Abstract
Alu repeats contribute to phylogenetic novelties in conserved regulatory networks in primates. Our study highlights how exonized Alus could nucleate large-scale mRNA–miRNA interactions. Using a functional genomics approach, we characterize a transcript isoform of an orphan gene, CYP20A1 (CYP20A1_Alu-LT) that has exonization of 23 Alus in its 3′UTR. CYP20A1_Alu-LT, confirmed by 3′RACE, is an outlier in length (9 kb 3′UTR) and widely expressed. Using publically available data sets, we demonstrate its expression in higher primates and presence in single nucleus RNA-seq of 15,928 human cortical neurons. miRanda predicts ∼4,700 miRNA recognition elements (MREs) for ∼1,000 miRNAs, primarily originated within these 3′UTR-Alus. CYP20A1_Alu-LT could be a potential multi-miRNA sponge as it harbors ≥10 MREs for 140 miRNAs and has cytosolic localization. We further tested whether expression of CYP20A1_Alu-LT correlates with mRNAs harboring similar MRE targets. RNA-seq with conjoint miRNA-seq analysis was done in primary human neurons where we observed CYP20A1_Alu-LT to be downregulated during heat shock response and upregulated in HIV1-Tat treatment. In total, 380 genes were positively correlated with its expression (significantly downregulated in heat shock and upregulated in Tat) and they harbored MREs for nine expressed miRNAs which were also enriched in CYP20A1_Alu-LT. MREs were significantly enriched in these 380 genes compared with random sets of differentially expressed genes (P = 8.134e-12). Gene ontology suggested involvement of these genes in neuronal development and hemostasis pathways thus proposing a novel component of Alu-miRNA-mediated transcriptional modulation that could govern specific physiological outcomes in higher primates.
Keywords: Cytochrome P450 20A1 (CYP20A1), miRNA recognition elements (MREs), Alu-miRNA, neurocoagulopathy, multi-miRNA sponge, 3 prime UnTranslated Region (3′UTR) extension
Significance
Insertion of repeats can contribute to variability in conserved gene networks. Primate-specific Alu repeats in the 3′UTRs of transcripts are reported as miRNA regulatory sites. However, studies on their role in modulating large-scale mRNA–miRNA networks have been limiting. In this study, we report a unique 3′UTR (9 kb) transcript isoform of CYP20A1 in humans that contains 23 Alus, that is, CYP20A1_Alu-LT. This provides more than 3,000 potential binding sites for 140 different types of miRNAs. Differential expression of this transcript could sponge an ensemble of miRNAs that can govern specific cellular outcomes. We demonstrate this in primary human neurons and propose how such lineage-specific events could govern pathways linked to coagulation and hemostatic outcomes in neuronal lineages.
Introduction
Nearly half of the human genome is occupied by transposable elements (Lander et al. 2001). These have been shown to fine-tune conserved gene regulatory networks in a lineage-specific manner (Chen et al. 2017; Wang et al. 2017; Trizzino et al. 2017). Depending upon the context, they contribute to gene expression divergence through large-scale transcriptional rewiring (Lynch et al. 2011; Rebollo et al. 2012; Chuong et al. 2016; Trizzino et al. 2017). Primate-specific Alu retrotransposons, which occupy ∼11% of the human genome, are one of the major players modulating gene expression (Lander et al. 2001; Grover et al. 2005). These provide noncanonical transcription factor binding and other regulatory sites that govern epigenetic modifications as well as cryptic splice sites that lead to alternative splicing or differential mRNA stability (Sorek et al. 2002; An et al. 2004; Polak and Domany 2006; Xie et al. 2009; Lynch et al. 2011; Bakshi et al. 2016; Tristán‐Flores et al. 2018; Payer et al. 2017). Alu-derived exons exhibit lineage specificity with high transcript inclusion levels and have relatively higher rates of evolution (Lin et al. 2008; Sorek 2009; Shen et al. 2011).
Nearly, 14% of the human transcripts contain at least one exonized Alu (Mandal et al. 2013). Exonization is frequently reported in genes that have arisen de novo in primates, with most of the events in 3′UTRs (Toll-Riera et al. 2009; Mandal et al. 2013). Such exonized Alus can increase the regulatory possibilities of a transcript, in a spatiotemporal manner, through antisense, miRNAs, Alu-miRNA recognition elements (MREs), A-to-I RNA editing, alternative splicing, and enhancers. We have earlier shown how a crosstalk between these events could govern transcript isoform dynamics and modulate cellular outcomes (Mandal et al. 2013; Pandey et al. 2016). Besides, Alus provide substrates for other regulatory events such as gain of poly-A sites, AU-rich motifs, and MREs that can lead to alternative polyadenylation, mRNA decay or translation stalling, and formation of specific secondary structures (Sobczak and Krzyzosiak 2002; An et al. 2004; Roy-Engel et al. 2005; Häsler and Strub 2006; Häsler et al. 2007; Lee et al. 2008).
In our earlier study on 3,177 Alu-exonized genes, we reported co-occurrence of cis-Alu antisense and A-to-I RNA editing marks at the level of single Alu exons in 319 genes (Mandal et al. 2013). Among these genes, during mapping of lineage-specific events, we observed a transcript isoform of CYP20A1 that has acquired an unusually long 3′UTR through exonization of 23 Alus. We report here a unique biological role of this Alu-exonized transcript isoform (CYP20A1_Alu-LT), through its extensive characterization. We report for the first time that this isoform that has originated primarily from Alu elements could potentially function as a multi-miRNA sponge as its 3′UTR contains predicted target sites for ∼1,000 miRNAs. Through RNA-seq analysis, we demonstrate the potential of this sponge activity in the presence of miRNAs in primary neurons in two different stress conditions where this transcript has opposite expression. CYP20A1_Alu-LT expression correlates with a set of 380 genes that share cognate MREs. These genes are enriched in the process of neurocoagulopathy, suggesting that the synergistic role of an ensemble of miRNAs function could be modulated by CYP20A1_Alu-LT. This adds to the growing repertoire of lineage-specific regulatory functions that are contributed by Alu elements.
Results
CYP20A1 Contains a Unique 3′UTR with Alu-Driven Divergence
Literature mining revealed that transcripts of 91 genes out of the 319, that have conjoint Alu exonization, editing and antisense events, map to apoptosis, and nearly 75% of them cluster around three discrete hubs: cell cycle-DNA damage response (p53 hub; 31 genes), mitochondrial events (mito hub; 22 genes), and proteostasis (ubi hub; 15 genes) (supplementary information S1 and table S1, Supplementary Material online). As the majority of these exonization events occur in the 3′UTRs, we further focused our study in the 3′UTRs of these genes.
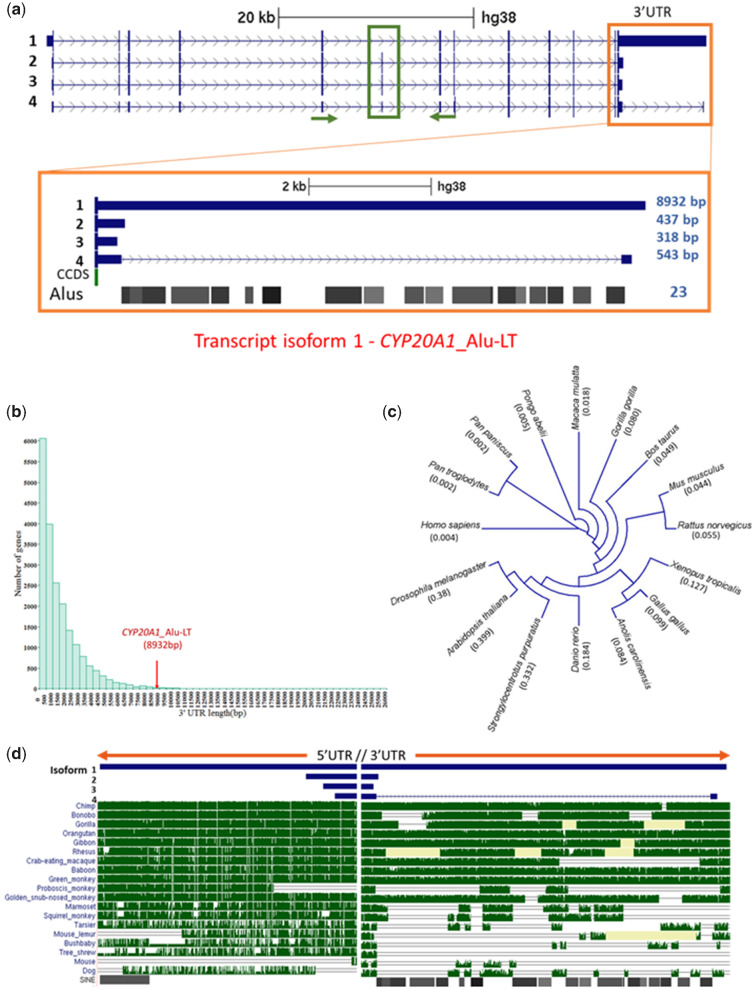
In the mitochondrial hub, we observed a transcript isoform of CYP20A1 gene (referred to as CYP20A1_Alu-LT hereafter) with a 8.93-kb long 3′UTR, 65% of which is derived from the exonization of 23 Alus (fig. 1a). Because this density of Alus across the length of the UTR was unusual, we characterized it further for regulatory potential. We observed a considerable disparity in the annotation of CYP20A1 transcript isoforms: ten, nine, and four annotated isoforms in Ensembl, NCBI, and UCSC, respectively (supplementary information S1, Supplementary Material online). Among the nine transcripts of human CYP20A1 annotated in NCBI, experimental evidence (from RNAseq or microarray) is available only for CYP20A1_Alu-LT (NM_177538.2), the longest isoform (10.94 kb). This isoform is an outlier in terms of its 3′UTR length as it occupies the 85th position in the genome-wide length distribution of 3′UTRs (fig. 1b). Less than 3% exonized transcripts have UTRs longer than 6 kb (supplementary fig. S1, Supplementary Material online). However, the length and enrichment does not seem to correlate with the density of exonized Alus (r = 0.25) compared with the genomic average of 5.42 exonization events/3′UTR. We ruled out the possibility of all these Alus arising due to self-expansion of one ancestral Alu in this UTR because the 23 Alus belong to different subfamilies that have arisen at different periods in primate evolution (Jurka and Smith 1988; Batzer and Deininger 1991; Richard Shen et al. 1991). Moreover, 17 of these Alus are full length (>250 bp). This also seems to be an early event as the Alus are conserved at orthologous positions in chimpanzee and other great apes.
Fig. 1.
