Abstract
Background
Nicotinamide phosphoribosyltransferase (NAMPT) and the transforming growth factor-β (TGF-β) signaling pathway play important roles in colorectal tumorigenesis and progress. However, the underlying regulatory mechanisms between NAMPT and TGF-β signaling in colorectal cancer (CRC) remain poorly understood.
Methods
Public data were extracted from the Oncomine database and the PrognoScan database to investigate the mRNA expression and the prognostic value of NAMPT, respectively, in CRC. Western blot tests were performed to detect Smad2, Smad3, p-Smad2, p-Smad3, Smad4 expression in CRC cells transfected with human NAMPT-siRNA or NAMPT-overexpressing plasmid. TGF-β1 concentrations in culture supernatants were assayed using ELISA kits. The effect of TGF-β1 on NAMPT expression was evaluated by quantitative real-time PCR and Western blot. The dual-luciferase reporter assay was employed to confirm the binding of miR-1-3p to NAMPT 3ʹ-UTR. Subsequently, NAMPT levels in HCT116 cells transfected with the mimics and inhibitors of miR-1-3p were detected by quantitative real-time PCR and Western blot.
Results
NAMPT was overexpressed in human CRC and was correlated with short overall survival. NAMPT increased the protein expression levels of components in the TGF-β signaling pathway including Smad2, Smad3, and Smad4. Moreover, NAMPT promoted TGF-β1 secretion. Intriguingly, the TGF-β1 treatment down-regulated NAMPT expression at mRNA and protein levels in CRC cells which were partly through the up-regulation of miR-1-3p that directly bound to the NAMPT 3ʹ-UTR. These outcomes demonstrated that NAMPT was a downstream target of miR-1-3p and there was a negative association between NAMPT and miR-1-3p in CRC.
Conclusion
There is a negative feedback loop between NAMPT and the TGF-β signaling pathway in CRC cells, providing new insight into the mechanism underlying the regulatory pathways in CRC.
Keywords: miR-1-3p, nicotinamide phosphoribosyltransferase, regulatory mechanism, Smad, tumorigenesis
Introduction
Colorectal cancer (CRC) is the third most common malignant tumor with an incidence of 6.1% and a mortality rate of 9.2% globally.1,2 Despite the great advances in clinical diagnosis and treatment in the last few decades, the effective therapeutic targets of CRC are still under challenge due to the complexity of its pathogenesis.3,4 Therefore, it is of particular importance to identify specific molecular targets of CRC and clarify the related mechanisms to improve the prognosis of CRC patients.
Nicotinamide phosphoribosyltransferase (NAMPT), also known as pre-B-cell colony-enhancing factor (PBEF) and visfatin, is a rate-limiting enzyme in the salvage pathway of nicotinamide adenine dinucleotide (NAD) synthesis in mammals.5 NAMPT was overexpressed in various types of cancers including CRC, which was associated with vascular invasion, advanced TNM stage, and poor prognosis of CRC patients.6,7 Our previous study has shown that NAMPT as a potential progression marker of CRC promotes CRC cell viability.8 In addition to its effect on impairment of cellular energy metabolism in cancer cells, NAMPT is also involved in non-metabolic functions such as sirtuin function, DNA repair machinery, redox homeostasis, and tumor-related immune suppression.9 It has been shown that NAMPT is a molecular target of potent anticancer agents and its inhibitors have been applied in several early phase clinical trials for cancer therapy.10 However, the molecular mechanism underlying NAMPT functions in CRC progression requires further clarification.
The transforming growth factor-β (TGF-β) is a superfamily of structurally and functionally related proteins and regulates numerous cellular functions including proliferation, apoptosis, differentiation, migration, and epithelial-mesenchymal transition (EMT).11,12 Paradoxically, TGF-β exhibits tumor suppressive and promoting effects in cancer depending on the molecular and cellular context.13,14 Activation of the receptors by TGF-β ligands phosphorylates downstream signaling transducer proteins Smad2 and Smad3, which then assemble into complexes with Smad4. Ultimately, Smad-complex shuttles to the nucleus and regulates transcription of TGF-β target genes, such as Inhibitor of Differentiation 1 (ID1).15–17 Although NAMPT and the TGF-β signaling pathway are involved in tumor progression, their interrelationship in carcinogenesis is not fully elucidated.
MicroRNAs (miRNAs) are small non-coding RNAs with a length of about 22 nucleotides that could suppress a gene by binding to the 3ʹ-untranslated region (3ʹ-UTR) of mRNAs, resulting in the degradation of mRNA and the suppression of translation. Regarding molecular mechanisms underlying cancer, miRNAs are critical gene expression regulators acting as oncogenic or tumor suppressor, therefore play crucial roles in tumorigenesis including CRC.18–20 Using an online TargetScan tool (http://www.targetscan.org/vert_72/), miR-1-3p is predicted to be one of the miRNAs that might bind to the 3′-UTR of the NAMPT gene. The bioinformatics analysis based on 645 cases from 9 microarray datasets showed that miR-1-3p was significantly down-regulated in CRC and may suppress CRC through multiple biological approaches.21 In primary tumors, miR-1-3p is associated with age, mucinous component, tumor grade, and overall survival of CRC patients.22 However, the effect of miR-1-3p on NAMPT and the related mechanism in CRC remains largely unknown.
In this study, we examined the expression of NAMPT in colorectal adenoma and carcinoma tissues by bioinformatics analyses and investigated the relationship between NAMPT and the TGF-β signaling pathway in CRC cells. The association of NAMPT and miR-1-3p in CRC cells and the possible mechanism underlying TGF-β1-mediated NAMPT regulation via miR-1-3p were also clarified.
