynamical properties of viruses. Examples of potential new physical virology approaches in the experimental fields include developments in optical microscopy that allow for nanometre resolution imaging158, or in AFM, allowing the acquisition of millisecond to microsecond height spectroscopy data159 for real-time assembly studies43,160. Beyond using such techniques for an improved understanding of assembly and steady-state dynamics, it also would be interesting to perform assembly kinetics experiments in crowded environments, to mimic the actual situation in cells and to reveal whether depletion attraction and other crowding effects alter the results.
Numerical simulations of coarse-grained models of viral capsids and their genomes have played an important role in improving our physical understanding of the dynamics of viruses. All-atom MD simulations of small viruses are possible, and they provide important insights into the soft-mode dynamics and kinetic trapping of assembled viruses. Still, all-atom MD simulations of viral assembly (and disassembly) are not practical in the foreseeable future because of the prohibitively long timescales of viral assembly (seconds to hours). To the extent that this is due to the presence of large energy activation barriers, metadynamics methods161 could make such simulations tractable. Separately, the combination of MD with Markov state modelling could provide a systematic road to the development of coarse-grained models of viral assembly162. Additionally, the development of kinetic models for the assembly kinetics of genome-filled particles, as already available for the case of empty capsids23,24, will be useful. Apart from the general physical insights that this research generates, in the wake of the crisis caused by COVID-19 the importance of fundamental knowledge of viral properties has been stressed again163–166. Physical virology approaches will help to pave the way to in-depth knowledge of the mechanisms behind viral reproduction and infection; they also promise to help in developing applications of viruses and VLPs in nanotechnology and nanomedicine.
Acknowledgements
The authors thank C. Knobler for a critical reading of the manuscript, and C. Uetrecht and S. Maity for their input. W.H.R. is supported by a VIDI grant from the Nederlandse Oganizatie voor Wetenschappelijk Onderzoek. R.B. is supported by the NSF-DMR under CMMT grant 1836404.
Glossary
- Capsids
Protein shells that surround the viral genome.
- Triangulation numbers
Classification system, developed by Caspar and Klug, for icosahedral viruses. T-numbers are integers and contain information on the number of protein subunits that make up a capsid.
- Virions
Viral particles, composed of both capsid proteins and the viral genome, that can successfully infect cells.
- Kinetic traps
Kinetically trapped assembly intermediates that are dead-end products and do not lead to the formation of correctly assembled viral particles.
- Bacteriophage
Also called phages, these are viruses that infect bacteria.
- Procapsids
Immature and precursor viral capsid structures of certain viruses.
- Host cells
Cells infected by virions.
- Archaeal
Next to bacteria and eukaryotes, archaea represent one of the three domains of life.
- Prokaryotic
A prokaryote is a single cell organism lacking a nucleus, that is, bacteria or archaea.
- Eukaryotic
A eukaryote is an organism built up from eukaryotic cells, that is, from cells that possess a nucleus. Animals, plants and fungi are examples of eukaryotes.
- Nucleotides
Building blocks of nucleic acids (of DNA or RNA).
- Secondary structure
The set of formed base pairs in a nucleic acid strand; in other words, the set of links between nucleotides in the strand.
- PolyU
Nucleic acid strand consisting solely of copies of the uracil (U) nucleotide.
- PolyA
Nucleic acid strand consisting solely of copies of the adenine (A) nucleotide.
- Langmuir adsorption theory
Model describing key physics of molecular interactions at interfaces.
- Gag protein
Group-specific antigen protein. The Gag polyprotein makes up the inner shell of immature retroviral particles, and during maturation Gag is proteolytically cleaved into MA (matrix), CA (capsid) and NC (nucleocapsid) domains leading to the formation of a mature virion.
- Nucleocapsid domain
Viral protein coat surrounding the viral genome. One of the three domains resulting from Gag protein cleavage.
- Retroviruses
Group of viruses with a viral RNA genome. During infection, DNA is produced using the viral RNA.
- Maturation
Process of conformational changes and/or proteolytic cleavage of viral proteins transforming the immature particle into a mature, infectious particle.
- Cytoplasmic membrane
Membrane enclosing the cellular cytoplasm. It is the outer membrane of eukaryotic cells and the first cellular contact point for viruses during infection.
- Endosome
Intracellular vesicles. Certain viruses are internalized into their host cell in an endosome and subsequently leave this endosome to continue the infectious pathway.
Author contributions
All authors contributed to all aspects of the article.
Competing interests
G.J.L.W. declares financial interest in fluorescence optical tweezers and acoustic force spectroscopy approaches which are patented and licensed to LUMICKS B.V.