CYP20A1 contains a unique 3′UTR with Alu-driven divergence. (a) UCSC tracks representing the four transcript isoforms of CYP20A1 with varying 3′UTR length. Only isoform 1 (NM_177538) contains the full-length 8,932 bp 3′UTR (CYP20A1_Alu-LT). The RepeatMasker track shows this 3′UTR harbors 23 Alu repeats from different subfamilies. (b) Genome-wide analysis of length distribution of 3′UTR reveals CYP20A1_Alu-LT to be an outlier. Mean and median 3′UTR lengths were 1,553 and 1,007 bp, respectively. (c) Cladogram of CYP20A1 protein sequence divergence among different classes of vertebrates. At the protein level, this gene seems to have diverged minimally. Values within the parentheses represent branch length (unit: substitutions per site) (d) DNA level conservation analysis of 5′UTR and 3′UTR among 20 mammals reveals that 5′UTR is well conserved among all primate lineages, suggesting that divergence is unique to 3′UTR. Repeat masker track shows the position of Alu elements in the UTR region (also see supplementary fig. S3, Supplementary Material online).
Genomic Region Proximal to CYP20A1_Alu-LT 3′UTR Is Relatively Well Conserved
The coding region of CYP20A1_Alu-LT is remarkably well conserved among vertebrates, both at the sequence level and at the length of the mature protein (table 1). The chimpanzee, macaque, and mouse CYP20A1 code for the conserved 462–470aa protein as in humans although their annotated transcript orthologs range between 1 and 3kb. Multiple sequence alignment across vertebrates reveals a strong conservation at both the N and the C terminals (supplementary fig. S2, Supplementary Material online). This is also corroborated by the minimal evolutionary divergence across vertebrate CYP20A1 proteins (fig. 1c) and a strong purifying selection in coding sequences (Ka/Ks ∼ 0.2 in mammals and <0.1 in nonmammalian vertebrates) (table 1).
Table 1.
CYP20A1 Protein Sequence Conservation across Vertebrates
| Organism | Scientific Name | Tax ID | % Identity | Query Cover | Protein ID | Length (aa) | Ka/Ks | P Value (Fisher) |
|---|---|---|---|---|---|---|---|---|
| Chimpanzee | Pan troglodytes | 9598 | 99 | 100 | XP_516042.2 | 462 | 0.711 | 0.499867 |
| Bonobo | Pan paniscus | 9597 | 99 | 100 | XP_003820821.1 | 462 | 0.497 | 0.266782 |
| Gorilla | Gorillla gorilla | 9595 | 82 | 100 | XP_004033127.1 | 417 | 1.014 | 1 |
| Orangutan | Pongo abelii | 9601 | 99 | 100 | XP_002812812.1 | 462 | 0.242 | 0.0078154 |
| Rhesus macaque | Macaca mulatta | 9544 | 96 | 100 | EHH21600.1 | 470 | 0.370 | 0.0042967 |
| House mouse | Mus musculus | 10090 | 83 | 100 | NP_084289.1 | 462 | 0.216 | 4.77E-35 |
| Rat | Rattus norvegicus | 10116 | 82 | 100 | NP_955433.1 | 462 | 0.194 | 1.19E-41 |
| Cow | Bos taurus | 9913 | 91 | 100 | NP_001015644.1 | 462 | 0.163 | 1.28E-27 |
| Grey wolf | Canis lupus familiaris | 9615 | 91 | 96 | XP_003434295.2 | 613 | 0.184 | 4.38E-29 |
| Chicken | Gallus gallus | 9031 | 74 | 99 | XP_426572.2 | 463 | 0.064 | 1.58E-207 |
| Anole lizard | Anolis carolinensis | 28377 | 76 | 99 | XP_003223588.2 | 557 | 0.080 | 2.87E-172 |
| Xenopus | Xenopus (Silurana) tropicalis | 8364 | 73 | 99 | NP_001039140.1 | 463 | 0.054 | 0 |
| Zebra fish | Danio rerio | 7955 | 64 | 99 | NP_998497.1 | 462 | 0.080 | 0 |
| Drosophila | Drosophila melanogaster | 7227 | 23 | 97 | NP_573003.2 | 495 | 0.490 | 8.73E-100 |
| Sea urchin | Strongylocentrotus purpuratus | 7668 | 32 | 99 | XP_792896.2 | 475 | 0.320 | 0 |
| Arabidopsis | Arabidopsis thaliana | 3702 | 22 | 94 | BAA28539.1 | 500 | 0.559 | 1.08E-49 |
Note.—CYP20A1 protein sequences among different vertebrate classes were compared using NCBI PBlast. Drosophila, sea urchin, and Arabidopsis were used as the evolutionary outgroups. All the pairwise comparisons are done with respect to the 462aa human CYP20A1 protein (NP_803882) and a query cover of ∼95% was obtained in each case. CYP20A1 is well conserved among vertebrates. Except for the three great apes (chimpanzee, bonobo, and gorilla), all the Ka/Ks comparisons are significant (Fisher’s exact test; P < 0.01). Lesser the value of Ka/Ks, the more stringent is the negative selection operative on the protein, that is, fewer nonsynonymous substitutions are tolerated in it.
On the other hand, 3′UTR extension in CYP20A1_Alu-LT seems to be majorly contributed by Alu insertion (65%). Its orthologs in mouse, rat, and zebrafish are short within 1 kb. In mouse, we observe a sparse presence of two B1 SINEs, one each of simple and low complexity repeat, whereas the zebrafish 3′UTR lacks repeats. The longest annotated CYP20A1 transcripts for mouse (NM_030013.3), rat (NM_199401.1), and zebrafish (NM_213332.2) are 2.27, 2.03, and 1.79 kb, respectively. The divergence in 3′UTR across the primate lineage, except lemur and proboscis monkey, as evident from Jukes Cantor measure, increases as we move from the great apes to rhesus macaque, with the breakpoints mostly coinciding with an Alu insertion (fig. 1d). Mouse was considered for the nonprimate evolutionary outgroup in this analysis. The 10-kb downstream of transcription end site of CYP20A1 was also found to be conserved among the higher primates, except for some New World monkeys (supplementary fig. S3, Supplementary Material online). Taken together, these suggest that Alu insertion might have contributed to the divergence of this 3′UTR, that is otherwise conserved, at least among the higher primates.
Characterization of CYP20A1_Alu-LT
We next investigated whether the full-length transcript containing this 3′UTR is actually transcribed. As two-thirds of this 3′UTR comprise Alus, it was challenging to capture the full-length transcript in expression arrays or map it uniquely from sequencing reads. Moreover, there are differences in annotations regarding the full-length 3′UTR-containing isoform (supplementary information S1, Supplementary Material online). Therefore, we designed 11 pairs of primers to experimentally confirm the expression of CYP20A1_Alu-LT. We validated three of its amplicons by Sanger sequencing to negate spurious amplification from other Alu-rich loci (fig. 2a and supplementary information S1, Supplementary Material online).
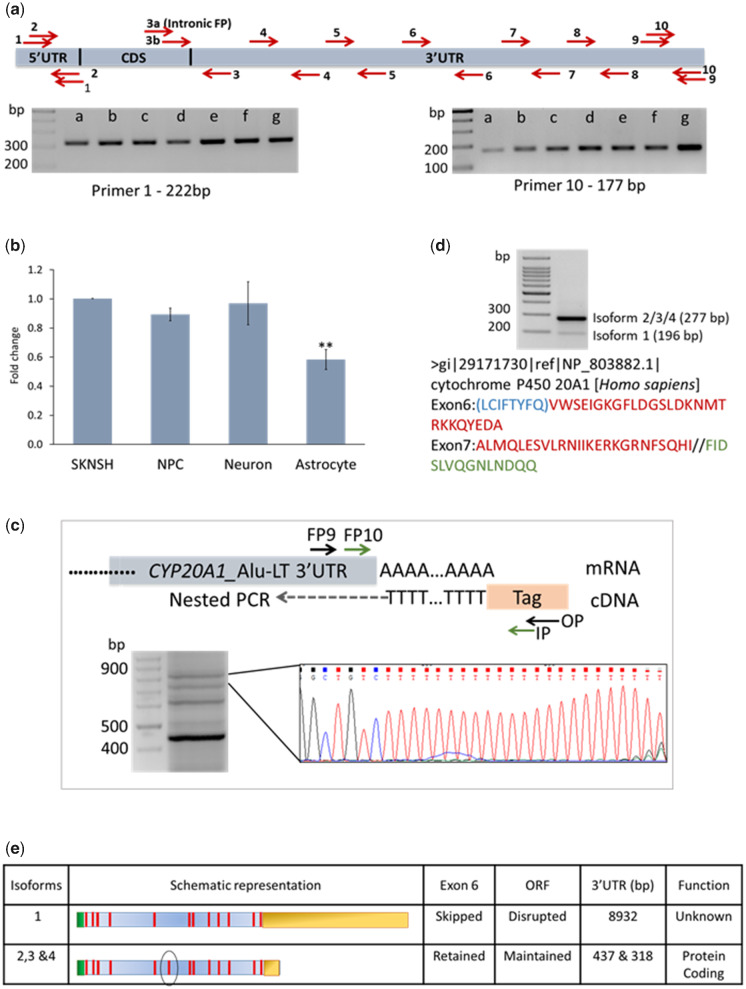
Fig. 2.
CYP20A1_Alu-LT is expressed and may be a long noncoding RNA. (a) A schematic representation of the primers designed on the CYP20A1_Alu-LT to encompass 5′UTR and full-length 3′UTR. To check for full-length expression of transcript, cDNA from multiple cell lines of different tissue origin was used for amplification. Lanes-a–g are from Hela-S3, A549, HeLa, HEK293, MCF7, SK-N-SH, and gDNA (positive control), respectively. Representative gel images of this isoform expression, via amplification from the starting of 5′UTR and the end of 3′UTR, are shown by primer pairs 1 and 10, respectively. Amplicons (1, 5, and 10) were also confirmed by Sanger sequencing. (b) RT-qPCR for CYP20A1_Alu-LT expression in cancerous and noncancerous cell types of neuronal origin. Fold change was calculated with respect to SK-N-SH, after normalization with the geometric mean of expression values from β-actin, GAPDH, and 18S rRNA. The error bars represent the SD of three biological replicates and the average of three technical replicates were taken for each biological replicate (**P < 0.01; Student’s t-test). (c) 3′RACE confirms the expression of the full-length transcript. The schematic depicts the oligo(dT) (attached to a tag sequence) primed reverse transcription, followed by nested PCR. The amplification products corresponding to the bands below 900 bp and above 700 bp mapped to CYP20A1_Alu-LT 3′UTR, suggesting that the full-length transcript is expressed in untreated MCF-7 cells (n = 3). (d) Differentiating the CYP20A1_Alu-LTR transcript from other isoforms. The schematic in figure 1a highlights the skipped exon 6 and the position of flanking primers on shared exons in green color. The presence of at least two different types of transcripts was confirmed. A 277-bp amplicon corresponds to isoform(s) that contain exon 6 but have shorter 3′UTRs (isoforms 2 and 3 in e) and 196-bp amplicon corresponds to the long-3′UTR isoform (isoform 1). None of the six translation frames of the long 3′UTR isoform matches with the annotated protein. The amino acids marked in red are common to both isoforms 2 and 3, blue exclusive to isoform 3 and green represents the sequence from isoform 1. (e) Schematic representation summarizing the differences between CYP20A1 transcript isoform 1 (CYP20A1_Alu-LT) and isoforms 2 and 3.
