Abstract
Introduction:
Capnocytophaga species are recognized as part of human oral microbiota and implicated as periodontal pathogens associated with various periodontal diseases. The three original Capnocytophaga species - Capnocytophaga ochracea, Capnocytophaga sputigena and Capnocytophaga gingivalis were initially isolated from periodontitis in adults, but subsequent studies demonstrated their presence also at periodontally healthy sites in both children and adults. Their association with periodontal disease is a matter of controversy. Considering the differing virulence features of the respective isolate, it is crucial to identify these isolates to species level.
Aims:
The aim of this study is to investigate the prevalence of Capnocytophaga species by polymerase chain reaction (PCR) through restriction fragment length polymorphism in healthy individuals and patients with periodontal disease.
Material and Methods:
The study included a total of 300 individuals, 100 each with Gingivitis, Chronic periodontitis, and Healthy individuals. The plaque samples were collected using sterile curette in reduced transport fluid. DNA extraction was carried out for PCR analysis.
Results:
Of 300 individuals, Capnocytophaga species were identified from 237 (79%) participants in all groups. The prevalence was statistically analyzed using Chi-square test. The prevalence was more in males in gingivitis and healthy individuals (42% and 49% respectively), and females in periodontitis (40%). C. ochracea was observed in a higher proportion (36.33%), followed by C. granulosa (32.66%) and C. gingivalis (10%). They were identified more in the age group of 30–40 years in gingivitis and periodontitis, (30 and 21 individuals, respectively) and 39 individuals in 18–29 years in healthy individuals. They were present in 87% in healthy individuals, 77% in gingivitis and 73% in periodontitis.
Conclusion:
Capnocytophaga species are commonly present in healthy individuals and may be associated with periodontal disease. There is a need for further study to know the prevalence of other species of Capnocytophaga in health and disease.
Keywords: Capnocytophaga, gingivitis, polymerase chain reaction via restriction fragment length polymorphism, periodontitis
INTRODUCTION
Chronic periodontitis is defined as the destruction of periodontal ligament and loss of the adjacent supporting bone, which may be due to inflammation of the gingiva and other adjacent attachment apparatus.[1] Periodontal diseases are very common, affecting up to 90% of the global population. Several Gram-negative anaerobic bacteria, including Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia, are frequently isolated from dental plaques in periodontal patients and were initially considered as periodontal pathogens.[1] Many other species are considered to be closely associated with periodontitis, such as Prevotella intermedia, Fusobacterium nucleatum, Capnocytophaga species, and Campylobacter rectus. The presence of these microorganisms in the periodontal pocket can be considered as a marker for the development of periodontitis or an indicator for the progression of inflammation.[2]
Capnocytophaga species are recognized as part of human oral microbiota. They have been implicated as putative periodontal pathogens associated with various periodontal diseases. They have been repeatedly recovered from a number of infectious diseases, such as septicemia, osteomyelitis, abscesses and keratitis. The three original Capnocytophaga species-Capnocytophaga ochracea, Capnocytophaga sputigena and Capnocytophaga gingivalis were initially isolated from periodontitis in adults, but subsequent studies demonstrated their presence at periodontally healthy sites in both children and adults. Thus, their association with periodontal disease is a matter of controversy.[3] Although the genus is known to contain five human oral isolates, accurate identification of these organisms to species level has been hampered by the lack of reliable methods. Hence, most studies to date have reported these isolates as Capnocytophaga species.[4]
The detection of these putative pathogens is usually carried out by culture method, which is not always sensitive enough and requires special skill.[5] Previous studies have attempted to distinguish between these species by biochemical tests, protein profiles, multilocus enzyme electrophoresis and serotyping and DNA probes. However, the results were inconclusive. Considering the differing virulence features of the respective isolate, it is crucial to identify these isolates at the species level. The universal nature of 16SrRNA gene has provided an accurate method for bacterial identification.[4] Hence, the aim of this study was to investigate the prevalence of Capnocytophaga species using PCR via restriction fragment length polymorphism (PCR-RFLP) in healthy individuals and patients with periodontal disease.
MATERIALS AND METHODS
The study sample was obtained from individuals who were diagnosed with Gingivitis, Chronic Periodontitis and Healthy Gingiva. The study was approved by the Ethical committee of the Institute. The patients were from both genders and of age range between 18 and 55 years. The duration of the study was between June 2015 and 2016. The study included a total of 300 individuals, 100 each with Gingivitis, Periodontitis and Healthy gingiva.
Inclusion criteria for the Gingivitis group were – Generalized presence of clinical signs of gingival inflammation, probing depth ≤3 mm and no clinical attachment loss.