Footnotes
Peer review information
Nature Reviews Physics thanks Kazunori Matsuura and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 1999;63:862. doi: 10.1128/MMBR.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caspar DLD, Klug A. Physical principles in construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/SQB.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 3.Roos WH, Bruinsma R, Wuite GJL. Physical virology. Nat. Phys. 2010;6:733–743. doi: 10.1038/nphys1797. [DOI] [Google Scholar]
- 4.Grunewald K, et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 5.Wan W, et al. Structure and assembly of the Ebola virus nucleocapsid. Nature. 2017;551:394–397. doi: 10.1038/nature24490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai XH, et al. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature. 2017;541:112–116. doi: 10.1038/nature20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beren C, et al. Genome organization and interaction with capsid protein in a multipartite RNA virus. Proc. Natl Acad. Sci. USA. 2020;117:10673. doi: 10.1073/pnas.1915078117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraenkel-Conrat H, Williams RC. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Natl Acad. Sci. USA. 1955;41:690–698. doi: 10.1073/pnas.41.10.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler PJG, Klug A. Assembly of a virus. Sci. Am. 1978;239:62–69. doi: 10.1038/scientificamerican1178-62. [DOI] [PubMed] [Google Scholar]
- 10.Bancroft, J. B. in Advances in Virus Research Vol. 16 (eds Smith, K. M. et al.) 99–134 (Academic, 1970). [DOI] [PubMed]
- 11.Adolph KW, Butler PJG. Assembly of a spherical plant virus. Phil. Trans. R. Soc. Lond. B. 1976;276:113–122. doi: 10.1098/rstb.1976.0102. [DOI] [PubMed] [Google Scholar]
- 12.Bancroft JB, Hiebert E. Formation of an infectious nucleoprotein from protein and nucleic acid isolated from a small spherical virus. Virology. 1967;32:354–356. doi: 10.1016/0042-6822(67)90284-X. [DOI] [PubMed] [Google Scholar]
- 13.Klug A. The tobacco mosaic virus particle: structure and assembly. Phil. Trans. R. Soc. Lond. B. 1999;354:531–535. doi: 10.1098/rstb.1999.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prinsen P, van der Schoot P, Gelbart WM, Knobler CM. Multishell structures of virus coat proteins. J. Phys. Chem. B. 2010;114:5522–5533. doi: 10.1021/jp911040z. [DOI] [PubMed] [Google Scholar]
- 15.Kegel WK, van der Schoot P. Physical regulation of the self-assembly of tobacco mosaic virus coat protein. Biophys. J. 2006;91:1501–1512. doi: 10.1529/biophysj.105.072603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceres P, Zlotnick A. Weak protein-protein interactions are sufficient to drive assembly of hepatitis B virus capsids. Biochemistry. 2002;41:11525–11531. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- 17.McPherson A. Micelle formation and crystallization as paradigms for virus assembly. BioEssays. 2005;27:447–458. doi: 10.1002/bies.20196. [DOI] [PubMed] [Google Scholar]
- 18.Prevelige PE, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.del Alamo M, Mateu MG. Electrostatic repulsion, compensatory mutations, and long-range non-additive effects at the dimerization interface of the HIV capsid protein. J. Mol. Biol. 2005;345:893–906. doi: 10.1016/j.jmb.2004.10.086. [DOI] [PubMed] [Google Scholar]
- 20.Casini GL, Graham D, Heine D, Garcea RL, Wu DT. In vitro papillomavirus capsid assembly analyzed by light scattering. Virology. 2004;325:320–327. doi: 10.1016/j.virol.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 21.Zandi R, van der Schoot P, Reguera D, Kegel W, Reiss H. Classical nucleation theory of virus capsids. Biophys. J. 2006;90:1939–1948. doi: 10.1529/biophysj.105.072975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medrano M, et al. Imaging and quantitation of a succession of transient intermediates reveal the reversible self-assembly pathway of a simple icosahedral virus capsid. J. Am. Chem. Soc. 2016;138:15385–15396. doi: 10.1021/jacs.6b07663. [DOI] [PubMed] [Google Scholar]
- 23.Zlotnick A. Distinguishing reversible from irreversible virus capsid assembly. J. Mol. Biol. 2007;366:14–18. doi: 10.1016/j.jmb.2006.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morozov AY, Bruinsma RF, Rudnick J. Assembly of viruses and the pseudo-law of mass action. J. Chem. Phys. 2009;131:155101. doi: 10.1063/1.3212694. [DOI] [PubMed] [Google Scholar]
- 25.Uetrecht C, Barbu IM, Shoemaker GK, van Duijn E, Heck AJR. Interrogating viral capsid assembly with ion mobility-mass spectrometry. Nat. Chem. 2011;3:126–132. doi: 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]
- 26.Lutomski CA, et al. Hepatitis B virus capsid completion occurs through error correction. J. Am. Chem. Soc. 2017;139:16932–16938. doi: 10.1021/jacs.7b09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comas-Garcia M, Cadena-Nava RD, Rao ALN, Knobler CM, Gelbart WM. In vitro quantification of the relative packaging efficiencies of single-stranded RNA molecules by viral capsid protein. J. Virol. 2012;86:12271–12282. doi: 10.1128/JVI.01695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varrelmann M, Maiss E. Mutations in the coat protein gene of Plum pox virus suppress particle assembly, heterologous encapsidation and complementation in transgenic plants of Nicotiana benthamiana. J. Gen. Virol. 2000;81:567–576. doi: 10.1099/0022-1317-81-3-567. [DOI] [PubMed] [Google Scholar]