Materials and Methods
Bioinformatics Analyses
The Oncomine database (http://www.oncomine.org) was applied to investigate the mRNA expression of NAMPT in CRC and normal tissues. The screening criteria were determined as follows: P-value = 0.05; fold-change = 1.5, mRNA data type, and 10% gene ranking. For the prognostic value of NAMPT in CRC, the expression data of GSE17536 in the PrognoScan database were obtained and the overall survival (OS) curve was plotted with the probe 243296_at.23 For exploring the relationship between NAMPT and Smad2, Smad4, TGFB1, the starBase online tool (http://starbase.sysu.edu.cn/panCancer.php) was used in TCGA-COAD data.24
Cell Culture and TGF-β1 Treatment
HT29, Caco2, and HCT116 human CRC cell lines were obtained from the Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). HT29 and HCT116 cells were cultured in RPMI 1640 medium (HyClone, Thermo Fisher Scientific Inc., Beijing, China) and Caco2 cells were cultured in MEM (HyClone) supplemented with 10% fetal bovine serum (HyClone) at 37°C in a humidified 5% CO2 atmosphere. After seeding at the cell density of 3×105 cells per well in 6-well plates for 24 h, the CRC cells were treated with TGF-β1 (R&D Systems, Minneapolis, MN, USA) at different doses (0, 2.5, 5, and 10 ng/mL) for 24 h.
NAMPT-siRNA, NAMPT Plasmid, miRNA Mimics, and Inhibitors Transfection
The cells were first seeded in 6-well plates for 24 h and then transiently transfected with human NAMPT-siRNA which was a pool of 3 target-specific siRNAs used to knock down gene expression (sc-45,843, Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Non-targeting siRNA (sc-37,007, Santa Cruz) was used as a negative control. Lipofectamine RNAiMAX (Invitrogen Life Technologies, Carlsbad, CA, USA) was used for transfection according to the manufacturer’s instruction.
For plasmid transfection, HCT116 cells were transfected using the Lipo8000™ Transfection Reagent (Beyotime, Haimen, Jiangsu, China) in the exponential phase with 2.5 μg FLAG-tagged plasmid carrying NAMPT cDNA (EX-A1275-M12, GeneCopoeia, Inc., Rockville, MD, USA) and empty vector control plasmid (EX-NEG-M12, GeneCopoeia).
The miR-1-3p mimic, miR-negative control (miR-NC), miR-1-3p inhibitor (anti-miR-1-3p), and the negative control of inhibitor (anti-miR-NC) were purchased from GenePharm (Shanghai, China) and their sequences were shown in Supplementary Table S1. After seeding cells into 6-well plates for 24 h, cells were transfected with miR-1-3p mimic or miR-NC and anti-miR-1-3p or anti-miR-NC using Lipo8000™ Transfection Reagent and incubated for 48 h.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from cells using the RNAeasy™ Animal RNA Isolation Kit with Spin Column (Beyotime) according to the manufacturer’s instruction. The miRNA and mRNA were reversely transcribed for PCR using the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science, Indianapolis, IN, Germany) with the reaction conditions as 25°C for 10 min, 50°C for 60 min, 95°C for 5 min, 4°C for 60 min. PCR primer sequences (Supplementary Table S2) were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). PCR amplification was performed at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 60°C for 31 s using an SYBR Green Master kit (Roche Applied Science). Each experiment was conducted in triplicate and repeated three times. The expression level of NAMPT mRNA normalized to GAPDH as an endogenous control was calculated using 2ΔΔCt, in which the threshold cycle (Ct) was obtained using the Sequence Detection Software V1.4 (7300 Real-Time PCR System, Applied Biosystems, Foster City, CA, USA).
Western Blot Analysis
Cells were lysed in sodium dodecyl sulfate (SDS) lysis buffer (Beyotime) containing 1% phenylmethylsulfonyl fluoride (PMSF), protease, and phosphatase inhibitor cocktail (Beyotime) followed by sonication. The protein concentration was determined by the BCA Protein Assay kit (Beyotime). An equal amount of protein (30 µg) was separated by 10% or 12% SDS-polyacrylamide gel electrophoresis and transferred to a PVDF membrane (Millipore, Billerica, MA, USA) with the transfer buffer (Beyotime). After blocking with 5% nonfat dry milk in Tris-buffered saline containing Tween-20 (TBS-T), the membrane was incubated with a primary antibody overnight at 4°C, followed by the incubation of horseradish peroxidase-conjugated goat anti-rabbit IgG or anti-mouse IgG (1:5000 dilution, Absin Bioscience Inc., Shanghai, China) for 1 h at room temperature. The unbound antibodies were eluted with TBS-T for 10 min x 3 times. The following primary antibodies were used: rabbit anti-NAMPT (1:1000 dilution), mouse anti-ID1 (1:1000 dilution) (Sigma Chemical Co., St. Louis, MO, USA), mouse anti-GAPDH (1:5000 dilution, Absin), rabbit anti-phospho-Smad2 (1:1000 dilution), rabbit anti-phospho-Smad3 (1:1000 dilution), rabbit anti-Smad2 (1:2000 dilution), rabbit anti-Smad3 (1:2000 dilution), rabbit anti-Smad4 (1:2000 dilution) (Cell Signaling Technology, Inc., Danvers, MA, USA). Signals were detected using the Immobilon™ Western Chemiluminescent HRP Substrate (Millipore) and quantified using the Tanon-4500 Gel Imaging System with GIS ID Analysis Software v4.1.5 (Tanon Science & Technology Co., Ltd., Shanghai, China).
ELISA
Cellular supernatants of HCT116 and Caco2 cells after transfection for 48 h were assayed for the detection of TGF-β1 concentration using commercially available human TGF-β1 ELISA kits (ImmunoWay Biotechnology Company, Plano, TX, USA), according to the manufacturer’s protocol. The colorimetric reaction was measured at 450 nm.
Dual-Luciferase Reporter Assay
The whole 3ʹ-UTR of NAMPT that contains the binding site of miR-1-3p was predicted by the TargetScan. The wild-type NAMPT clone and corresponding 3′-UTR-mutated NAMPT clone were synthesized simultaneously by GENEWIZ, Inc (Suzhou, Jiangsu, China). A partial 3′-UTR of NAMPT (514 bp) that contains the predicted binding site of miR-1-3p was amplified from genomic DNA and inserted into the reporter vector pmirGLO. All clones were verified by restriction enzymes NheI and XhoI digestion and DNA sequencing.
HCT116 cells were cultured in 24-well plates and cotransfected with 0.5 μg wild-type or mutated NAMPT reporter plasmid and 50 nM miR-1-3p mimics or negative controls using Roche X tremeGENE siRNA Transfection Reagent (Roche Applied Science). After transfection for 24 h, cells were lysed and luciferase activities were detected using the Luc-Pair™ Duo-Luciferase Assay Kit (GeneCopoeia) following the manufacturer’s instructions. Renilla luciferase activity was applied for standardization.