Inclusion criteria for periodontitis group were – Generalized presence of clinical signs of gingival inflammation, generalized probing depth ≥5 mm and generalized clinical attachment loss of ≥3 mm.
Inclusion criteria for healthy groups were-Absence of any clinical sign of gingival inflammation, probing depth ≤3 mm and no clinical attachment loss.
Exclusion criteria were - Patients with any systemic disease, smokers, pregnant or lactating women, cervical or subgingival caries or restorations, periodontal or antimicrobial therapy within 3 months before sampling.
After meeting all inclusion and exclusion criteria, plaque samples were collected. The plaque samples from these individuals were collected using sterile curettes and transferred to reduced transport fluid (RTF). The RTF was then sent to the Laboratory of Department of Molecular biology and Immunology. It was then vortexed to release the organisms in broth and DNA extraction was carried out for PCR analysis.
Procedure for DNA extraction
DNA extraction was carried out by the Modified proteinase-K method. The samples were first transferred to the tube containing T.E buffer, and then, it was homogenized by vortexed for a few seconds. Samples were then centrifuged at 5000 rpm for 5 min. The supernatant was removed and again washed with a fresh TE buffer. Supernatant was discarded and 50 μl lysis buffer I (1M Tris buffer: 500 μl, Triton X-100: 500 μl, 0.5M EDTA: 100 μl, Distilled Water: Made to 50 ml) was added. This was vortexed and kept for 5 min at room temperature. Followed by the addition of 50 μl lysis buffer II (Tris HCL: 50 mM (pH 8.0), KCL: 50 mM, MgCl2: 2.5 mM, Tween 20: 0.45%, Nodient P-40: 0.45%) and Proteinase – K (10 mg/ml) was added. Tubes were incubated at 60°C for 2 h, followed by enzyme deactivation by keeping in boiling water bath for 10 min. Later, samples were centrifuged at 5000 rpm for 5 min and supernatant containing DNA was collected in fresh tube and stored at −20°C till further use.[6]
PCR procedure
PCR was carried out in 25 μl total volume. Amplicon red master mix was used which contained Tris-HCL pH 8.5, (NH4) 2SO4, 3 mM MgCl2, 0.2% Tween 20, 0.4 mM of each dNTP, 0.2 units/μl Amplicon Taq DNA Polymerase. Primers targeting 16SrRNA conserved region of Capnocytophaga species was used; Forward primer27f (5′ AGAGTTTGATCMTGGCTCAG 3') and Reverse primer1492r (5′ TACGGYTACCTTGTTACGACTT 3′) and 1 μl of 15 pmole of each primer and 3 μl of 100 μg/ml of DNA was added to the mixture. Thermal cycling conditions were performed in verity thermal cycler (Applied biosystems, USA) which is as follows:
95°C for 5 min, followed by 30 cycles at 94°C for 1 min, 54°C for 1 min and 72°C for 1 min. Final extension was done at 72°C for 5 min. Amplified product of 1500 bp was detected on 2% Agarose gel electrophoresis. PCR amplified samples were digested with restriction enzyme Hhal (thermofisher Scientific, Massachsetts, USA). 10 μl of PCR amplicons were digested with 1u of fast digest restriction enzyme Hhal. Incubation was done at 37°C for 5 min. The mixture was then loaded on 2.5% Agarose for electrophoresis at 80v for 1 h. 100 bp DNA ladder was added simultaneously with all the samples in each gel. Gel was stained with 0.5 μg/ml of ethidium bromide and was viewed and captured using gel documentation system (Major Science, Saratoga, USA). Analysis of different DNA banding pattern was done using total lab software (Newcastle, Upontyne, England).[4]
RESULTS
Three hundred subgingival plaque samples with an equal distribution of healthy, gingivitis and periodontitis were analyzed by PCR to detect the presence of various Capnocytophaga species. Among the participants, 150 (50%) were male and 150 (50%) were female [Table 1]. The prevalence of Capnocytophaga species was statistically analyzed using Chi-square test. From total of 7 species, only 3 species, (C. ochracea, Capnocytophaga granulosa and C. gingivalis) were detected by PCR [Figure 1]. C. sputigena, C. haemolytica, C. genospecies and C. leadbetteri were not detected in any of the three groups. C. ochracea was observed in a higher proportion (36.33%), followed by C. granulosa (32.66%) and C. gingivalis (10%) [Table 1]. We found that the prevalence of these species was more in males in gingivitis and healthy individuals (42% and 49%, respectively), while the prevalence of these species was more in females in periodontitis group (40%), compared to males [Table 2] and the difference was statistically not significant. Out of 300 subjects, Capnocytophaga species were identified from 237 (79%) participants in all groups. Prevalence of individual Capnocytophaga species in these three groups was analyzed, which showed the prevalence of C. ochracea was higher than the other 2 species. C. ochracea was found to be 40% in healthy individuals, 32% in gingivitis and 37% in periodontitis. C. granulosa was found to be 39% in healthy individuals, 33% in gingivitis and 26% in periodontitis. C. gingivalis was found to be 8% in healthy individuals, 12% in gingivitis and 10% in periodontitis. There was no significant difference found between the species of Capnocytophaga in three groups [Figure 2].