- 29.Tang JH, et al. The role of subunit hinges and molecular “switches” in the control of viral capsid polymorphism. J. Struct. Biol. 2006;154:59–67. doi: 10.1016/j.jsb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Lutomski CA, et al. Multiple pathways in capsid assembly. J. Am. Chem. Soc. 2018;140:5784–5790. doi: 10.1021/jacs.8b01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endres D, Zlotnick A. Model-based analysis of assembly kinetics for virus capsids or other spherical polymers. Biophys. J. 2002;83:1217–1230. doi: 10.1016/S0006-3495(02)75245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagan MF, Elrad OM, Jack RL. Mechanisms of kinetic trapping in self-assembly and phase transformation. J. Chem. Phys. 2011;135:104115. doi: 10.1063/1.3635775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaels TCT, Bellaiche MMJ, Hagan MF, Knowles TPJ. Kinetic constraints on self-assembly into closed supramolecular structures. Sci. Rep. 2017;7:12295. doi: 10.1038/s41598-017-12528-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levinthal C. Are there pathways for protein folding? J. Chim. Phys. 1968;65:44–45. doi: 10.1051/jcp/1968650044. [DOI] [Google Scholar]
- 35.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein-folding — a synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 36.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: a kinetic approach to the sequence–structure relationship. Proc. Natl Acad. Sci. USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asor R, et al. Assembly reactions of hepatitis B capsid protein into capsid nanoparticles follow a narrow path through a complex reaction landscape. ACS Nano. 2019;13:7610–7626. doi: 10.1021/acsnano.9b00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tuma R, Tsuruta H, French KH, Prevelige PE. Detection of intermediates and kinetic control during assembly of bacteriophage P22 procapsid. J. Mol. Biol. 2008;381:1395–1406. doi: 10.1016/j.jmb.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tresset G, et al. Norovirus capsid proteins self-assemble through biphasic kinetics via long-lived stave-like intermediates. J. Am. Chem. Soc. 2013;135:15373–15381. doi: 10.1021/ja403550f. [DOI] [PubMed] [Google Scholar]
- 40.Law-Hine D, Zeghal M, Bressanelli S, Constantin D, Tresset G. Identification of a major intermediate along the self-assembly pathway of an icosahedral viral capsid by using an analytical model of a spherical patch. Soft Matter. 2016;12:6728–6736. doi: 10.1039/C6SM01060A. [DOI] [PubMed] [Google Scholar]
- 41.Chen B. HIV capsid assembly, mechanism, and structure. Biochemistry. 2016;55:2539–2552. doi: 10.1021/acs.biochem.6b00159. [DOI] [PubMed] [Google Scholar]
- 42.Valbuena A, Mateu MG. Kinetics of surface-driven self-assembly and fatigue-induced disassembly of a virus-based nanocoating. Biophys. J. 2017;112:663–673. doi: 10.1016/j.bpj.2016.11.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valbuena A, Maity S, Mateu MG, Roos WH. Visualization of single molecules building a viral capsid protein lattice through stochastic pathways. ACS Nano. 2020;14:8724–8734. doi: 10.1021/acsnano.0c03207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ando T, Uchihashi T, Scheuring S. Filming biomolecular processes by high-speed atomic force microscopy. Chem. Rev. 2014;114:3120–3188. doi: 10.1021/cr4003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maity S, et al. VPS4 triggers constriction and cleavage of ESCRT-III helical filaments. Sci. Adv. 2019;5:eaau7198. doi: 10.1126/sciadv.aau7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruan Y, et al. Structural titration of receptor ion channel GLIC gating by HS-AFM. Proc. Natl Acad. Sci. USA. 2018;115:10333–10338. doi: 10.1073/pnas.1805621115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou JS, et al. Characterization of virus capsids and their assembly intermediates by multicycle resistive-pulse sensing with four pores in series. Anal. Chem. 2018;90:7267–7274. doi: 10.1021/acs.analchem.8b00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kondylis P, et al. Competition between normative and drug-induced virus self-assembly observed with single-particle methods. J. Am. Chem. Soc. 2019;141:1251–1260. doi: 10.1021/jacs.8b10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brändén, C.-I. & Tooze, J. Introduction to Protein Structure 2nd edn (Garland, 1999).
- 50.Rao, V. B. & Feiss, M. Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu. Rev. Virol.2, 351–378 (2015). [DOI] [PMC free article] [PubMed]
- 51.Smith DE, et al. The bacteriophage phi 29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 52.Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proc. Natl Acad. Sci. USA. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keller N, Grimes S, Jardine PJ, Smith DE. Single DNA molecule jamming and history-dependent dynamics during motor-driven viral packaging. Nat. Phys. 2016;12:757. doi: 10.1038/nphys3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tafoya S, et al. Molecular switch-like regulation enables global subunit coordination in a viral ring ATPase. Proc. Natl Acad. Sci. USA. 2018;115:7961–7966. doi: 10.1073/pnas.1802736115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ordyan M, Alam I, Mahalingam M, Rao VB, Smith DE. Nucleotide-dependent DNA gripping and an end-clamp mechanism regulate the bacteriophage T4 viral packaging motor. Nat. Commun. 2018;9:5434. doi: 10.1038/s41467-018-07834-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newcomb WW, Cockrell SK, Homa FL, Brown JC. Polarized DNA ejection from the Herpesvirus capsid. J. Mol. Biol. 2009;392:885–894. doi: 10.1016/j.jmb.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanhijarvi KJ, Ziedaite G, Pietila MK, Haeggstrom E, Bamford DH. DNA ejection from an archaeal virus — a single-molecule approach. Biophys. J. 2013;104:2264–2272. doi: 10.1016/j.bpj.2013.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mangenot S, Hochrein M, Radler J, Letellier L. Real-time imaging of DNA ejection from single phage particles. Curr. Biol. 2005;15:430–435. doi: 10.1016/j.cub.2004.12.080. [DOI] [PubMed] [Google Scholar]