Statistical Analysis
All data were presented as the mean ± the standard error of the mean (SEM) from at least three independent experiments as indicated. Statistical analyses were carried out using GraphPad Prism 6 (La Jolla, CA, USA) and SPSS v16.0 (Chicago, IL, USA) software. Student’s t-test or one-way ANOVA test was applied to calculate the significance of differences between two groups or among multiple groups. Differences with the value of p<0.05 were considered to be statistically significant.
Results
NAMPT is Overexpressed in CRC Patients
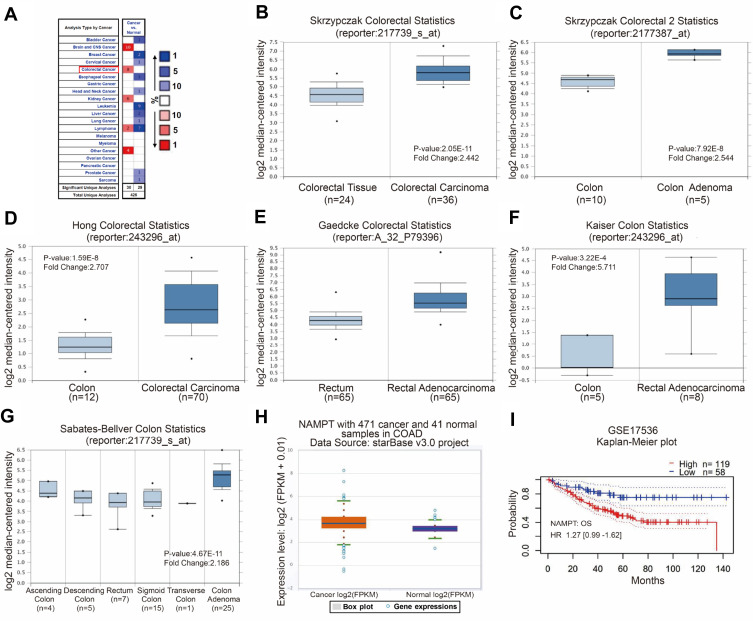
Cancer data analyses of the Oncomine database revealed that several types of cancer have an increased expression of NAMPT, including colorectal cancer (Figure 1A). A high level of NAMPT mRNA expression was found in adenoma and carcinoma tissues compared with normal colorectal tissues from different groups in the microarray datasets with fold change >2 (Figure 1B–G). A similar result was obtained from the data in COAD with 471 colorectal cancer and 41 normal samples (Figure 1H). These results indicated that NAMPT might be involved in CRC progression. Besides, the survival plots from GSE17536 revealed that the overexpression of NAMPT mRNA was associated with worse OS of patients with CRC (Figure 1I).
Figure 1.
Bioinformatics analysis of NAMPT mRNA expression and the related survival plot in colorectal tissues. (A–G) Data were obtained from the microarray datasets of the Oncomine database. NAMPT was overexpressed in various types of cancers including colorectal cancer in A. High levels of NAMPT mRNA expression were observed in colorectal cancer compared with normal colorectal tissues in the microarray datasets of Skrzypczak in B and C, Hong in D, Gaedcke in E, Kaiser in F, Sabates-Bellver in G, respectively. (H) TCGA-COAD data was used to compare the NAMPT expression in colorectal cancer tissues with normal tissues. (I) Survival plot. The data from GSE17536 showed that high expression of NAMPT mRNA was associated with short overall survival (OS) in CRC patients.
Knockdown of NAMPT Inhibits the TGF-β Signaling Pathway
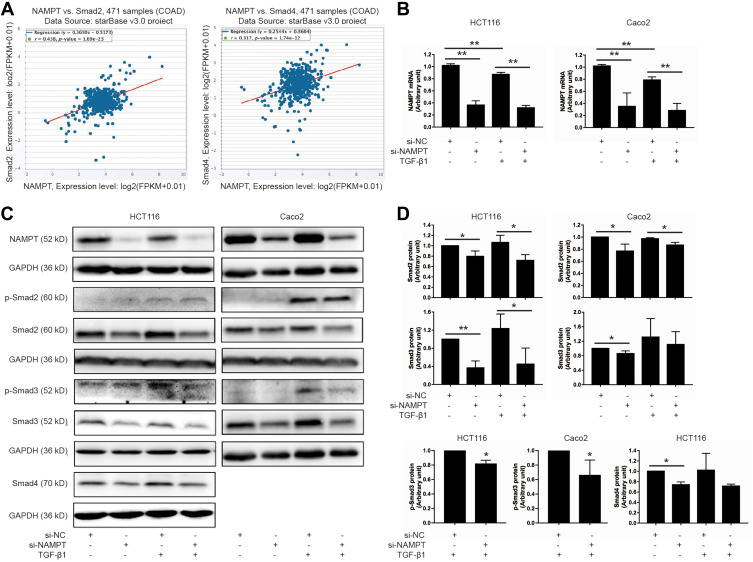
Since NAMPT and the TGF-β signaling pathway play an important part in colorectal tumorigenesis, we speculated that NAMPT might regulate the TGF-β/Smad signaling pathway in CRC. The starBase online tool was used in TCGA-COAD data to explore the relationship between NAMPT and the TGF-β signaling mediators (Smad2 and Smad4). The results revealed that the expression of NAMPT was positively correlated with Smad2 and Smad4 (Figure 2A).
Figure 2.
NAMPT silencing inhibited the TGF-β signaling pathway. (A) The starBase online tool in TCGA-COAD data was applied to explore the relationship of NAMPT with Smad2 (left panel) and Smad4 (right panel). (B) HCT116 and Caco2 cells were transfected with si-NAMPT or si-NC for 48 h and incubated in the presence or absence of TGF-β1 (5 ng/mL) for 24 h. The effect of NAMPT knockdown after si-NAMPT transfection was detected by qRT-PCR. (C) The influence of NAMPT silencing on the protein expression of transforming growth factor-β (TGF-β) signaling cascades (p-Smad2, p-Smad3, Smad2, Smad3, and Smad4) was analyzed by Western blot in HCT116 and Caco2 cells. GAPDH was used as a loading control. (D) Histograms showed a semi-quantitative analysis of the relative optical density of protein bands in C. Data were presented as mean ± SEM from three independent experiments. *P < 0.05; **P < 0.01.