We observed variable expression of CYP20A1_Alu-LT in the six cell lines (fig. 2a). To delineate whether this could be a consequence of aberrant expression often reported in cancerous cell lines (Suzuki et al. 2014), we also compared its expression in a neuroblastoma cell line (SK-N-SH) with those in primary neuron, glia (astrocyte), and neural progenitor cells (NPCs). Neuroblastoma shares features with both mature neurons and NPCs but is distinct from glia. We found that CYP20A1_Alu-LT expression differs significantly only between glia and SK-N-SH but not in neurons or NPCs (fig. 2b). This corroborates with our observations in the cancerous cell lines.
We selected MCF-7, a breast adenocarcinoma cell line, for some of our subsequent experiments as it has been extensively used for screening effect of drugs and other xenobiotics on different CYP family genes (Coumoul et al. 2001; Ptak et al. 2010). The copy number of CYP20A1 is not altered in this line (2n = 2) (Pan et al. 2016). We performed 3′RACE to determine the exact transcription termination site for CYP20A1_Alu-LT and further confirmed the transcript by nested polymerase chain reaction (PCR) and amplicon sequencing (fig. 2c and supplementary information S3, Supplementary Material online). Our findings corroborate with TargetScan (release 7.2) which builds on the longest Gencode 3′UTR. The algorithm calculates 3′UTR length based on 3P-seq tags, accounting for the usage of mRNA cleavage and splice sites, normalized across multiple tissues.
Exon Skipping Differentiates CYP20A1_Alu-LT from the Protein Coding Isoforms
We observed that the expression of CYP20A1_Alu-LT is low although CYP20A1 protein is relatively abundant (supplementary fig. S4, Supplementary Material online), suggesting that other isoforms may contribute to protein levels. When we compare CYP20A1_Alu-LT with the shorter 3′UTR-containing isoforms, we observe a skipping of the sixth exon in this transcript. Using primers encompassing the sixth exon, we could distinguish between the transcripts; with the larger isoform (i.e., CYP20A1_Alu-LT) corresponding to the 196-bp amplicon and shorter one to 277 bp that also shows a relatively higher expression (fig. 2d).
In order to assess the relative contribution of different isoforms to the overall expression of CYP20A1, we used publicly available RNA-seq data from 15,928 single nuclei derived from the different layers of the human cerebral cortex (Hodge et al. 2019). NM_177538 (CYP20A1_Alu-LT) is expressed in 75% of the nuclei whereas all the other RefSeq isoforms are found in <1% (cutoff CPM ≥ 50). There are 7,038, 5,134, and 1,841 single nuclei in which NM_177538 (but no other isoform) is expressed with ≥10, 50, and 100 reads, respectively (supplementary table S2, Supplementary Material online). Interestingly, it is expressed in rosehip neurons—a highly specialized cell type in humans (supplementary table S3, Supplementary Material online) (Boldog et al. 2018).
Although the long 3′UTR transcript is annotated as the principal isoform, its expression level did not correlate with CYP20A1 protein which is relatively abundant in MCF-7 cells (supplementary fig. S4, Supplementary Material online). Thus, we performed in silico translation of all CYP20A1 isoforms in six reading frames and compared them with the annotated human CYP20A1 protein. The two short 3′UTR isoforms matched—one perfectly and another with an additional amino acid stretch (fig. 2d), but the CYP20A1_Alu-LT was out of frame in the sixth and seventh exon and BLAST analysis of the human proteome did not report any hits with the truncated 24 amino acid peptide. Taken together, these data suggest that the CYP20A1_Alu-LT is unlikely to be coding for CYP20A1 protein and may represent a novel transcript isoform originating from the same locus (fig. 2e).
CYP20A1_Alu-LT Expression in Nonhuman Primates
Among the nonhuman primates, we did not find any annotated transcripts beyond 3 kb from this locus. We further checked CYP20A1_Alu-LT expression in the reference transcriptomes of nonhuman primates (http://www.nhprtr.org, last accessed November 14, 2020) (Pipes et al. 2013). Total RNA reads derived from 157 libraries of 14 nonhuman primate species show consistent mapping patterns on CYP20A1 3′UTR. In chimpanzee, reads are evenly distributed across the length of the entire 3′UTR; however, distribution is patchy in the other Old world monkeys (with peaks mostly in the nonrepeat regions). Expression is minimal in New world monkeys (marmoset, squirrel monkey) and completely absent in lemur, although the adjoining coding exons show comparable expression, suggesting that CYP20A1_Alu-LT is expressed in the higher primates (supplementary fig. S5, Supplementary Material online).
CYP20A1_Alu-LT 3′UTR as an Evolving miRNA Regulatory Hub
Based on our earlier knowledge of 3′UTR exonized Alus providing novel miRNA binding sites, we explored whether the Alu-rich 3′UTR of CYP20A1_Alu-LT is also targeted by miRNAs (Pandey et al. 2016). A query in miRTarBase (release 6.0) (Hsu et al. 2011) revealed that CYP20A1_Alu-LT 3′UTR had predicted target sites for 169 miRNAs, of which 46 were listed as functional miRNAs in FuncMir (miRDB) (supplementary table S4, Supplementary Material online). Interestingly, ∼50% of these are either primate-specific or human-specific miRNAs (microRNAviewer) (Kiezun et al. 2012). The occurrence of target sites for human-specific miRNAs in this recently evolved UTR made us carry out further in-depth analysis of MREs.
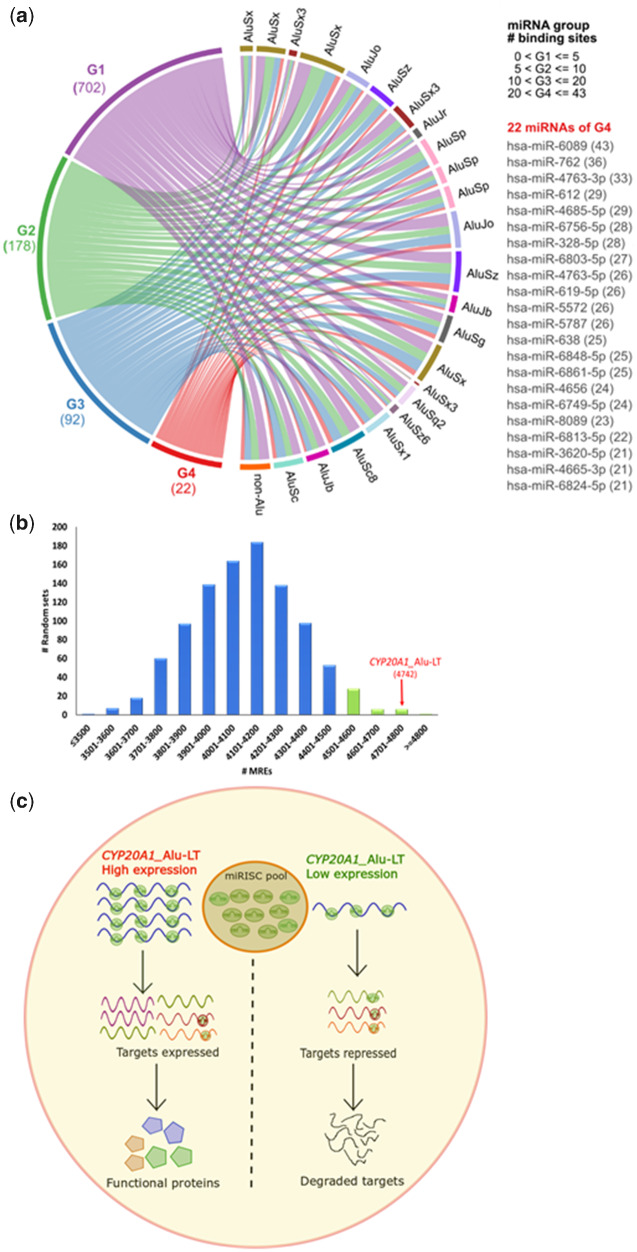
Using stringent cutoff criteria, we obtained a total of 4,742 MREs for 994 miRNAs, 4,500 of which overlap with Alus (4,382 MREs, if a conservative estimate of >50% overlap is considered) (supplementary table S5, Supplementary Material online). The MREs overlapping with Alu elements are considered as Alu-MREs. These 4,742 MREs span the entire length of the 3′UTR along with several high density pockets in Alu regions (fig. 3a). The 23 exonized Alus belong to Alu S and J family and are from 13 subfamilies—AluSx, AluSp, AluSc, AluSz6, AluSq2, AluSx3, AluSc8, AluSx1, AluSz, AluSg, AluJo, AluJb, and AluJr. Their presence in the 3′UTR of CYP20A1_Alu-LT in 5′ to 3′ direction is represented in figure 3a from top to bottom of the circos plot. The 994 miRNAs were grouped on the basis of numbers of MREs in CYP20A1_Alu-LT. MREs are grouped for 1–5, 6–10, 11–20, and 21–43, for group 1 (G1), group 2 (G2), group 3 (G3), and group 4 (G4), respectively. The total numbers of miRNAs in each group are 702, 178, 92, and 22 for G1, G2, G3, and G4, respectively. Only 2% of total miRNAs have MREs more than 20 (group G4), whereas ∼70% of the miRNAs were in the group G1 with ≤5 MREs. The miRNAs present in G4 are shown in figure 3a, with their number of MREs on 3′UTR written in brackets. The connections in circos plot show the presence of binding sites in each Alu. Non-Alu region is grouped as one and shown at the bottom of the circos. Majority of sites are present in Alu as all the connections from each of the groups fall in all the Alu elements. Only ∼5% of miRNA binding sites fall in non-Alu regions (fig. 3a).
Fig. 3.
CYP20A1_Alu-LT has the potential to act as a miRNA sponge. (a) Circos plot representing the MREs for the 994 miRNAs on CYP20A1_Alu-LT 3′UTR. miRNAs are grouped on the basis of the number of MREs. Twenty-three Alus in this 3′UTR contribute to 65% of its length and are distributed throughout the UTR. Only 11% of miRNAs have MREs > 10 (92 and 22 in G3 and G4, respectively). (b) Distribution of MREs for these 994 miRNAs on 1,000 random sets of 23 length and subfamily-matched Alu repeats. Only six sets contain MREs in range of 4,701–4,800 suggesting this is a nonrandom phenomenon and MREs are created post-Alu exaptations. Highlighted in green are sets with more than 4,500 MREs. (c) Proposed model to demonstrate the effect of potential sponge activity of CYP20A1_Alu-LT. In the condition where it is highly expressed, it will recruit multiple miRISC complexes which could relieve the repression of cognate targets leading to their translation, whereas in case of its reduced expression, those miRISC complexes remain free to load on the cognate targets and affect translational repression or promote mRNA degradation. CYP20A1_Alu-LT has the potential to sponge multiple miRNAs at the same time thereby regulating a large repertoire of transcripts.