- 59.Tsukamoto H, et al. Evidence that SV40VP1-DNA interactions contribute to the assembly of 40-nm spherical viral particles. Genes Cells. 2007;12:1267–1279. doi: 10.1111/j.1365-2443.2007.01134.x. [DOI] [PubMed] [Google Scholar]
- 60.Cadena-Nava RD, et al. Self-assembly of viral capsid protein and RNA molecules of different sizes: requirement for a specific high protein/RNA mass ratio. J. Virol. 2012;86:3318–3326. doi: 10.1128/JVI.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perlmutter JD, Hagan MF. Mechanisms of viral assembly. Annu. Rev. Phys. Chem. 2015;66:217–239. doi: 10.1146/annurev-physchem-040214-121637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu YF, Zandi R, Anavitarte A, Knobler CM, Gelbart WM. Packaging of a polymer by a viral capsid: the interplay between polymer length and capsid size. Biophys. J. 2008;94:1428–1436. doi: 10.1529/biophysj.107.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beren C, Dreesens LL, Liu KN, Knobler CM, Gelbart WM. The effect of RNA secondary structure on the self-assembly of viral capsids. Biophys. J. 2017;113:339–347. doi: 10.1016/j.bpj.2017.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu T, Zhang R, Shkovskii BI. Electrostatic theory of viral self-assembly. Phys. A. 2008;387:3059–3064. doi: 10.1016/j.physa.2008.01.010. [DOI] [Google Scholar]
- 65.Garmann RF, et al. Role of electrostatics in the assembly pathway of a single-stranded RNA virus. J. Virol. 2014;88:10472–10479. doi: 10.1128/JVI.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gopal, A. et al. Viral RNAs are unusually compact. PLoS ONE9, e105875 (2014). [DOI] [PMC free article] [PubMed]
- 67.Perlmutter JD, Perkett MR, Hagan MF. Pathways for virus assembly around nucleic acids. J. Mol. Biol. 2014;426:3148–3165. doi: 10.1016/j.jmb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu T, Shklovskii BI. Kinetics of viral self-assembly: role of the single-stranded RNA antenna. Phys. Rev. E. 2007;75:051901. doi: 10.1103/PhysRevE.75.051901. [DOI] [PubMed] [Google Scholar]
- 69.Bruinsma RF, Comas-Garcia M, Garmann RF, Grosberg AY. Equilibrium self-assembly of small RNA viruses. Phys. Rev. E. 2016;93:032405. doi: 10.1103/PhysRevE.93.032405. [DOI] [PubMed] [Google Scholar]
- 70.Garmann RF, Goldfain AM, Manoharan VN. Measurements of the self-assembly kinetics of individual viral capsids around their RNA genome. Proc. Natl Acad. Sci. USA. 2019;116:22485–22490. doi: 10.1073/pnas.1909223116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dragnea B. Watching a virus grow. Proc. Natl Acad. Sci. USA. 2019;116:22420–22422. doi: 10.1073/pnas.1915986116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marchetti M, et al. Real-time assembly of viruslike nucleocapsids elucidated at the single-particle level. Nano Lett. 2019;19:5746–5753. doi: 10.1021/acs.nanolett.9b02376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Rosmalen MGM, et al. Revealing in real-time a multistep assembly mechanism for SV40 virus-like particles. Sci. Adv. 2020;6:eaaz1639. doi: 10.1126/sciadv.aaz1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sandalon Z, DalyotHerman N, Oppenheim AB, Oppenheim A. In vitro assembly of SV40 virions and pseudovirions: vector development for gene therapy. Hum. Gene Ther. 1997;8:843–849. doi: 10.1089/hum.1997.8.7-843. [DOI] [PubMed] [Google Scholar]
- 75.van Rosmalen MGM, Li C, Zlotnick A, Wuite GJL, Roos WH. Effect of dsDNA on the assembly pathway and mechanical strength of SV40-VP1 virus-like particles. Biophys. J. 2018;115:1656–1665. doi: 10.1016/j.bpj.2018.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kler S, et al. RNA encapsidation by SV40-derived nanoparticles follows a rapid two-state mechanism. J. Am. Chem. Soc. 2012;134:8823–8830. doi: 10.1021/ja2110703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chevreuil M, et al. Nonequilibrium self-assembly dynamics of icosahedral viral capsids packaging genome or polyelectrolyte. Nat. Commun. 2018;9:3071. doi: 10.1038/s41467-018-05426-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rein A. RNA packaging in HIV. Trends Microbiol. 2019;27:715–723. doi: 10.1016/j.tim.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Comas-Garcia M, et al. Efficient support of virus-like particle assembly by the HIV-1 packaging signal. eLife. 2018;7:e38438. doi: 10.7554/eLife.38438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Keane SC, et al. Structure of the HIV-1 RNA packaging signal. Science. 2015;348:917. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ding P, et al. Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc. Natl Acad. Sci. USA. 2020;117:17737. doi: 10.1073/pnas.2008519117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Webb JA, Jones CP, Parent LJ, Rouzina I, Musier-Forsyth K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging. RNA. 2013;19:1078–1088. doi: 10.1261/rna.038869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang W, Tang L. Atomic cryo-EM structures of viruses. Curr. Opin. Struct. Biol. 2017;46:122–129. doi: 10.1016/j.sbi.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luque D, Caston JR. Cryo-electron microscopy for the study of virus assembly. Nat. Chem. Biol. 2020;16:231–239. doi: 10.1038/s41589-020-0477-1. [DOI] [PubMed] [Google Scholar]