Abbreviations: si-NC, negative control of siRNA; si-NAMPT, NAMPT-siRNA.
Smads are the key signal transducers of the TGF-β signaling pathway. To test the effects of NAMPT on Smads, we first detected the influence of NAMPT knockdown on the expression of Smads in HCT116 and Caco2 cells. qRT-PCR and Western blot tests confirmed that both mRNA and protein expression levels of NAMPT were reduced in cells transfected with NAMPT-siRNA compare with the control (Figure 2B and C). As shown in Figure 2C and D, Smad2 and Smad3 were phosphorylated upon 5 ng/mL TGF-β1 stimulation for 24 h. However, TGF-β1-induced phosphorylation of Smad3 was significantly suppressed after transfection with si-NAMPT. No significant variation of the p-Smad2 level was found between the TGF-β1 + si-NC group and TGF-β1 + si-NAMPT group both in HCT116 and Caco2 cells. Moreover, knockdown of NAMPT significantly decreased the protein levels of Smad2, Smad3, and Smad4. Treatment with TGF-β1 did not attenuate the effect of NAMPT silencing on Smads. These results indicated that knockdown of NAMPT repressed the TGF-β1 signaling pathway proteins and decreased TGF-β1-mediated phosphorylation of Smad3.
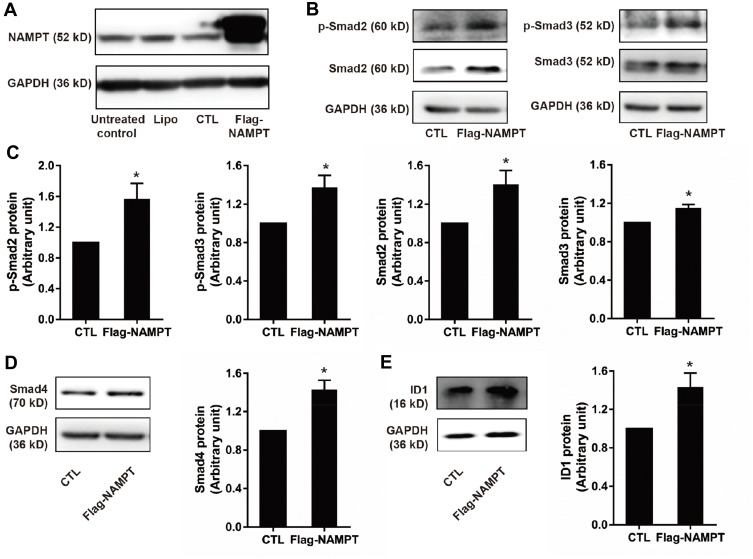
NAMPT Overexpression Activates the TGF-β Signaling Pathway
As shown in Figure 3A, the NAMPT protein level was increased in HCT116 cells transfected with the NAMPT-overexpressing plasmid. Western blot was applied to evaluate the effect of NAMPT overexpression on the TGF-β signaling pathway. NAMPT overexpression increased the protein levels of Smad2, Smad3, Smad4 as well as phosphorylation of Smad2 and Smad3 compared to the control plasmid transfected cells (all P < 0.05) (Figure 3B–D). ID1, as one of the targeted genes directly regulated by the TGF-β signaling pathway, was also significantly up-regulated in HCT116 cells transfected NAMPT-overexpressing plasmid (Figure 3E).
Figure 3.
NAMPT overexpression activated the TGF-β signaling pathway and up-regulates ID1 in HCT116 cells. (A) Western blot analysis showed NAMPT overexpression in cells transfected with a plasmid for 72 h. (B) Western blot analysis showed the expression status of the components in the TGF-β signaling (p-Smad2, p-Smad3, Smad2, Smad3. (C) Semi-quantitative analysis of the relative optical density of protein expression in B (n=3). (D) The effect of NAMPT overexpression on Smad4 was detected by Western blot and semi-quantitatively analyzed. (E) ID1 protein expression was detected by Western blot and the relative optical density of protein bands was quantitatively analyzed in cells transfected with a plasmid for 72 h. Data were presented as mean±SEM. Lipo, control cells treated with Lipo8000™ transfection reagent; CTL, cells transfected with empty vector control plasmid; Flag-NAMPT, cells transfected with FLAG-tagged plasmid carrying NAMPT cDNA. *P < 0.05 compared to vector.
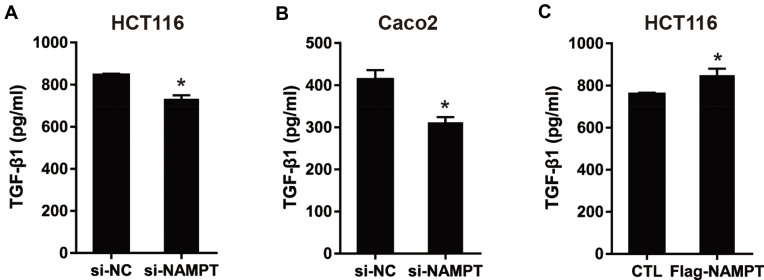
NAMPT Promotes the Secretion Level of TGF-β1
To evaluate the influence of NAMPT on the secretion of TGF-β1 in CRC cells, we measured the TGF-β1 concentrations in cell supernatants from NAMPT-overexpressing and NAMPT-silencing cells by ELISA. TGF-β1 concentration in supernatants from NAMPT-silencing HCT116 and Caco2 cells was lower compared with the control cells (Figure 4A and B). However, transfection with NAMPT-overexpressing plasmid in HCT116 cells promoted the TGF-β1 production in the supernatants compared with the respective control (Figure 4C).
Figure 4.
NAMPT promoted the secretion level of TGFβ-1. (A and B) HCT116 and Caco2 cells were transfected with si-NAMPT or si-NC. (C) HCT116 cells were transfected with NAMPT cDNA plasmid or vector control. After treatment for 48 h, cell supernatants were harvested and TGF-β1 concentrations were determined by ELISA. *P < 0.05.