It is plausible that the accumulation of so many Alu-MREs (miRNA binding sites within Alu) in this 3′UTR has been due to the retrotransposition or recombination of Alus with preexisting target sites. To test whether the MREs originated later to retrotransposition (Britten et al. 1988), we carried out analysis on 1,000 sets of 23 Alus taken randomly from the genome with matched length, composition and subfamily. We did not observe a similar distribution of Alu-MREs—only 0.5% and 4.2% of these random sets had MREs ≥4,742 and 4,500, respectively (fig. 3b). All the 23 Alus on CYP20A1_Alu-LT UTR have diverged from the consensus sequences of their respective subfamilies; some of these substituted bases might have aided the creation of MREs within this UTR. Next, we sampled the distribution of MREs for each subfamily randomly picking 2,000 members from the genome and queried for distribution of CYP-Alus. We found CYP-Alus as outliers for all 13 subfamilies. However, when a similar distribution is plotted only for 3′UTR-Alus, CYP-Alus lie above the median (but within the distribution) for eight (out of 13) subfamilies, namely, AluJo, Sc, Sc8, Sg, Sx, Sx1, Sx3 (1 out of 3), and Sz (supplementary information S4, Supplementary Material online). Taken together, these data suggest that there are certain subsets of 3′UTR-Alus, at least for the 13 subfamilies analyzed, that have accumulated MREs. We also calculated the probability of finding MREs for each of the 23 CYP20A1 3′UTR Alu elements (compared with MRE per base from all subfamily-matched, 3′UTR resident Alus) (supplementary information S4, Supplementary Material online). This suggests that the chance of Alus having retro-transposed into 3′UTRs with preexisting MREs is extremely low and these MREs might have been created within Alus post exaptation. An alternative, albeit less likely, possibility is greater retention of MRE sites within 3′UTR-Alus than elsewhere in the genome. However, even among the 3′UTRs, the propensity of Alu elements with high MRE content to occur in tandem in a single UTR is low, that is, CYP20A1_Alu-LT UTR contains many instances of such high MRE containing Alus, whereas other UTRs have relatively fewer such instances. Taken together, there is a possibility that accumulation of MREs could potentiate its function as miRNA sponge for a regulatory network (fig. 3c).
CYP20A1_Alu-LT Isoform Has the Potential to Function as a miRNA Sponge
To determine if CYP20A1_Alu-LT can be a potential miRNA sponge, we characterized this 3′UTR further using bioinformatics and experimental approaches. Because enrichment of Alu repeats in long RNAs could drive their nuclear localization, we first checked this for CYP20A1_Alu-LT isoform (Lubelsky and Ulitsky 2018). Using reverse transcription quantitative PCR (RT-qPCR) in both nuclear and cytosolic fractions, we found that it is predominantly localized to the cytosol—a feature observed in most sponges (fig. 4a). A sponge RNA also typically contains 4–10 low binding energy MREs for a particular miRNA that are separated by a few nucleotides and is generally devoid of destabilizing RNA elements. In CYP20A1_Alu-LT, using a stringent cutoff for MRE prediction (binding energy ≤−25 kcal/mol), we observed that out of the 994 miRNAs, 140 have ≥10 MREs and are distributed across the length of the UTR. Some of the binding energy predicted were as low as −47 kcal/mol and there were as many as 43 MREs for some miRNAs (fig. 3a and supplementary information S4, Supplementary Material online).
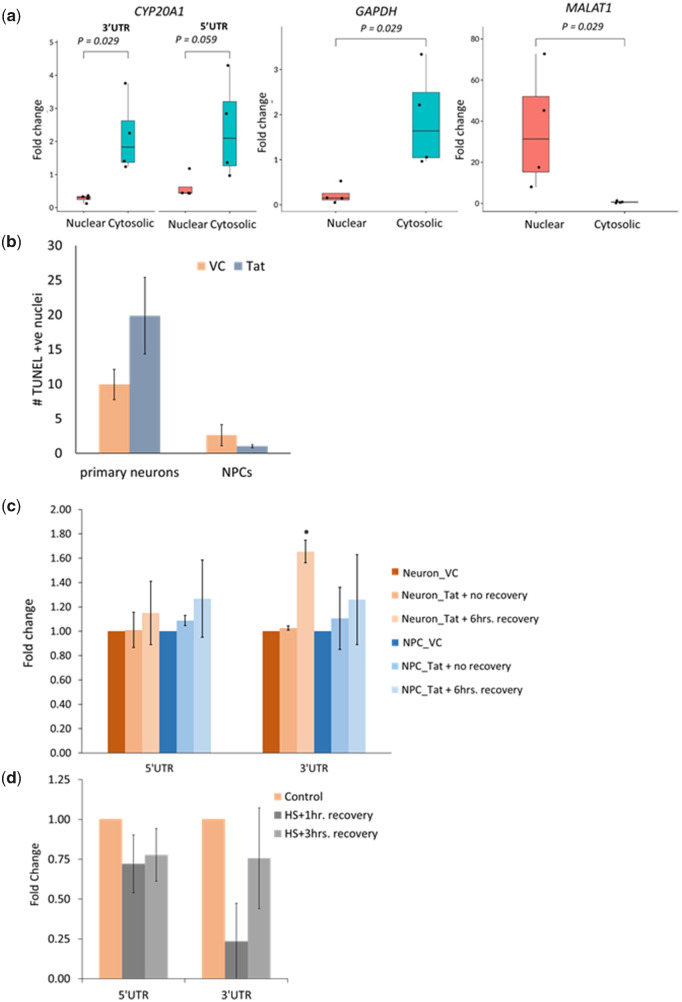
Fig. 4.
Features of CYP20A1_Alu-LT for being a potential sponge RNA. (a) Cytosolic localization of CYP20A1_Alu-LT confirmed by RT-qPCR. Fold change was calculated with respect to total RNA, after internal normalization using the primers against spiked-in control. The error bars represent the SD of four independent experiments and the average of two technical replicates was used for each experiment. Quality controls for assessing the purity of cytosolic (GAPDH) and nuclear (MALAT1) fractions are also shown. The RT-qPCR data were analyzed in accordance with the MIQE guidelines (Bustin et al. 2009) (supplementary information S3, Supplementary Material online). (b) Late apoptotic cells in primary neurons and NPCs in response to HIV1-Tat treatment were scored by the number of TUNEL positive nuclei. Tat is neurotoxic and kills ∼50% more neurons compared with the vehicle control (VC, i.e., saline), whereas the difference is not statistically significant for NPCs (P values 0.04 and 0.21 for primary neurons and NPCs, respectively, for Student’s t-test assuming equal variance). The data represent the mean and SD of three independent experiments and >1,000 nuclei were scored per condition for each experiment. (c, d) Expression of CYP20A1_Alu-LT in response to HIV1-Tat (c) and heat shock (d) treatment was assessed by RT-qPCR using both 5′ and 3′UTR primers. The 3′UTR was found to be upregulated following 6 h recovery after Tat treatment in neurons (P value = 0.035; *P value < 0.05, Student’s t-test), but not in NPCs (P value = 0.348) (c). It was also strongly downregulated in neurons (P value = 0.031) immediately after heat shock (HS + 1 h recovery). This difference was not significant during recovery (P value = 0.310; HS + 3 h recovery) (d). In both these cases, the 5′UTR primer exhibits the same trend as the 3′UTR but does not qualify the statistical significance cutoff of P < 0.05. Fold change was calculated with respect to saline (vehicle) treatment, after internal normalization with the geometric mean of GAPDH, ACTB, and 18S rRNA in (c) and with respect to control (no heat shock treatment) cells, after internal normalization with the geomean of GAPDH and ACTB (d). The error bars represent the SD of three independent experiments and the average of 2–3 technical replicates was taken for each experiment.
Potential Sponge Activity of CYP20A1_Alu-LT in Primary Neurons in Response to Heat Shock and HIV1-Tat
To probe if the alteration in CYP20A1_Alu-LT level could affect expression of transcripts containing cognate MREs, we looked for conditions where it is likely to be altered. In these conditions, the miRNA that targets these MREs should also be expressed. We anticipated that in conditions where there is a higher expression of the potential “sponge” (CYP20A1_Alu-LT), the abundant MREs would sequester the miRNAs. This potentially would relieve its other cognate targets resulting in higher expression of those genes. Whereas in conditions where CYP20A1_Alu-LT is downregulated, the miRNA would be free to bind its cognate targets, thereby potentially reducing their expression (fig. 3c).
We first queried for the expression of the 994 miRNAs having potential MREs in CYP20A1_Alu-LT from publically available miRNA expression profiles. These experiments, mostly microarray based, showed low concordance across replicates and high variability across experiments (supplementary information S1, Supplementary Material online). So, we tested this experimentally in MCF-7 and primary neurons. Because primary neurons preferentially express longer 3′UTRs, we hypothesized that it would be a good model to study miRNA-mediated regulation events (Hilgers et al. 2011; Miura et al. 2013; Wang and Yi 2014; Wehrspaun et al. 2014; Tushev et al. 2018). We carried out small RNA-seq and using a cutoff of at least ten MREs on CYP20A1_Alu-LT 3′UTR and TPM value of 50, we obtained a set of 21 and 9 miRNAs in MCF-7 and neurons, respectively, of which seven were common to both (table 2).
Table 2.
List of 23 Prioritized miRNAs
| miRNA | Expression (TPMa) |
MREs with Binding Energy ≤ −25 kcal/mol | |
|---|---|---|---|
| MCF-7 | Pr. Neurons | ||
| miR-941 | 119,639.3 | 950.33 | 10 |
| miR-3677-3p | 2,892.33 | 51.67 | 12 |
| miR-1304-3p | 1,922.5 | 80.33 | 10 |
| miR-4446-3p | 1,839.33 | — | 13 |
| miR-296-3p | 1,406.5 | 70.33 | 10 |
| miR-1254 | 1,235 | — | 10 |
| miR-6724-5p | 330.17 | — | 20 |
| miR-619-5p | 193.17 | 89 | 26 |
| miR-1908-3p | 191.67 | — | 16 |
| miR-3944-3p | 158.67 | — | 14 |
| miR-6842-3p | 158.17 | 175.67 | 10 |
| miR-4767 | 129.83 | 18 | |
| miR-5096 | 98.5 | 72.33 | 14 |
| miR-7703 | 97.17 | — | 13 |
| miR-939-5p | 81.83 | — | 19 |
| miR-3620-5p | 81.33 | — | 21 |
| miR-1226-5p | 81.17 | — | 18 |
| miR-1915-5p | 78.5 | — | 14 |
| miR-6732-3p | 57.5 | — | 13 |
| miR-1273g-3p | 56.67 | — | 11 |
| miR-4707-3p | 53 | — | 10 |
| miR-668-3p | — | 175.33 | 10 |
| miR-370-3p | — | 244.33 | 14 |
Note.—miRNAs were prioritized based on their expression level (≥50 TPM), number of MREs ≥ 10 with binding energy ≤25 kcal/mol.