- 85.Tang L, et al. The structure of Pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 2001;8:77–83. doi: 10.1038/83089. [DOI] [PubMed] [Google Scholar]
- 86.Koning RI, et al. Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat. Commun. 2016;7:12524. doi: 10.1038/ncomms12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Twarock R, Leonov G, Stockley PG. Hamiltonian path analysis of viral genomes. Nat. Commun. 2018;9:2021. doi: 10.1038/s41467-018-03713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dykeman EC, et al. Simple rules for efficient assembly predict the layout of a packaged viral RNA. J. Mol. Biol. 2011;408:399–407. doi: 10.1016/j.jmb.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 89.Rudnick J, Bruinsma R. Icosahedral packing of RNA viral genomes. Phys. Rev. Lett. 2005;94:038101. doi: 10.1103/PhysRevLett.94.038101. [DOI] [PubMed] [Google Scholar]
- 90.Twarock, R. & Stockley, P. G. RNA-mediated virus assembly: mechanisms and consequences for viral evolution and therapy. Annu. Rev. Biophys. 48, 495–514 (2019).. [DOI] [PMC free article] [PubMed]
- 91.Coffin, J. M., Hughes, S. H. & Varmus, H. E. Retroviruses (Cold Spring Harbor Laboratory Press, 1997). [PubMed]
- 92.Dharmavaram S, She SB, Lázaro G, Hagan MF, Bruinsma R. Gaussian curvature and the budding kinetics of enveloped viruses. PLoS Comput. Biol. 2019;15:e1006602. doi: 10.1371/journal.pcbi.1006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pornillos O, Ganser-Pornillos BK. Maturation of retroviruses. Curr. Opin. Virol. 2019;36:47–55. doi: 10.1016/j.coviro.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perilla JR, Gronenborn AM. Molecular architecture of the retroviral capsid. Trends Biochem. Sci. 2016;41:410–420. doi: 10.1016/j.tibs.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mattei S, Schur FKM, Briggs JAG. Retrovirus maturation — an extraordinary structural transformation. Curr. Opin. Virol. 2016;18:27–35. doi: 10.1016/j.coviro.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 96.Kubo Y, Hayashi H, Matsuyama T, Sato H, Yamamoto N. Retrovirus entry by endocytosis and cathepsin proteases. Adv. Virol. 2012;2012:640894. doi: 10.1155/2012/640894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deshmukh L, Ghirlando R, Clore GM. Conformation and dynamics of the Gag polyprotein of the human immunodeficiency virus 1 studied by NMR spectroscopy. Proc. Natl Acad. Sci. USA. 2015;112:3374–3379. doi: 10.1073/pnas.1501985112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gupta S, Louis JM, Tycko R. Effects of an HIV-1 maturation inhibitor on the structure and dynamics of CA-SP1 junction helices in virus-like particles. Proc. Natl Acad. Sci. USA. 2020;117:10286–10293. doi: 10.1073/pnas.1917755117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Briggs JAG. Structural biology in situ — the potential of subtomogram averaging. Curr. Opin. Struct. Biol. 2013;23:261–267. doi: 10.1016/j.sbi.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 100.Schur FKM, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 101.Dick RA, et al. Inositol phosphates are assembly co-factors for HIV-1. Nature. 2018;560:509–512. doi: 10.1038/s41586-018-0396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marchetti M, Wuite GJL, Roos WH. Atomic force microscopy observation and characterization of single virions and virus-like particles by nano-indentation. Curr. Opin. Virol. 2016;18:82–88. doi: 10.1016/j.coviro.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 103.de Pablo PJ, Mateu MG. Mechanical properties of viruses. Subcell. Biochem. 2013;68:519–551. doi: 10.1007/978-94-007-6552-8_18. [DOI] [PubMed] [Google Scholar]
- 104.Buzón P, Maity S, Roos WH. Physical virology: from virus self-assembly to particle mechanics. WIREs Nanomed. Nanobiotechnol. 2020;12:e1613. doi: 10.1002/wnan.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ivanovska IL, et al. Bacteriophage capsids: tough nanoshells with complex elastic properties. Proc. Natl Acad. Sci. USA. 2004;101:7600–7605. doi: 10.1073/pnas.0308198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Snijder J, et al. Probing the biophysical interplay between a viral genome and its capsid. Nat. Chem. 2013;5:502–509. doi: 10.1038/nchem.1627. [DOI] [PubMed] [Google Scholar]
- 107.Kol N, et al. Mechanical properties of murine leukemia virus particles: effect of maturation. Biophys. J. 2006;91:767–774. doi: 10.1529/biophysj.105.079657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kol N, et al. A stiffness switch in human immunodeficiency virus. Biophys. J. 2007;92:1777–1783. doi: 10.1529/biophysj.106.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pang HB, et al. Virion stiffness regulates immature HIV-1 entry. Retrovirology. 2013;10:4. doi: 10.1186/1742-4690-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jacques DA, et al. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature. 2016;536:349–353. doi: 10.1038/nature19098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rankovic S, Varadarajan J, Ramalho R, Aiken C, Rousso I. Reverse transcription mechanically initiates HIV-1 capsid disassembly. J. Virol. 2017;91:e00289-17. doi: 10.1128/JVI.00289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rouzina I, Bruinsma R. DNA confinement drives uncoating of the HIV virus. Eur. Phys. J. Spec. Top. 2014;223:1745–1754. doi: 10.1140/epjst/e2014-02223-x. [DOI] [Google Scholar]