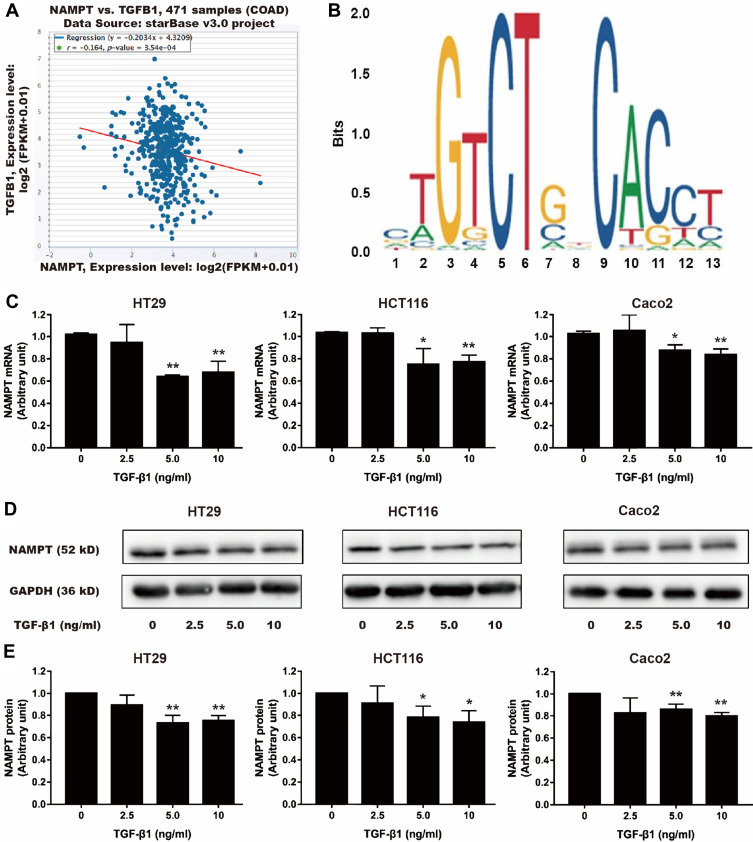
TGF-β1 Suppresses NAMPT Expression in CRC Cells
Using bioinformatics analysis, we found a negative correlation between NAMPT and TGFB1 (Figure 5A). Furthermore, the sequence of potential Smad2/3 binding sites was found within the NAMPT promoter in the JASPAR database (Figure 5B). Therefore, we speculated that the NAMPT expression might be regulated by TGF-β1. After TGF-β1 treatment, NAMPT expression in HT29, HCT116, and Caco2 cells was significantly decreased at the mRNA (Figure 5C) and protein (Figure 5D and E) levels (P <0.05), when the concentration of TGF-β1 was higher than 5 ng/mL. These results indicated that there was a negative feedback loop between NAMPT and the TGF-β signaling pathway.
Figure 5.
Effect of TGF-β1 on NAMPT expression in colorectal cells. (A) The starBase online tool in TCGA-COAD data was used to explore the relationship between NAMPT and TGFB1. (B) The sequence logo of potential Smad2/3 binding site in JASPAR. (C) HT29, HCT116 and Caco2 cells were treated with TGF-β1 at different concentrations (0, 2.5, 5, and 10 ng/mL) for 24 h. NAMPT mRNA expression was detected by qRT-PCR. (D) NAMPT protein expression was detected by Western blot. (E) Histograms showed the semi-quantitative analysis of the gels from D after densitometry. NAMPT expression was decreased by TGF-β1 treatment at a dose of 5 and 10 ng/mL. Data were presented as mean ± SEM. *P < 0.05; **P < 0.01. n = 3 independent experiments.
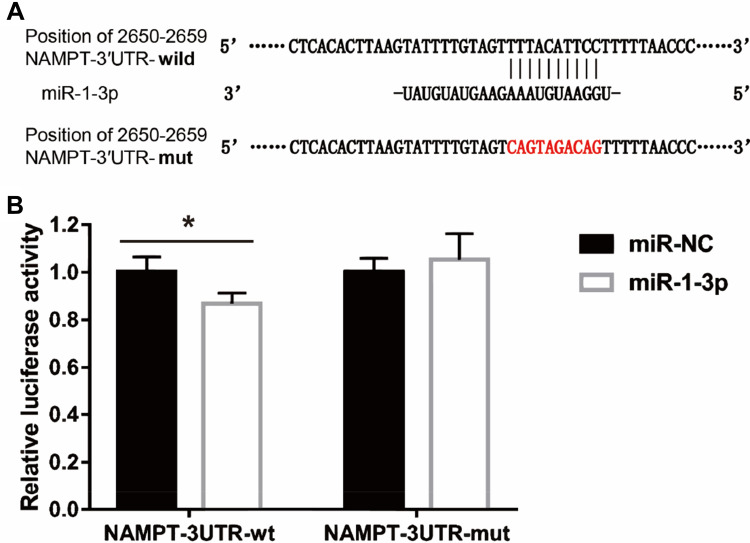
MiR-1-3p Binds to the 3ʹ-UTR of NAMPT
To investigate the molecular mechanism of the effect of TGF-β1 on NAMPT expression, we performed the bioinformatics analysis which indicated that the 3′-UTR of NAMPT contains a potential binding site of miR-1-3p. To determine whether miR-1-3p directly binds to the 3ʹ-UTR of NAMPT, we constructed two plasmids: a wild-type NAMPT 3′-UTR (NAMPT-3ʹUTR-wt) with the miR-1-3p binding site (position at 2650–2659 of 3′-UTR) and a mutated NAMPT 3′-UTR (NAMPT-3ʹUTR-mut) in which the binding site was changed from TTTACATTCC to CAGTAGACAG (Figure 6A). The dual-luciferase reporter assay revealed that the luciferase activity was significantly decreased after the cotransfection of NAMPT-3ʹUTR-wt plasmids with miR-1-3p mimics compared with the miR-NC (P<0.05). However, no significant difference in luciferase activity was observed between miR-1-3p mimics and miR-NC groups after cells cotransfected with NAMPT-3ʹUTR-mut plasmids (Figure 6B). These findings suggest that miR-1-3p directly targets NAMPT 3′-UTR.
Figure 6.