Expression values <50 TPM have not been represented.
To screen for MREs that would efficiently dock miRNA without degrading the CYP20A1_Alu-LT transcript, we next checked for the presence of bulge within the MREs of prioritized 23 miRNAs (table 2) using miRanda with default parameters. We used twin criteria—a complete match in 6-mer (2–7 nt) seed site and presence of mismatch or insertion at 9–12 nt position. The 6-mer sites with wobble base pairing were also retained as two wobble-pairs were maximally present in some of the MREs. We found five such sites for miR-6724-5p, two each for miR-1254, miR-4767, and miR-3620-5p and one each for miR-941, miR-4446-3p, miR-296-3p, miR-619-5p, miR-6842-3p, and miR-1226-5p (table 3). At all these sites, we observed insertion in CYP20A1_Alu-LT, which suggests the possibility of a bulge formation in the transcript. This can potentially prevent the transcript from miRNA directed degradation and increase its efficiency to sequester miRNA molecules.
Table 3.
Features of MREs with Seed Site Match and Presence of Bulge
| miRNA | Total No. of MREs on CYP20A1 3′UTR | Average of Overall Complementarity of miRNA with CYP20A1 3′UTR | MRES with Seed Match (2–7 nt) Including Wobble | Number of MREs with Mismatch or Insertion at Bulge Position | Features of MREs |
||
|---|---|---|---|---|---|---|---|
| Forward Score | Binding Energy | Alignment Length | |||||
| miR-941 | 29 | 59.37 | 2 | 1 | 146 | −22.05 | 21 |
| miR-3677-3p | 57 | 54.14 | 3 | ||||
| miR-1304-3p | 28 | 59.57 | 12 | ||||
| miR-4446-3p | 34 | 60.56 | 3 | 1 | 120 | −27.05 | 22 |
| miR-296-3p | 42 | 54.76 | 5 | 1 | 146 | −25.23 | 25 |
| miR-1254 | 45 | 59.72 | 18 | 2 | 126,128 | −22.05, −23.75 | 25, 23 |
| miR-6724-5p | 105 | 44.18 | 19 | 5 | 9,999,103,107,126 | −21.14, −23.87, −26.94, −22.16, −23.6 | 22, 22, 22, 21, 18 |
| miR-619-5p | 45 | 67.17 | 27 | 1 | 115 | −21.76 | 20 |
| miR-1908-3p | 43 | 52.60 | 1 | ||||
| miR-3944-3p | 64 | 54.82 | 23 | ||||
| miR-6842-3p | 39 | 59.20 | 11 | 1 | 104 | −24.14 | 29 |
| miR-4767 | 72 | 54.71 | 9 | 2 | 101,124 | −25.3, −23.93 | 27, 16 |
| miR-5096 | 22 | 65.80 | 8 | ||||
| miR-7703 | 51 | 57.20 | 4 | ||||
| miR-939-5p | 63 | 51.32 | 9 | ||||
| miR-3620-5p | 77 | 53.71 | 32 | 2 | 108,130 | −21.24, −22.89 | 22, 20 |
| miR-1226-5p | 73 | 56.95 | 6 | 1 | 124 | −26.76 | 29 |
| miR-1915-5p | 40 | 54.88 | 1 | ||||
| miR-6732-3p | 35 | 60.37 | 0 | ||||
| miR-1273g-3p | 30 | 67.30 | 12 | ||||
| miR-4707-3p | 30 | 58.03 | 6 | ||||
| miR-668-3p | 38 | 52.86 | 12 | ||||
| miR-370-3p | 56 | 56.57 | 2 | ||||
Because CYP20A1 was identified from a set of Alu-exonized genes that map to apoptosis, we tried to ascertain its involvement in two stress conditions for studying this further. HIV1-Tat is a potent neurotoxin that kills ∼50% more neurons compared with the vehicle control (fig. 4b). Upon treating primary human neurons with HIV1 full-length Tat protein, followed by 6 h recovery, CYP20A1_Alu-LT was found to be significantly upregulated (1.65-fold). However, progenitor cells, which are immune to Tat (fig. 4b), did not show any such trend (fig. 4c). We also found CYP20A1_Alu-LT to be significantly downregulated (2.68-fold) in primary neurons upon heat shock (HS), followed by 1 h recovery (fig. 4d).
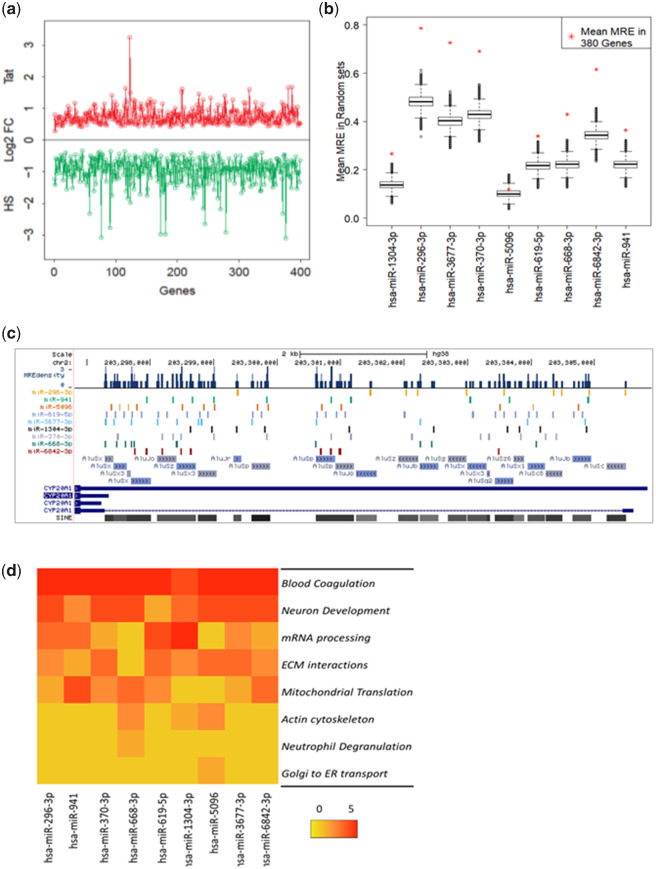
In order to query the expression of the other cognate targets of these miRNAs in these two conditions, we performed strand-specific RNA-seq of primary neurons after these treatments. The expression of CYP20A1_Alu-LT in RNA-seq showed similar patterns of expression as observed in RT-qPCR, significantly downregulated by 2.68-fold (log2FC = −1.42) upon HS recovery and 1.21-fold upregulated (log2FC = 0.28) during Tat response. The latter, however, did not cross the stringent statistical significance threshold of adjusted P value < 0.05. Out of the 3,876 genes differentially expressed in HS or Tat, expression of 380 positively correlates with that of CYP20A1_Alu-LT (fig. 5a and supplementary table S6, Supplementary Material online). All the 380 genes contain MRE sites for at least one or more for the nine prioritized miRNAs and the majority of their MREs are canonical and not Alu derived. There is a significant enrichment of MRE sites (nine prioritized miRNAs) in this set of 380 genes compared with all expressed genes (complete transcriptome; FPKM > 2) or those with a significant differential expression (FPKM > 2; FDR < 0.05) (table 4; Kolmogorov–Smirnov test). The MRE distribution in expressed genes and significantly differentially expressed genes were not significantly different. Similar results were obtained by comparing the median distribution of MREs in the three sets using Mann–Whitney U test (supplementary fig. S6, Supplementary Material online, and table 4). The abundance of MREs (of nine miRNAs), plotted against the FPKM values in significantly differentially expressed genes, exhibit a normal distribution (Shapiro–Wilk test, P < 2.2e-16). Genes with high MRE counts have lower expression values and vice versa (supplementary fig. S7, Supplementary Material online). To further validate the enrichment of MREs in the 380 genes, we performed Monte-Carlo simulation using one million random sets of 380 genes. The MRE densities in the 380 genes with expression pattern correlated to CYP20A1_Alu-LT, were outliers when plotted with the distributions derived from random sets (P value 9.99999e-07), for all the miRNAs except miR-5096 (fig. 5b). The nine miRNAs studied had nearly similar distribution of MREs across genes (supplementary fig. S8, Supplementary Material online). The CYP20A1_Alu-LT contains a total of 116 MREs for all these nine miRNAs. The MREs for the prioritized nine miRNAs from neurons had high density in few regions of the UTR with majority mapping to Alu (fig. 5c). Taken together these data suggest that the set of 380 genes represents potential cognate targets whose expression levels can be modulated by CYP20A1_Alu-LT through competing for the miRNAs targeting them. Further highly localized presence of MREs for these nine miRNA in a particular condition might increase the effectiveness of the miRNA sponging activity (fig. 5c).
Fig. 5.
Fold change (log2FC values) of 380 genes. (a) Figure represents log2FC of a set of 380 genes upregulated in response to Tat treatment (red) and downregulated during heat shock recovery (green) in primary neurons, resonating with the trend exhibited by CYP20A1_Alu-LT. All the transcripts contain one or more MREs for the nine miRNAs that can be potentially titrated by sponge activity of CYP20A1_Alu-LT in neurons. These represent potential cognate targets whose expression can be regulated by CYP20A1_Alu-LT perturbation. Genes are plotted in order as supplementary table S6, Supplementary Material online. (b) Enrichment of MRE sites in the 380 gene set compared with 1 million random sets of equal number of genes (Monte-Carlo simulations, P = 9.99999e-07). (c) The distribution of nine prioritized MREs on the CYP20A1 3′UTR, their overlap with Alu elements, and the MREs dense regions are shown. The orientation and subfamily of the 23 Alus present in this 3'UTR are also represented. (d) The heat map represents the top five biological processes targeted by each miRNA from pathway enrichment of 380 genes. Scale 0–5 is an arbitrary scale where 5 being the most targeted process.
Table 4.