- 113.Veesler D, Johnson JE. Virus maturation. Annu. Rev. Biophys. 2012;41:473–496. doi: 10.1146/annurev-biophys-042910-155407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hernando-Perez M, Lambert S, Nakatani-Webster E, Catalano CE, de Pablo PJ. Cementing proteins provide extra mechanical stabilization to viral cages. Nat. Commun. 2014;5:4520. doi: 10.1038/ncomms5520. [DOI] [PubMed] [Google Scholar]
- 115.Roos WH, et al. Mechanics of bacteriophage maturation. Proc. Natl Acad. Sci. USA. 2012;109:2342–2347. doi: 10.1073/pnas.1109590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiang W, et al. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- 117.Juhala RJ, et al. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 118.Lander GC, et al. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kant R, et al. Changes in the stability and biomechanics of P22 bacteriophage capsid during maturation. Biochim. Biophys. Acta Gen. Subj. 2018;1862:1492–1504. doi: 10.1016/j.bbagen.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 120.Hernando-Perez M, et al. The interplay between mechanics and stability of viral cages. Nanoscale. 2014;6:2702–2709. doi: 10.1039/C3NR05763A. [DOI] [PubMed] [Google Scholar]
- 121.Mangel WF, Martin CSan. Structure, function and dynamics in adenovirus maturation. Viruses. 2014;6:4536–4570. doi: 10.3390/v6114536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Perez-Berna AJ, et al. The role of capsid maturation on adenovirus priming for sequential uncoating. J. Biol. Chem. 2012;287:31582–31595. doi: 10.1074/jbc.M112.389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ortega-Esteban A, et al. Mechanics of viral chromatin reveals the pressurization of human adenovirus. ACS Nano. 2015;9:10826–10833. doi: 10.1021/acsnano.5b03417. [DOI] [PubMed] [Google Scholar]
- 124.Denning D, et al. Maturation of adenovirus primes the protein nano-shell for successful endosomal escape. Nanoscale. 2019;11:4015–4024. doi: 10.1039/C8NR10182E. [DOI] [PubMed] [Google Scholar]
- 125.Ortega-Esteban A, et al. Monitoring dynamics of human adenovirus disassembly induced by mechanical fatigue. Sci. Rep. 2013;3:1434. doi: 10.1038/srep01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lindert S, Silvestry M, Mullen TM, Nemerow GR, Stewart PL. Cryo-electron microscopy structure of an adenovirus–integrin complex indicates conformational changes in both penton base and integrin. J. Virol. 2009;83:11491–11501. doi: 10.1128/JVI.01214-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Snijder J, et al. Integrin and defensin modulate the mechanical properties of adenovirus. J. Virol. 2013;87:2756–2766. doi: 10.1128/JVI.02516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Müller, K. A. & Thomas, H. Structural Phase Transitions I. Topics in Current Physics Vol. 23 (Springer, 1981).
- 129.Guerin T, Bruinsma R. Theory of conformational transitions of viral shells. Phys. Rev. E. 2007;76:061911. doi: 10.1103/PhysRevE.76.061911. [DOI] [PubMed] [Google Scholar]
- 130.Widom M, Lidmar J, Nelson DR. Soft modes near the buckling transition of icosahedral shells. Phys. Rev. E. 2007;76:031911. doi: 10.1103/PhysRevE.76.031911. [DOI] [PubMed] [Google Scholar]
- 131.Lidmar J, Mirny L, Nelson DR. Virus shapes and buckling transitions in spherical shells. Phys. Rev. E. 2003;68:051910. doi: 10.1103/PhysRevE.68.051910. [DOI] [PubMed] [Google Scholar]
- 132.Klug WS, et al. Failure of viral shells. Phys. Rev. Lett. 2006;97:228101. doi: 10.1103/PhysRevLett.97.228101. [DOI] [PubMed] [Google Scholar]
- 133.May ER, Feng J, Brooks CL. Exploring the symmetry and mechanism of virus capsid maturation via an ensemble of pathways. Biophys. J. 2012;102:606–612. doi: 10.1016/j.bpj.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bothner B, et al. Crystallographically identical virus capsids display different properties in solution. Nat. Struct. Biol. 1999;6:114–116. doi: 10.1038/5799. [DOI] [PubMed] [Google Scholar]
- 135.Speir JA, et al. Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics. J. Virol. 2006;80:3582–3591. doi: 10.1128/JVI.80.7.3582-3591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Speir JA, Munshi S, Wang GJ, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryoelectron microscopy. Structure. 1995;3:63–78. doi: 10.1016/S0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Milner ST, Safran SA. Dynamical fluctuations of droplet microemulsions and vesicles. Phys. Rev. A. 1987;36:4371–4379. doi: 10.1103/PhysRevA.36.4371. [DOI] [PubMed] [Google Scholar]
- 138.Wilts BD, Schaap IAT, Schmidt CF. Swelling and softening of the cowpea chlorotic mottle virus in response to pH shifts. Biophys. J. 2015;108:2541–2549. doi: 10.1016/j.bpj.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kononova O, et al. Structural transitions and energy landscape for cowpea chlorotic mottle virus capsid mechanics from nanomanipulation in vitro and in silico. Biophys. J. 2013;105:1893–1903. doi: 10.1016/j.bpj.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dykeman, E. C. & Sankey, O. F. Atomistic modeling of the low-frequency mechanical modes and Raman spectra of icosahedral virus capsids. Phys. Rev. E81, 021918 (2010). [DOI] [PubMed]