Interaction between NAMPT and miR-1-3p transcripts. (A) Illustration of the sequences of the putative miR-1-3p binding site in the NAMPT 3′-UTR and its mutant. (B) The interaction of miR-1-3p with the NAMPT mRNA at 3′-UTR in HCT116 cells was confirmed by the dual-luciferase reporter assay. Data were presented as the mean ± SEM. *P < 0.05.
Abbreviations: 3′-UTR, 3′-untranslated region; miR-NC, negative control of miRNA.
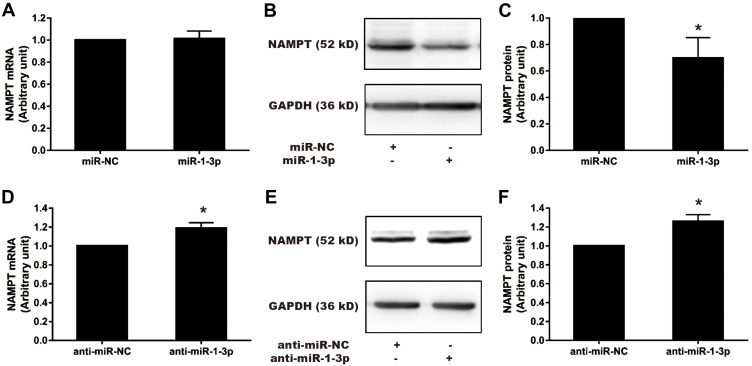
NAMPT is Mediated by miR-1-3p in HCT116 Cells
To confirm that miR-1-3p would negatively regulate NAMPT expression, gain-of-function and loss-of-function approaches were applied. The expression of NAMPT was detected by qRT-PCR and Western blot in HCT116 cells transfected with miR-1-3p mimics or inhibitors. Although miR-1-3p mimics had no effect on the mRNA level of NAMPT (Figure 7A), NAMPT expression at the protein level was significantly down-regulated after miR-1-3p mimics transfection (Figure 7B and C). The down-regulation of miR-1-3p by its inhibitor led to an increase in the expression of NAMPT mRNA and protein (Figure 7D–F).
Figure 7.
Effect of miR-1-3p mimics and inhibitors on NAMPT mRNA and protein expression in HCT116 cells. Cells were transfected with 50 nM miR-1-3p mimics, anti-miR-1-3p, or their corresponding NC. NAMPT mRNA was detected by qRT-PCR and protein was detected by Western blot at 48 h post-transfection. (A) Detection of NAMPT mRNA after miR-1-3p mimics treatment. (B and C) NAMPT protein expression and densitometry analysis after miR-1-3p mimics treatment. (D) NAMPT mRNA detection after transfection with anti-miR-1-3p. (E and F) NAMPT protein expression and densitometry analysis after transfection with anti-miR-1-3p. n=3; *P < 0.05.
Abbreviations: miR, microRNA; NC, negative control.
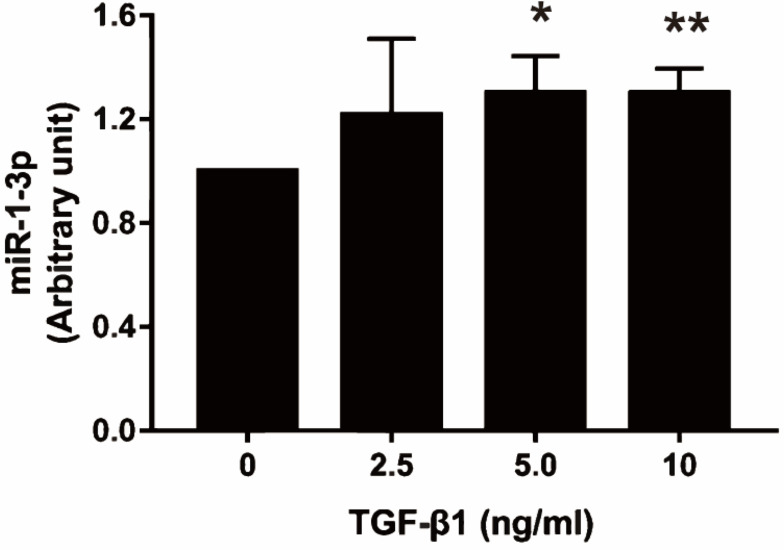
MiR-1-3p Expression is Regulated by TGF-β1
The present study revealed that NAMPT expression was down-regulated by TGF-β1 and miR-1-3p, so we speculated that the effect of TGF-β1 on NAMPT expression might be mediated by miR-1-3p. To confirm this hypothesis, we investigated whether TGF-β1 affects miR-1-3p expression. The qRT-PCR revealed that miR-1-3p expression was significantly increased in HCT116 after treatment with 5 ng/mL of TGF-β1 (P < 0.05) and 10 ng/mL of TGF-β1 (P < 0.01) for 24 h compared with the control (Figure 8). These data indicated that TGF-β-regulated NAMPT expression was at least in part mediated via miR-1-3p.
Figure 8.
Effect of TGF-β1 on miR-1-3p expression in HCT116 cells. The miR-1-3p expression was detected by qRT-PCR after treatment with TGF-β1 (0, 2.5, 5, and 10 ng/mL) for 24 h. Data were presented as mean ± SEM. n=3; *P<0.05, **P<0.01.