Three Hundred and Eighty Genes (Expression Correlated with CYP20A1_Alu-LT) Are Significantly Enriched in MREs Compared with All Expressed Genes and Those with Significant Differential Expression
| Test | Kolmogorov–Smirnov Test |
Mann–Whitney U test |
||
|---|---|---|---|---|
| Gene Set | 380 Genes | Expressed Genes (FPKM > 2) | 380 Genes | Expressed Genes (FPKM > 2) |
| Expressed genes (FPKM > 2) | 5.102e-07 | 4.716e-12 | ||
| Differentially expressed genes (FPKM > 2; FDR < 0.05) | 4.311e-07 | 0.9925 | 8.134e-12 | 0.7505 |
Gene ontology analysis for these 380 genes set using Toppfun (FDR < 0.05) revealed the top five processes mapping to hemostasis (28 genes), axon guidance (25 genes), neutrophil degranulation (23 genes), platelet signaling, activation and aggregation (18 genes), and ECM organization (18 genes). Other processes include mRNA processing and mitochondria translation, metabolism, amino acid and nucleotide synthesis, and antigen presentation (supplementary table and fig. S9, Supplementary Material online). For the nine miRNAs, analysis of target genes in the set of 380 genes and their pathways revealed blood coagulation to be the major biological process targeted by almost all miRNAs followed by neuron development (fig. 5d). This implies that CYP20A1_Alu-LT might be important in maintaining the homeostasis and fine tuning the neurological pathways.
Discussion
Exonized Alus Create a Unique 3′UTR in CYP20A1_Alu-LT
We report a putative miRNA sponge from a unique transcript isoform of CYP20A1 gene derived through exonization of 23 Alus that make a major fraction of this transcript. This makes this isoform an outlier in terms of the length of its 9-kb long 3′UTR as the average length of 3′UTRs in the transcriptome is 5.42. None of the longer 3′UTRs also has matching repeat content. CYP20A1_Alu-LT isoform seems to have been neofunctionalized from a protein coding locus and its expression is detected in the higher primates. This could result in inclusion of the CYP20A1 gene into new regulatory networks. Interestingly, our study in primary human neurons suggests that differential expression of this RNA could modulate expression of multiple modules of a regulatory network and synchronize specific outcomes in response to environmental cues.
Although the CYP20A1 protein is highly conserved across the vertebrate phylogeny, we observe that the CYP20A1_Alu-LT isoform has a skipped exon that results in an out-of-frame coding sequences. Interestingly this isoform is expressed not only in 75% of single nuclei derived from different layers of the human cerebral cortex but also in rosehip neurons—a highly specialized cell type in humans.
CYP20A1_Alu-LT: A Potential miRNA Sponge
The presence of ≥10 MREs for as many as 140 miRNAs makes CYP20A1_Alu-LT distinct from the other naturally occurring sponges that have been mechanistically characterized. Most of them contain MREs for a single or a few related miRNA species (Franco-Zorrilla et al. 2007; Poliseno et al. 2010; Hu et al. 2018). Among transposable elements, Alus have been reported to have the maximum number of MREs (Spengler et al. 2014). We also observed that out of the 4,742 total MREs in CYP20A1_Alu-LT more than 80% are within Alu. Contribution of Alus in modulating as well as modifying existing miRNA regulatory networks in a lineage-specific manner is being increasingly reported (Lynch et al. 2011; Hoffman et al. 2014; Spengler et al. 2014; Trizzino et al. 2017; Hu et al. 2018). Our group has also reported the functional significance of MREs within 3′UTR-Alus in fine tuning the p53 regulatory network during stress response (Pandey et al. 2016). Also some Alu-derived miRNAs for example miR-1285-1 targets exonized Alus in transcripts (Spengler et al. 2014) and some like miR-661 targets specific genes like MDM2 and MDM4 (Hoffman et al. 2014). Sequestration of miR-566 by elevated levels of free Alu RNA have been shown to correlate with cancer progression (Di Ruocco et al. 2018). Our observations suggest that multiple exonized Alus in CYP20A1_Alu-LT 3′UTR can alter bioavailability of multiple miRNA that govern different genes of a specific pathway. Binding of miRNAs to MREs could also alter secondary structure formation of the 3′UTR and further affect the availability and accessibility of other MREs in a condition-specific manner.
CYP20A1_Alu-LT Sponge Activity Could Modulate mRNA–miRNA Networks in Neurocoagulopathy
Expression of competing endogenous (ce)RNAs, such as sponge RNAs, is tightly regulated and often specific to tissue, development stage, or stress conditions (Cesana et al. 2011; Tay et al. 2014). Because sponge RNA could titrate miRISC complexes, their expression could correlate with expression of mRNA having shared targets (Ebert et al. 2007; Ebert and Sharp 2010) (fig. 3c). We also observe that expression of CYP20A1_Alu-LT is inducible in response to HIV1-Tat and decreased during HS response in primary human neurons. Though we have not looked at its turnover rates, nevertheless we make similar observations in 380 genes which correlate with the expression pattern of CYP20A1_Alu-LT, that is, downregulated during HS and upregulated upon Tat treatment. This 380 gene set shows a significant, nonrandom enrichment of MREs for the nine prioritized miRNAs whose sites are also abundant in CYP20A1_Alu-LT. Also, these are otherwise not enriched in all the transcribing or differentially expressed genes under these conditions. This substantiates that the 380 set could be cognate target genes whose expression could be modulated through titration of the miRNAs by presence or absence of CYP20A1_Alu-LT.
Exposure to HIV1-Tat is known to cause axonal damage, loss of blood–brain barrier integrity, changes in neurite outgrowth, etc. These are mediated by astrocyte activation, inflammatory cytokine expression, inducing mitochondrial injury and rearrangement of microtubules (Robinson et al. 2007; Dickens et al. 2017; Fatima et al. 2017; Leibrand et al. 2017; Liu et al. 2018; Santerre et al. 2019). The set of 380 genes which correlate with the expression pattern of CYP20A1_Alu-LT were also enriched in similar pathways like axon guidance, hemostasis, platelet activation and aggregation, ECM organization, regulation of actin cytoskeleton, antigen presentation, Golgi to ER transport, and mitochondrial translation. In the light of our observations, it is possible that the changes observed upon Tat exposure could partly be mediated and synergized by the sponging effect of CYP20A1_Alu-LT.
The 380 genes map to processes that are involved in blood coagulation and neuronal pathways. Blood coagulation factors have been reported to affect pathophysiology of central nervous system (CNS) via coagulation protein mediated signal transduction (De Luca et al. 2017). This process is being linked to several neurodegenerative diseases including multiple sclerosis, cancer of the CNS, addiction, and mental health (De Luca et al. 2017). Although the exact biological role of CYP20A1_Alu-LT remains to be mechanistically elucidated, yet enrichment of coagulation pathways in gene set showing correlated expression with this transcript suggests that it may be involved in fine-tuning outcomes in neurocoagulopathy, a possibility for future studies.
This RNA isoform could be evolving as a hub of Alu mediated posttranscriptional regulatory events as earlier, sites for antisense and editing events have also been reported in this transcript (Mandal et al. 2013). The impact of these events in case of CYP20A1_Alu-LT remains a possibility for future investigation.
Conclusion
In this study, we postulate a novel dimension of its regulatory potential—that of creation of a miRNA sponge through Alu exaptations in the 3′UTR regions. CYP20A1 provides an interesting model for studying Alu-derived novel transcripts that can function as ceRNAs and coregulate multiple genes in a network or cellular process. Thus, the addition of a lineage-specific sponge could be a top-up on existing networks that modulate intermediate phenotypes such as neurocoagulation. These could act as regulatory switches and in response to biological cues rapidly release or sequester miRNAs to govern specific cellular outcomes.
Materials and Methods
Bioinformatics
Characterization of a Novel Transcript Isoform of CYP20A1
Extensive annotation of different transcript isoforms of CYP20A1 was carried out using Ensembl, NCBI, and UCSC. Details are provided in supplementary information S1, Supplementary Material online.
Length Comparison of the 3′UTR of CYP20A1_Alu-LT with Other 3′UTRs at Genome-Wide Scale
The coordinates for human transcripts (NM and XM IDs) were downloaded from NCBI RefSeq version 74 (hg38). For every gene, only the longest 3′UTR was considered. The summary statistics for size distribution were calculated using R scripts.
DNA Conservation Analysis
DNA sequence conservation across different species was checked with UCSC genome browser using multiple alignment across 20 species generated by multiz (Blanchette 2004). Both gaps and unaligned sequences were treated as “missing” data.
Protein Conservation Analysis
CYP20A1 protein sequences from different species were taken from the top hits obtained in NCBI PBlast by using the human protein as a reference. Multiple sequence alignment was performed using Clustal Omega (O 1.2.2). As described in Gautam et al. (2015), Ka/Ks ratio was calculated (see supplementary information S1, Supplementary Material online, for details).
CYP20A1_Alu-LT Expression in Nonhuman Primates
We used publicly available chimp and macaque RNA-seq data sets from GEO (GSM1432846, 55, 65 [SRR1510158, 167, 177]; GSM2265102, 4, 6 [SRR4012405, 08, 09, 13]). Reads were mapped to both human and chimp/macaque 3′UTR to increase fidelity and mapping on housekeeping genes like ACTB, GAPDH, and EIF4A2 was also checked to control for data quality and mapping parameters. To query more expression data sets, we took advantage of the sequence differences in this transcript due to skipping of sixth exon. We performed BLAST against human data sets in SRA using a 289-bp sequence reconstructed by joining exons 5 and 7. The hits were reconfirmed by alignment of reads to the 3′UTR.
RNA-seq Reads from nonhuman primate reference transcriptome mapped on hg19 were exported as UCSC genome browser tracks. We additionally incorporated the stranded RNA-seq data generated as a part of this study to compare the expression level of this transcript between human and other nonhuman primates.
miRNA Target Prediction in CYP20A1_Alu-LT
miRNA target sites (MREs) on CYP20A1 3′UTR were predicted using miRanda (version 3.3a) (36), with the parameters set as follows: score threshold(-sc): 100, gap opening penalty(-go): −8, gap extension penalty(-ge): −2, binding energy(-en): −25 kcal/mol, and “strict” (i.e., G:U pairs and gaps were not tolerated in the seed region). miRanda uses miRBase (which contains ∼2,500 miRNAs) for annotation. For bulge analysis, target prediction for 23 miRNAs on CYP20A1 3′UTR was performed using miRanda offline version 3.2a with default parameters (gap opening penalty = −8, gap extension = −2, score threshold = 50, energy threshold = −20kcal/mol, and scaling parameter = 4).
RNA-Seq
Fastq files were checked using FASTQC and overall Q score was >20 with no adapter contamination. Overrepresented sequences were not removed. Reads were mapped on hg38 using Tophat, followed by isoform quantification (Cufflinks) and collation (Cuffmerge). Overall read mapping rate was between 59% and 86.3% and concordant pair alignment ranged between 53.1% and 81.1%. Cuffdiff was used to calculate the differential expression (D.E.; calculated for each experimental condition against untreated). Summary of sample-wise RNAseq data is provided in supplementary information S3, Supplementary Material online.