- 141.Hadden JA, et al. All-atom molecular dynamics of the HBV capsid reveals insights into biological function and cryo-EM resolution limits. eLife. 2018;7:e32478. doi: 10.7554/eLife.32478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu C, et al. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat. Commun. 2016;7:10714. doi: 10.1038/ncomms10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Quinn CM, et al. Dynamic regulation of HIV-1 capsid interaction with the restriction factor TRIM5α identified by magic-angle spinning NMR and molecular dynamics simulations. Proc. Natl Acad. Sci. USA. 2018;115:11519–11524. doi: 10.1073/pnas.1800796115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van de Waterbeemd M, et al. Structural analysis of a temperature-induced transition in a viral capsid probed by HDX-MS. Biophys. J. 2017;112:1157–1165. doi: 10.1016/j.bpj.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lim XX, et al. Conformational changes in intact dengue virus reveal serotype-specific expansion. Nat. Commun. 2017;8:14339. doi: 10.1038/ncomms14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Perilla JR, Schulten K. Physical properties of the HIV-1 capsid from all-atom molecular dynamics simulations. Nat. Commun. 2017;8:15959. doi: 10.1038/ncomms15959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hadden JA, Perilla JR. All-atom virus simulations. Curr. Opin. Virol. 2018;31:82–91. doi: 10.1016/j.coviro.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zink M, Grubmuller H. Mechanical properties of the icosahedral shell of southern bean mosaic virus: a molecular dynamics study. Biophys. J. 2009;96:1350–1363. doi: 10.1016/j.bpj.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao G, et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Andoh Y, et al. All-atom molecular dynamics calculation study of entire poliovirus empty capsids in solution. J. Chem. Phys. 2014;141:165101. doi: 10.1063/1.4897557. [DOI] [PubMed] [Google Scholar]
- 151.Freddolino PL, Arkhipov AS, Larson SB, McPherson A, Schulten K. Molecular dynamics simulations of the complete satellite tobacco mosaic virus. Structure. 2006;14:437–449. doi: 10.1016/j.str.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 152.Singh AR, Kosmrlj A, Bruinsma R. Finite temperature phase behavior of viral capsids as oriented particle shells. Phys. Rev. Lett. 2020;124:158101. doi: 10.1103/PhysRevLett.124.158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Battisti, A. J. Structural Studies of Pleomorphic Viruses. PhD thesis, Purdue Univ. (2011).
- 154.Overby AK, Pettersson RF, Grunewald K, Huiskonen JT. Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus. Proc. Natl Acad. Sci. USA. 2008;105:2375–2379. doi: 10.1073/pnas.0708738105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 156.Perotti LE, et al. Useful scars: physics of the capsids of archaeal viruses. Phys. Rev. E. 2016;94:012404. doi: 10.1103/PhysRevE.94.012404. [DOI] [PubMed] [Google Scholar]
- 157.Hochstein R, et al. Structural studies of Acidianus tailed spindle virus reveal a structural paradigm used in the assembly of spindle- shaped viruses. Proc. Natl Acad. Sci. USA. 2018;115:2120–2125. doi: 10.1073/pnas.1719180115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gwosch KC, et al. MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods. 2020;17:217–224. doi: 10.1038/s41592-019-0688-0. [DOI] [PubMed] [Google Scholar]
- 159.Heath GR, Scheuring S. High-speed AFM height spectroscopy reveals μs-dynamics of unlabeled biomolecules. Nat. Commun. 2018;9:4983. doi: 10.1038/s41467-018-07512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Maity S, et al. Caught in the act: mechanistic insight into supramolecular polymerization-driven self-replication from real-time visualization. J. Am. Chem. Soc. 2020;142:13709–13717. doi: 10.1021/jacs.0c02635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tiwary P, Parrinello M. From metadynamics to dynamics. Phys. Rev. Lett. 2013;111:230602. doi: 10.1103/PhysRevLett.111.230602. [DOI] [PubMed] [Google Scholar]
- 162.Perkett MR, Hagan MF. Using Markov state models to study self-assembly. J. Chem. Phys. 2014;140:214101. doi: 10.1063/1.4878494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mateu MG. Introduction: the structural basis of virus function. Sub-Cell. Biochem. 2013;68:3–51. doi: 10.1007/978-94-007-6552-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cong Y, Kriegenburg F, de Haan CAM, Reggiori F. Coronavirus nucleocapsid proteins assemble constitutively in high molecular oligomers. Sci. Rep. 2017;7:5740. doi: 10.1038/s41598-017-06062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yang J, et al. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Koch C, et al. Novel roles for well-known players: from tobacco mosaic virus pests to enzymatically active assemblies. Beilstein J. Nanotechnol. 2016;7:613–629. doi: 10.3762/bjnano.7.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Garmann RF, Comas-Garcia M, Gopal A, Knobler CM, Gelbart WM. The assembly pathway of an icosahedral single-stranded RNA virus depends on the strength of inter-subunit attractions. J. Mol. Biol. 2014;426:1050–1060. doi: 10.1016/j.jmb.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ryu, W.-S. in Molecular Virology of Human Pathogenic Viruses (ed. Ryu, W.-S.) 31–45 (Academic, 2017).