Discussion
NAMPT, as an adipokine and proinflammatory cytokine, is dramatically increased in several types of cancer and plays an important role in carcinogenesis.25–27 Consistent with our previous study,8 bioinformatics analyses in this study revealed NAMPT overexpression in colorectal adenoma and carcinoma tissues, providing extra evidence for the roles of NAMPT in CRC development. Besides, we found NAMPT overexpression might be associated with poor survival of CRC patients. Similarly, the results of immunohistochemical staining from previous studies indicated that high expression of NAMPT in CRC is correlated with invasion, advanced TNM stage, and short overall survival.6,8 Nevertheless, the underlying regulatory mechanisms of NAMPT in CRC are still limited. NAMPT is a potent oncogene that modulates cancer stem cell properties through Sirt1 and PARP in CRC.28 Another study also showed that NAMPT regulates Sirt1/P53 signaling during CRC cell growth.29 Moreover, the Wnt/β-catenin pathway is involved in the functions of NAMPT during CRC proliferation.30
The TGF-β signaling pathway is involved in the CRC progression and is also found to be inversely associated with prognosis.31,32 However, the interrelationship between NAMPT and the TGF-β signaling pathway in CRC remains unclear. Currently, only two studies have been published regarding the correlation between NAMPT and the TGF-β signaling pathway. The analysis results of pharmacogenomic databases from 43 CRC cell lines demonstrated that inactivating mutations and deletions in type II TGF-β receptors (TGFBR2) are significantly associated with increased NAMPT inhibitor FK866 sensitivity.33 Another study showed that in breast cancer cells, NAMPT promotes EMT by increasing TGF-β1 secretion and activating the TGF-β signaling pathway, as Smad3 is phosphorylated in response to recombinant human NAMPT.34 In the current study, we demonstrated that in CRC cells, NAMPT silencing decreased the secreted TGF-β1 level and the levels of transducer proteins in the TGF-β signaling pathway, including Smad2, Smad3, Smad4, and phosphorylated Smad3. Conversely, NAMPT overexpression increased the protein expression of Smads (Smad2, Smad3, Smad4, p-Smad2, p-Smad3) and TGF-β target protein ID1, as well as promoted the secretion of TGF-β1. These results indicated that NAMPT might regulate the TGF-β/Smad-dependent pathway. Intriguingly, TGF-β1 inhibited the expression of NAMPT at mRNA and protein levels, generating a negative feedback loop between NAMPT and the TGF-β signaling pathway in CRC cells. However, this feedback loop in CRC tissues remains unknown. Further study may be needed to investigate the functions of the negative feedback loop during the CRC progression.
Although Smad2 and Smad3 are closely related transducer proteins of the TGF-β signaling with 92% amino acid sequence similarity, they have differential sensitivities in relaying signaling.35,36 The present study revealed that NAMPT silencing down-regulated p-Smad3, total Smad2, and Smad3. However, no decrease in phosphorylated Smad2 was detected, probably due to the low sensitivity or expression level. Smad4 as a transcription factor for the TGF-β signaling is frequently mutated in CRC.37 Loss of Smad4 protein expression is found in approximately 20–40% of human CRCs and could disrupt canonical TGF-β/Smad signaling.38 In this study, the protein expression of Smad4 was failed to be detected in HT29 and Caco2 cells.
The current study demonstrated that the treatment of HCT116 cells with miR-1-3p mimics and inhibitors resulted in a decrease and an increase of NAMPT expression, indicating that NAMPT is a target of miR-1-3p. Furthermore, TGF-β1 up-regulated miR-1-3p in HCT116 cells. The luciferase reporter assay demonstrated that the regulatory outcome of miR-1-3p on NAMPT was partly mediated by binding of miR-1-3p to the NAMPT 3′-UTR. Collectively, these results suggested the regulation of NAMPT expression by TGF-β1 was at least in part via the miR-1-3p. It is predicted that 16 miRNAs might bind to the 3′-UTR of the NAMPT gene in TargetScan, implying that NAMPT is regulated by miRNAs, including miR-1-3p.6 Indeed, NAMPT can be targeted by miRNAs as a previous study showed that miR-26b serves as a tumor suppressor by targeting NAMPT in CRC cells.39
In conclusion, NAMPT is overexpressed in colorectal adenoma and carcinoma tissues, which is associated with worse survival, suggesting that NAMPT may be a potential biomarker of CRC. NAMPT increases the transducer protein expression of TGF-β signaling and promotes TGF-β1 secretion, while TGF-β1 down-regulates NAMPT expression through the up-regulation of miR-1-3p expression. These findings provide a novel regulatory mechanism by which NAMPT and TGF-β signaling mutually affect each other, generating a negative feedback loop in CRC cells. Targeting this loop may have therapeutic potential for CRC.
Acknowledgment
This work was supported by grants from Jinshan District Commission of Health and Family Planning (#JSKJ-KTMS-2018-05), “Qi Hang” Project of Jinshan Hospital, Fudan University (#2018-JSYYQH-05), Fourth Training Program for the Outstanding Young Talents, Jinshan Health Commission (#JSYQ201904), Key Construction Project on Clinical Pharmacy of Shanghai (#2019-1229), and the National Natural Science Foundation of China (#81872121).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Wrobel P, Ahmed S. Current status of immunotherapy in metastatic colorectal cancer. Int J Colorectal Dis. 2019;34(1):13–25. doi: 10.1007/s00384-018-3202-8 [DOI] [PubMed] [Google Scholar]
- 4.Wang N, Lu Y, Khankari NK, et al. Evaluation of genetic variants in association with colorectal cancer risk and survival in Asians. Int J Cancer. 2017;141(6):1130–1139. doi: 10.1002/ijc.30812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garten A, Schuster S, Penke M, Gorski T, de Giorgis T, Kiess W. Physiological and pathophysiological roles of NAMPT and NAD metabolism. Nat Rev Endocrinol. 2015;11(9):535–546. [DOI] [PubMed] [Google Scholar]
- 6.Li XQ, Lei J, Mao LH, et al. NAMPT and NAPRT, key enzymes in NAD salvage synthesis pathway, are of negative prognostic value in colorectal cancer. Front Oncol. 2019;9:736. doi: 10.3389/fonc.2019.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M, Wang Y, Li Y, et al. Association of plasma visfatin with risk of colorectal cancer: an observational study of Chinese patients. Asia Pac J Clin Oncol. 2016;12(1):e65–74. doi: 10.1111/ajco.12090 [DOI] [PubMed] [Google Scholar]