Small RNA-Seq
The data were quality checked using FastQC (version 0.11.2), followed by adapter trimming by cutadapt (version 1.18) and reads were not discarded. As expected, around 95% of the adapter trimming events happened at the 3′end of the reads. Filtering based on length and quality was carried out by cutadapt; Q30 reads with sequence length >15 but <35 nt were retained for mapping. Nearly, 80% of the reads were retained after these filtering steps. Size distribution of the reads and k-mer position was (21–25, 28–32) and 26–28, respectively. Subsequently, these reads were mapped onto hg38 using Bowtie2. On average, 61% of the reads were uniquely mapped. miRDeep2 was run to obtain the read counts as TPM. Summary of sample-wise small RNA-seq data is provided in supplementary information S3, Supplementary Material online.
MRE-Enrichment Analysis
The abundance of MREs for nine prioritized miRNAs in the 380 CYP20A1_Alu-LT coregulated genes (listed in supplementary table S6, Supplementary Material online; correspond to 399 unique transcripts) was compared with that in all expressed genes (cutoff: FPKM > 2) and in genes with significant differential expression (FPKM > 2, FDR < 0.05) in our RNA-seq data sets. The MRE counts for each of the individual nine miRNAs were added to obtain a single value (total MREs) against each transcript 3′UTR, followed by statistical tests for the median (Mann–Whitney U test) as well as for the distribution (Kolmogorov–Smirnov test). P values < 0.05 were considered statistically significant. Expressed genes were also binned by MRE count—individually for each miRNA—to evaluate if the presence of MRE sites was correlated to their expression. The MRE density in 380 genes was compared with the distribution in one million random subsets (of 380 genes each) using Monte-Carlo simulation. All of these analyses (averaged expression values, MRE prediction) were done at the transcript level and subsequently mapped to the gene.
Experiments
Expression Analysis of CYP20A1_Alu-LT across Diverse Cell Lines
RNA Isolation, cDNA Synthesis, and RT-qPCR
Total RNA was isolated using TRIzol (Ambion, Cat. No. 15596-026) as per manufacturer’s protocol and its integrity was checked on 1% agarose gel followed by Nanodrop quantification (ND1000, Nanodrop Technologies, USA). cDNA was prepared from oligo(dT)-primed DNase-treated RNA (Invitrogen, Cat. No. AM1907) and SuperScript III RT (Invitrogen, Cat. No. 18080-044). RNA template was digested from the cDNA using two units of Escherichia coli RNaseH (Invitrogen, Cat. No. 18021071). Primers were designed using Primer3 (version 4.0.0) and were synthesized by Sigma (supplementary information S2, Supplementary Material online). To ensure there was no spurious amplification, we designed two pairs of overlapping primers both on the 5′ as well as 3′ends of our transcript of interest and included “minus-RT” controls in every reaction. Additionally, we sequenced three amplicons (1, 5, and 10) to check the specificity of amplification (supplementary information S1, Supplementary Material online). BlastN (NCBI; 2.4.0+) against the corresponding in silico predicted amplicons had revealed >95% sequence identity with an average query cover of 90%; BLAT against the whole genome (hg38) gave CYP20A1 as the top hit in every case. RT-qPCR was performed using 2X SYBR Green I master mix (Kapa Biosystems, Cat. No. KK3605) and the reaction was carried out in Roche LightCycler 480 (USA) (supplementary information S1, Supplementary Material online). Melting curves were confirmed to contain a single peak and the fold change was calculated by ΔΔCt method. MIQE guidelines were followed for data analysis.
3′RACE for Mapping the Full-Length Transcripts
cDNA for 3′RACE was prepared using RLM-RACE kit (Ambion, Cat. No. AM1700) with 1 µg MCF-7 total RNA as per the manufacturer’s recommendation. Nested PCR was performed with FP10 and an internal primer using the amplicon produced by FP9 and external primer. The product of this nested PCR was electrophoresed on 2% agarose gel and four major bands were observed, which were gel eluted using Qiaquick gel extraction kit (Qiagen, Cat. No. 28704) and subsequently sequenced. Details of the results are provided in the supplementary information S3, Supplementary Material online.
Cell Culture Studies
MCF-7 cell line was procured from National Centre for Cell Sciences (Pune, India) and cultured in GlutaMax-Dulbecco's Modified Eagle Medium (DMEM) high glucose (4.5 gm/l) (Gibco, Cat. No. 10569044) supplemented with 10% heat inactivated Fetal bovine serum (FBS) (Gibco, Cat. No. 10082147), HEPES (Gibco, Cat. No. 11560496), and 1× antibiotic-antimycotic (Gibco, Cat. No. 15240096). The culture was maintained at 70–80% confluency at 37 °C, 5% CO2. Cell line lineage was confirmed by STR profiling and cells were routinely screened for any contamination (supplementary information S1, Supplementary Material online).
Primary human neuron and astrocyte cultures comply with the guidelines approved by the Institutional Human Ethics Committee of NBRC as well as the Stem Cell and Research Committee of the Indian Council of Medical Research (Fatima et al. 2017). Briefly, NPCs derived from the telencephalon region of a 10–15-week-old aborted fetus were isolated, suspended into single cells and plated on poly-d-lysine (Sigma, Cat. No. P7886)-coated flasks. The cells were maintained in neurobasal media (Gibco, Cat. No. 21103049) containing N2 supplement (Gibco, Cat. No. 17502048), Neural Survival Factor 1 (Lonza, Cat. No. CC-4323), 25 ng/ml bovine fibroblast growth factor (Sigma, Cat. No. F0291), and 20 ng/ml human epidermal growth factor (Sigma, Cat. No.E9644) and allowed to proliferate over one or two passages. The stemness of NPCs was functionally assayed by 1) formation of neurospheres and 2) ability to differentiate into neurons or astrocytes. Additionally, NPCs were also checked for the presence of specific markers like Nestin. For commitment to the neuronal lineage, NPCs were starved of bovine fibroblast growth factor and Epidermal Growth Factor (EGF); with 10 ng/ml each of platelet derived growth factor (PDGF) (Sigma, Cat. No. P3326) and Brain derived neurotrophic factor (BDNF) (Sigma, Cat. No. B3795) added to the media cocktail. Differentiation of NPCs to astrocytes required Minimum Essential Medium (Sigma, Cat. No. M0268-10x) supplemented with 10% FBS. The process of neuronal differentiation completes in exactly 21 days; our experiments were completed within a week postdifferentiation. Differentiated cultures of primary neurons and astrocytes were also checked for specific markers by immunostaining to determine the efficiency of the differentiation process (supplementary fig. S10, Supplementary Material online).
Nuclear-Cytosolic Localization of CYP20A1_Alu-LT
Nuclear and cytosolic RNAs were isolated using PARIS kit (Ambion, Cat. No. AM1921) as per manufacturer’s protocol. Briefly, nearly 10 million cells were resuspended in a fractionation buffer, incubated on ice, and centrifuged at 4 °C to separate the nuclear and cytosolic fractions. The nuclear pellet was additionally treated with a cell disruption buffer before mixing with the 2× lysis/binding solution and absolute ethanol and passing through a column. The RNA was subsequently eluted in a hot elution buffer; quantified using Nanodrop and its integrity was checked on 1% agarose gel. Nuclear RNA contains an additional hnRNA band above the 28S rRNA band and is usually of lower yield than cytosolic RNA. RT-qPCR was done as described earlier using gene-specific primers from 5′UTR and 3′UTR region.
Induction of Stress
Cells were gently washed once with 1× phoshpate buffer saline (PBS) and fresh media was replenished before treatment for accurate quantification of stress response. HS was given at 45 °C (±0.2) for 30 min in a water bath. Subsequent to the treatment, cells were transferred to 37 °C/5% CO2 for recovery and harvested after 1r, 3, and 24 h. For Tat treatment, full-length lyophilized recombinant HIV1 Tat protein was purchased from ImmunoDX, LLC (Woburn, MA) and reconstituted in saline. The dosage for treatment was determined by drawing a “kill curve” using graded dose of Tat on neurons (supplementary fig. S11, Supplementary Material online). Treatment was performed for 6 h with 100 ng/ml Tat and cells were either harvested just after the treatment or allowed to recover at 37 °C and 5% CO2 for another 6 h prior to harvesting.
TUNEL Assay
The assay was performed with in situ Cell Death Detection kit, TMR red (Millipore Sigma, Cat. No. 12156792910). Nearly 20,000 cells were seeded per well (on coverslips) in 12-well plates. Post Tat treatment, cells were washed once with 1× PBS, and fixed with 4% paraformaldehyde (PFA), followed by three washes with 1× PBS, permeabilization and blocking with 4% BSA containing 0.5% Triton-X 100, incubation with Terminal deoxynucleotidyl transferase (TdT) for 1 h in the dark and three washes with 1× PBS. Coverslips were then mounted on clean glass slides using hardset mounting media containing DAPI (Vectashield, Cat. No. H-1500). Six to eight random fields were imaged for each experimental group using AxioImager, Z1 microscope (Carl Zeiss, Germany). Fixed cells treated with two units of DNaseI (for 10 min at RT, followed by the addition of ethylenediaminetetraacetic acid to stop the reaction) were used as a positive control in this experiment. TUNEL positive nuclei were scored using ImageJ software (NIH, USA). Minimum of 1,000 cells were scored for each replicate.
RNA Sequencing and Small RNA Sequencing
Detailed methods for library preparation for RNA-seq and small RNA-seq are provided in supplementary information S1, Supplementary Material online. Briefly, libraries for RNA-seq were made using 500 ng of total RNA per sample and three biological replicates were taken per experimental condition. Libraries were prepared following Illumina’s TruSeq stranded total RNA protocol. The final libraries were pooled, diluted, and denatured to a final concentration of 8 pM. Clusters were generated using TruSeq PE cluster kit V3-cBot-HS on cBot system, followed by paired end sequencing on HiSeq2000 using TruSeq SBS kit V3-HS (200 cycles). Libraries for small RNA sequencing were prepared using Illumina’s TruSeq small RNA library preparation kit from 1 µg total RNA. The libraries were normalized to 2 nM, denatured, and subjected to cluster generation on cBot using TruSeq SR cluster kit v3-cBOT-HS. Single read sequencing was performed on HiSeq2000 using TruSeq SBS kit v3-HS (50 cycles).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The work was supported by the Council of Scientific and Industrial Research (Grant No. MLP-901 to M.M.). Financial support from CSIR in the form of fellowships to A.B. and K.S. is acknowledged. V.J. was supported by Persistent Systems Ltd. G.C. and D.D. were supported by fellowships from the University Grants Commission (UGC) and Department of Biotechnology (DBT), respectively. M.F. and P.S. acknowledge the support of the facilities provided through the Distributed Information Centre at NBRC, Manesar, under the Biotechnology Information System Network (BTISNET) grant, DBT, India. M.F. was supported by a fellowship from CSIR, New Delhi and P.S. was partially supported by research grants from DBT and NBRC core funds. The authors acknowledge Chitra Mohindar Singh Singal, NBRC for her help with the TUNEL assay, Parashar Dhapola for his help with data visualization in the initial phase of this work, Madiha Haider for data visualization of miRNA-pathway enrichment, Dr Amit Chaurasia for Jukes Cantor divergence analysis, Dr Rakesh Dey for providing reagents and many fruitful discussions and Dr Gaurav Ahuja and Dr Debarka Sengupta (IIIT Delhi) for their help with the MRE-enrichment analyses. The authors would also like to acknowledge Mr Raghunandanan MV and Mr Amit Khulve at IT division CSIR-IGIB for their constant help and support for data upload in GEO.