- 170.Yin J, Redovich J. Kinetic modeling of virus growth in cells. Microbiol. Mol. Biol. Rev. 2018;82:e00066–00017. doi: 10.1128/MMBR.00066-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu S-L, Wang Z-G, Zhang Z-L, Pang D-W. Tracking single viruses infecting their host cells using quantum dots. Chem. Soc. Rev. 2016;45:1211–1224. doi: 10.1039/C5CS00657K. [DOI] [PubMed] [Google Scholar]
- 172.Batinovic S, et al. Bacteriophages in natural and artificial environments. Pathogens. 2019;8:100. doi: 10.3390/pathogens8030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ofir G, Sorek R. Contemporary phage biology: from classic models to new insights. Cell. 2018;172:1260–1270. doi: 10.1016/j.cell.2017.10.045. [DOI] [PubMed] [Google Scholar]
- 174.Bernal JD, Fankuchen I. Structure types of protein crystals from virus-infected plants. Nature. 1937;139:923–924. doi: 10.1038/139923a0. [DOI] [Google Scholar]
- 175.Rossmann MG. Structure of viruses: a short history. Q. Rev. Biophys. 2013;46:133–180. doi: 10.1017/S0033583513000012. [DOI] [PubMed] [Google Scholar]
- 176.Kausche GA, Pfankuch E, Ruska H. Die Sichtbarmachung von pflanzlichem virus im Übermikroskop. Naturwissenschaften. 1939;27:292–299. doi: 10.1007/BF01493353. [DOI] [Google Scholar]
- 177.Kaelber JT, Hryc CF, Chiu W. Electron cryomicroscopy of viruses at near-atomic resolutions. Annu. Rev. Virol. 2017;4:287–308. doi: 10.1146/annurev-virology-101416-041921. [DOI] [PubMed] [Google Scholar]
- 178.Jardetzky O, Akasaka K, Vogel D, Morris S, Holmes KC. Unusual segmental flexibility in a region of tobacco mosaic virus coat protein. Nature. 1978;273:564–566. doi: 10.1038/273564a0. [DOI] [PubMed] [Google Scholar]
- 179.Hemminga MA, Veeman WS, Hilhorst HW, Schaafsma TJ. Magic angle spinning carbon-13 NMR of tobacco mosaic virus. An application of the high-resolution solid-state NMR spectroscopy to very large biological systems. Biophys. J. 1981;35:463–470. doi: 10.1016/S0006-3495(81)84802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Quinn CM, et al. Magic angle spinning NMR of viruses. Prog. Nucl. Magn. Reson. Spectrosc. 2015;86-87:21–40. doi: 10.1016/j.pnmrs.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.DeBlois RW, Wesley RK. Sizes and concentrations of several type C oncornaviruses and bacteriophage T2 by the resistive-pulse technique. J. Virol. 1977;23:227–233. doi: 10.1128/jvi.23.2.227-233.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys. Rev. Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 183.Piontek MC, Roos WH. Atomic force microscopy: an introduction. Methods Mol. Biol. 2017;1665:243–258. doi: 10.1007/978-1-4939-7271-5_13. [DOI] [PubMed] [Google Scholar]
- 184.Ashkin A. Acceleration and trapping of particles by radiation pressure. Phys. Rev. Lett. 1970;24:156–159. doi: 10.1103/PhysRevLett.24.156. [DOI] [Google Scholar]
- 185.Hashemi Shabestari, M., Meijering, A. E. C., Roos, W. H., Wuite, G. J. L. & Peterman, E. J. G. in Methods in Enzymology Vol. 582 (eds Spies, M. & Chemla, Y. R.) 85–119 (Academic, 2017). [DOI] [PubMed]
- 186.Zlotnick A, Aldrich R, Johnson JM, Ceres P, Young MJ. Mechanism of capsid assembly for an icosahedral plant virus. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 187.Young G, et al. Quantitative mass imaging of single biological macromolecules. Science. 2018;360:423. doi: 10.1126/science.aar5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 189.Uetrecht C, et al. High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids. Proc. Natl Acad. Sci. USA. 2008;105:9216–9220. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dülfer, J., Kadek, A., Kopicki, J.-D., Krichel, B. & Uetrecht, C. in Advances in Virus Research Vol. 105 (ed. Rey, F. A.) 189–238 (Academic, 2019). [DOI] [PubMed]
- 191.Sitters G, et al. Acoustic force spectroscopy. Nat. Methods. 2015;12:47–50. doi: 10.1038/nmeth.3183. [DOI] [PubMed] [Google Scholar]
- 192.Tuukkanen AT, Spilotros A, Svergun DI. Progress in small-angle scattering from biological solutions at high-brilliance synchrotrons. IUCrJ. 2017;4:518–528. doi: 10.1107/S2052252517008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Roos WH, Wuite GJL. Nanoindentation studies reveal material properties of viruses. Adv. Mater. 2009;21:1187–1192. doi: 10.1002/adma.200801709. [DOI] [Google Scholar]
- 194.Zlotnick A. To build a virus capsid — an equilibrium-model of the self-assembly of polyhedral protein complexes. J. Mol. Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- 195.Hagan MF, Elrad OM. Understanding the concentration dependence of viral capsid assembly kinetics — the origin of the lag time and identifying the critical nucleus size. Biophys. J. 2010;98:1065–1074. doi: 10.1016/j.bpj.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Rolfsson O, et al. Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J. Mol. Biol. 2016;428:431–448. doi: 10.1016/j.jmb.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]