- 8.Lv X, Zhang L, Zhu Y, Said HM, Shi J, Xu G. Regulative effect of nampt on tumor progression and cell viability in human colorectal cancer. J Cancer. 2015;6(9):849–858. doi: 10.7150/jca.12341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heske CM. Beyond energy metabolism: exploiting the additional roles of NAMPT for cancer therapy. Front Oncol. 2020;9:1514. doi: 10.3389/fonc.2019.01514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi D, Imaizumi T, Yagi K, et al. Nicotinamide phosphoribosyltransferase is a molecular target of potent anticancer agents identified from phenotype-based drug screening. Sci Rep. 2019;9(1):7742. doi: 10.1038/s41598-019-43994-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Syed V. TGF-β signaling in cancer. J Cell Biochem. 2016;117(6):1279–1287. doi: 10.1002/jcb.25496 [DOI] [PubMed] [Google Scholar]
- 12.Zhao M, Mishra L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14(2):111–123. doi: 10.7150/ijbs.23230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahimi RA, Leof EB. TGF-beta signaling: a tale of two responses. J Cell Biochem. 2007;102(3):593–608. doi: 10.1002/jcb.21501 [DOI] [PubMed] [Google Scholar]
- 14.Seoane J, Gomis RR. TGF-β family signaling in tumor suppression and cancer progression. Cold Spring Harb Perspect Biol. 2017;9(12):a022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296(5573):1646–1647. doi: 10.1126/science.1071809 [DOI] [PubMed] [Google Scholar]
- 16.Shi Y, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/S0092-8674(03)00432-X [DOI] [PubMed] [Google Scholar]
- 17.Ma H, Wei Y, Leng Y, et al. TGF-β1-induced expression of Id-1 is associated with tumor progression in gastric cancer. Med Oncol. 2014;31(7):19. doi: 10.1007/s12032-014-0019-3 [DOI] [PubMed] [Google Scholar]
- 18.Masuda T, Hayashi N, Kuroda Y, Ito S, Eguchi H, Mimori K. MicroRNAs as biomarkers in colorectal cancer. Cancers (Basel). 2017;9(9):124. doi: 10.3390/cancers9090124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. [DOI] [PubMed] [Google Scholar]
- 20.Qu YL, Wang HF, Sun ZQ, et al. Up-regulated miR-155-5p promotes cell proliferation, invasion and metastasis in colorectal carcinoma. Int J Clin Exp Pathol. 2015;8(6):6988–6994. [PMC free article] [PubMed] [Google Scholar]
- 21.Wang JY, Huang JC, Chen G, Wei DM. Expression level and potential target pathways of miR-1-3p in colorectal carcinoma based on 645 cases from 9 microarray datasets. Mol Med Rep. 2018;17(4):5013–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno EC, Pascual A, Prieto-Cuadra D, et al. Novel molecular characterization of colorectal primary tumors based on miRNAs. Cancers (Basel). 2019;11(3):346. doi: 10.3390/cancers11030346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2(1):18. doi: 10.1186/1755-8794-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Databaseissue):D92–97. doi: 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gholinejad Z, Kheiripour N, Nourbakhsh M, et al. Extracellular NAMPT/Visfatin induces proliferation through ERK1/2 and AKT and inhibits apoptosis in breast cancer cells. Peptides. 2017;92:9–15. doi: 10.1016/j.peptides.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 26.Guo Q, Han N, Shi L, et al. NAMPT: a potential prognostic and therapeutic biomarker in patients with glioblastoma. Oncol Rep. 2019;42(3):963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ju HQ, Zhuang ZN, Li H, et al. Regulation of the Nampt-mediated NAD salvage pathway and its therapeutic implications in pancreatic cancer. Cancer Lett. 2016;379(1):1–11. doi: 10.1016/j.canlet.2016.05.024 [DOI] [PubMed] [Google Scholar]
- 28.Lucena-Cacace A, Otero-Albiol D, Jiménez-García MP, Muñoz-Galvan S, Carnero A. NAMPT is a potent oncogene in colon cancer progression that modulates cancer stem cell properties and resistance to therapy through Sirt1 and PARP. Clin Cancer Res. 2018;24(5):1202–1215. doi: 10.1158/1078-0432.CCR-17-2575 [DOI] [PubMed] [Google Scholar]
- 29.Pan JH, Zhou H, Zhu SB, et al. Nicotinamide phosphoribosyl transferase regulates cell growth via the Sirt1/P53 signaling pathway and is a prognosis marker in colorectal cancer. J Cell Physiol. 2019;234(4):4385–4395. doi: 10.1002/jcp.27228 [DOI] [PubMed] [Google Scholar]
- 30.Ye C, Qi L, Li X, et al. Targeting the NAD+ salvage pathway suppresses APC mutation-driven colorectal cancer growth and Wnt/β-catenin signaling via increasing axin level. Cell Commun Signal. 2020;18(1):16. doi: 10.1186/s12964-020-0513-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gulubova M, Manolova I, Ananiev J, Julianov A, Yovchev Y, Peeva K. Role of TGF-beta1, its receptor TGFbetaRII, and Smad proteins in the progression of colorectal cancer. Int J Colorectal Dis. 2010;25(5):591–599. doi: 10.1007/s00384-010-0906-9 [DOI] [PubMed] [Google Scholar]
- 32.Gachpazan M, Kashani H, Hassanian SM, et al. Therapeutic potential of targeting transforming growth factor-beta in colorectal cancer: rational and progress. Curr Pharm Des. 2019;25(38):4085–4089. [DOI] [PubMed] [Google Scholar]
- 33.Brandl L, Kirstein N, Neumann J, et al. The c-MYC/NAMPT/SIRT1 feedback loop is activated in early classical and serrated route colorectal cancer and represents a therapeutic target. Med Oncol. 2018;36(1):5. doi: 10.1007/s12032-018-1225-1 [DOI] [PubMed] [Google Scholar]
- 34.Soncini D, Caffa I, Zoppoli G, et al. Nicotinamide phosphoribosyltransferase promotes epithelial-to-mesenchymal transition as a soluble factor independent of its enzymatic activity. J Biol Chem. 2014;289(49):34189–34204. doi: 10.1074/jbc.M114.594721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown KA, Pietenpol JA, Moses HL. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J Cell Biochem. 2007;101(1):9–33. doi: 10.1002/jcb.21255 [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Liu X, Ren X, et al. Smad2 and Smad3 have differential sensitivity in relaying TGFβ signaling and inversely regulate early lineage specification. Sci Rep. 2016;6(1):21602. doi: 10.1038/srep21602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itatani Y, Kawada K, Sakai Y. Transforming growth factor-β signaling pathway in colorectal cancer and its tumor microenvironment. Int J Mol Sci. 2019;20(23):5822. doi: 10.3390/ijms20235822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, Tong J, Huang G, El-Rifai W. Nicotinamide phosphoribosyl transferase (Nampt) is a target of microRNA-26b in colorectal cancer cells. PLoS One. 2013;8(7):e69963. doi: 10.1371/journal.pone.0069963 [DOI] [PMC free article] [PubMed] [Google Scholar]