Author Contributions
M.M. and A.B. designed the study and cowrote the manuscript along with K.S .and R.P. A.B. performed conservation analyses, miRNA target prediction, cell culture, and molecular biology experiments and helped in RNA-seq data analysis. V.J. analyzed mRNA and small RNA-seq data, ran miRNA target prediction on miRanda, and helped in data visualization. K.S. performed molecular biology experiments, ran miRNA bulge analysis with D.D., and contributed to improving data visualization along with R.K. M.F. carried out primary human neuron and NPC culture under the supervision of P.S. D.S. performed some of the cellular assays. G.C. prepared NGS libraries and carried out sequencing, assisted by A.B. and K.S. T.E.B. analyzed the single nuclei RNA-seq data. B.P. contributed reagents, helped in troubleshooting experiments, and provided critical inputs. M.M. supervised the overall study.
Data Availability
The raw data for small RNA and mRNA sequencing generated in this study have been submitted to GEO (GSE132447).
dbGap link for the human temporal cortex (MTG) raw sequence reads https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi? study_id=phs001790.v1.p1.
Literature Cited
- An HJ, Lee D, Lee KH, Bhak J. 2004. The association of Alu repeats with the generation of potential AU-rich elements (ARE) at 3′ untranslated regions. BMC Genomics. 5(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi A, Herke SW, Batzer MA, Kim J. 2016. DNA methylation variation of human-specific Alu repeats. Epigenetics 11(2):163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. 1991. A human-specific subfamily of Alu sequences. Genomics 9(3):481–487. [DOI] [PubMed] [Google Scholar]
- Blanchette M. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14(4):708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog E, et al. 2018. Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat Neurosci. 21(9):1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Baron WF, Stout DB, Davidson EH. 1988. Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci U S A. 85(13):4770–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, et al. 2009. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem. 55(4):611–622. [DOI] [PubMed] [Google Scholar]
- Cesana M, et al. 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147(2):358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, et al. 2017. The exonization and functionalization of an Alu-J element in the protein coding region of glycoprotein hormone alpha gene represent a novel mechanism to the evolution of hemochorial placentation in primates. Mol Biol Evol. 34(12):3216–3231. [DOI] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351(6277):1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumoul X, Diry M, Robillot C, Barouki R. 2001. Differential regulation of cytochrome P450 1A1 and 1B1 by a combination of dioxin and pesticides in the breast tumor cell line MCF-7. Cancer Res. 61(10):3942–3948. [PubMed] [Google Scholar]
- De Luca C, Virtuoso A, Maggio N, Papa M. 2017. Neuro-coagulopathy: blood coagulation factors in central nervous system diseases. Int J Mol Sci. 18(10):2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruocco F, et al. 2018. Alu RNA accumulation induces epithelial-to-mesenchymal transition by modulating MiR-566 and is associated with cancer progression. Oncogene 37(5):627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens AM, et al. 2017. Chronic low-level expression of HIV-1 Tat promotes a neurodegenerative phenotype with aging. Sci Rep. 7 (1):7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 4(9):721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. 2010. MicroRNA sponges: progress and possibilities. RNA 16(11):2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima M, et al. 2017. Novel insights into role of MiR-320a-VDAC1 axis in astrocyte-mediated neuronal damage in NeuroAIDS. Glia 65(2):250–263. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, et al. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 39 (8):1033–1037. [DOI] [PubMed] [Google Scholar]
- Gautam P, et al. 2015. Population diversity and adaptive evolution in keratinization genes: impact of environment in shaping skin phenotypes. Mol Biol Evol. 32(3):555–573. [DOI] [PubMed] [Google Scholar]
- Grover D, Kannan K, Brahmachari SK, Mukerji M. 2005. ALU-ring elements in the primate genomes. Genetica 124(2–3):273–289. [DOI] [PubMed] [Google Scholar]
- Häsler J, Samuelsson T, Strub K. 2007. Useful ‘Junk’: Alu RNAs in the human transcriptome. Cell Mol Life Sci. 64(14):1793–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häsler J, Strub K. 2006. Alu elements as regulators of gene expression. Nucleic Acids Res. 34(19):5491–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, et al. 2011. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc Natl Acad Sci U S A. 108(38):15864–15869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, et al. 2019. Conserved cell types with divergent features in human versus mouse cortex. Nature 573(7772):61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman Y, Pilpel Y, Oren M. 2014. MicroRNAs and Alu elements in the P53-Mdm2-Mdm4 regulatory network. J Mol Cell Biol. 6(3):192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S-D, et al. 2011. MiRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 39(Suppl 1):D163–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, et al. 2018. Recently evolved tumor suppressor transcript TP73-AS1 functions as sponge of human-specific MiR-941. Mol Biol Evol. 35(5):1063–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Smith T. 1988. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci U S A. 85(13):4775–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, et al. 2012. MiRviewer: a multispecies microRNA homologous viewer. BMC Res Notes 5(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409(6822):860–921. [DOI] [PubMed] [Google Scholar]
- Lee JY, Ji Z, Tian B. 2008. Phylogenetic analysis of MRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res. 36(17):5581–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibrand CR, et al. 2017. HIV-1 Tat disrupts blood–brain barrier integrity and increases phagocytic perivascular macrophages and microglia in the dorsal striatum of transgenic mice. Neurosci Lett. 640:136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, et al. 2008. Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet. 4(10):e1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. 2018. HIV-1 protein Tat1–72 impairs neuronal dendrites via activation of PP1 and regulation of the CREB/BDNF pathway. Virol Sin. 33(3):261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Ulitsky I. 2018. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 555(7694):107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP. 2011. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. 43(11):1154–1159. [DOI] [PubMed] [Google Scholar]
- Mandal AK, Pandey R, Jha V, Mukerji M. 2013. Transcriptome-wide expansion of non-coding regulatory switches: evidence from co-occurrence of Alu exonization, antisense and editing. Nucleic Acids Res. 41(4):2121–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. 2013. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res. 23(5):812–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S-T, et al. 2016. Computational identification of the paralogs and orthologs of human cytochrome P450 superfamily and the implication in drug discovery. Int J Mol Sci. 17(7):1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, et al. 2016. Alu-MiRNA interactions modulate transcript isoform diversity in stress response and reveal signatures of positive selection. Sci Rep. 6(1):32348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer LM, et al. 2017. Structural variants caused by Alu insertions are associated with risks for many human diseases. Proc Natl Acad Sci U S A. 114(20):E3984–E3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes L, et al. 2013. The non-human primate reference transcriptome resource (NHPRTR) for comparative functional genomics. Nucleic Acids Res. 41(D1):D906–D914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Domany E. 2006. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 7(1):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, et al. 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465(7301):1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak A, et al. 2010. Induction of cytochrome P450 1A1 in MCF-7 human breast cancer cells by 4-chlorobiphenyl (PCB3) and the effects of its hydroxylated metabolites on cellular apoptosis. Environ Int. 36(8):935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 46 (1):21–42. [DOI] [PubMed] [Google Scholar]
- Richard Shen M, Batzer MA, Deininger PL. 1991. Evolution of the master Alu gene(s). J Mol Evol. 33(4):311–320. [DOI] [PubMed] [Google Scholar]
- Robinson B, Li Z, Nath A. 2007. Nucleoside reverse transcriptase inhibitors and human immunodeficiency virus proteins cause axonal injury in human dorsal root ganglia cultures. J Neurovirol. 13(2):160–167. [DOI] [PubMed] [Google Scholar]
- Roy-Engel AM, et al. 2005. Human retroelements may introduce intragenic polyadenylation signals. Cytogenet Genome Res. 110(1–4):365–371. [DOI] [PubMed] [Google Scholar]
- Santerre M, et al. 2019. HIV-1 Tat protein promotes neuronal dysregulation by inhibiting E2F transcription factor 3 (E2F3). J Biol Chem. 294(10):3618–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, et al. 2011. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci U S A. 108(7):2837–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, Krzyzosiak WJ. 2002. Structural determinants of BRCA1 translational regulation. J Biol Chem. 277(19):17349–17358. [DOI] [PubMed] [Google Scholar]
- Sorek R. 2009. When new exons are born. Heredity 103(4):279–280. [DOI] [PubMed] [Google Scholar]
- Sorek R, Ast G, Graur D. 2002. Alu-containing exons are alternatively spliced. Genome Res. 12(7):1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler RM, Oakley CK, Davidson BL. 2014. Functional microRNAs and target sites are created by lineage-specific transposition. Hum Mol Genet. 23(7):1783–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, et al. 2014. Aberrant transcriptional regulations in cancers: genome, transcriptome and epigenome analysis of lung adenocarcinoma cell lines. Nucleic Acids Res. 42(22):13557–13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. 2014. The multilayered complexity of CeRNA crosstalk and competition. Nature 505(7483):344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toll-Riera M, Castelo R, Bellora N, Mar Albà M. 2009. Evolution of primate orphan proteins. Biochem Soc Trans. 37(4):778–782. [DOI] [PubMed] [Google Scholar]
- Tristán‐Flores FE, et al. 2018. Liver X receptor–binding DNA motif associated with atherosclerosis‐specific DNA methylation profiles of Alu elements and neighboring CpG islands. J Am Heart Assoc. 7(3):e007686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, et al. 2017. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 27 (10):1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tushev G, et al. 2018. Alternative 3′ UTRs modify the localization, regulatory potential, stability, and plasticity of mRNAs in neuronal compartments. Neuron 98(3):495–511.e6. [DOI] [PubMed] [Google Scholar]
- Wang L, Rishishwar L, Mariño-Ramírez L, Jordan IK. 2017. Human population-specific gene expression and transcriptional network modification with polymorphic transposable elements. Nucleic Acids Res. 45(5):2318–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yi R. 2014. 3′UTRs take a long shot in the brain. BioEssays 36(1):39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrspaun CC, Ponting CP, Marques AC. 2014. Brain-expressed 3′UTR extensions strengthen miRNA cross-talk between ion channel/transporter encoding MRNAs. Front Genet. 5:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, et al. 2009. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Res. 37(13):4331–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data for small RNA and mRNA sequencing generated in this study have been submitted to GEO (GSE132447).
dbGap link for the human temporal cortex (MTG) raw sequence reads https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi? study_id=phs001790.v1.p1.