Table 1.
Descriptive statistics with frequencies and averages (n=300) of study volunteers
| Frequencies | Frequency (%) |
|---|---|
| Group | |
| Healthy | 100 (100) |
| Gingivitis | 100 (100) |
| Chronic periodontitis | 100 (100) |
| Gender | |
| Male | 150 (50) |
| Female | 150 (50) |
| Prevalence of Capnocytophagaspecies (%) | |
| Capnocytophaga gingivalis | 30 (10) |
| Capnocytophagaochracea | 109 (36.33) |
| Capnocytophagagranulosa | 98 (32.66) |
| Capnocytophagasputigena | 0 (0) |
| Capnocytophagahemolytica | 0 (0) |
| Capnocytophagagenospecies (AHN8471) | 0 (0) |
| Capnocytophagaleadbetteri (AHN8855) | 0 (0) |
Figure 1.
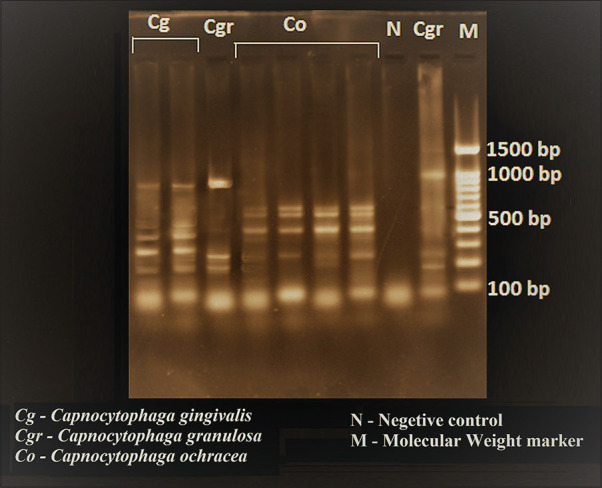
Photograph showing agarose gel electrophoresis with different banding patterns of Capnocytophaga species
Table 2.
Number of positive Capnocytophaga species in both genders in three groups
| Group | Number of samples | Gender | Total Capnocytophagaspecies isolated | Chi-square test (χ2, P) | |
|---|---|---|---|---|---|
| Male | Female | ||||
| Gingivitis | 100 | 42 | 35 | 77 | 3.148, 0.2072 (not significant) |
| Periodontitis | 100 | 33 | 40 | 73 | |
| Healthy individuals | 100 | 49 | 38 | 87 | |
Figure 2.
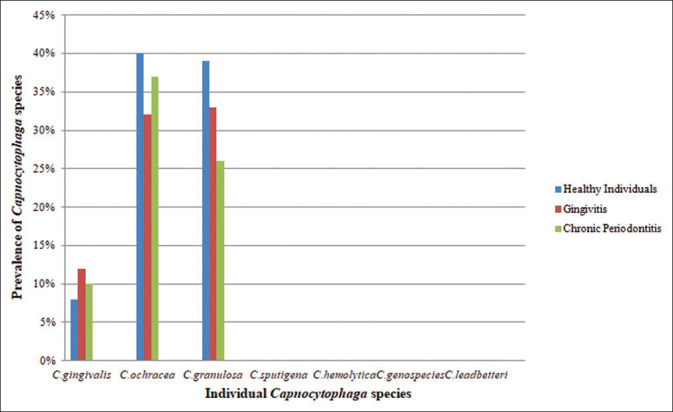
Graph showing the prevalence of Capnocytophaga species in healthy individuals, gingivitis and chronic periodontitis
The prevalence of Capnocytophaga species was also studied in different age groups from 18 to 55 years in all three groups. We found that Capnocytophaga species were identified more in the age group of 30–40 years in gingivitis and periodontitis that is 30 subjects in gingivitis and 21 individuals in periodontitis, while these species were identified more in age groups between 18 and 29 years in healthy individuals that is in 39 individuals [Table 3]. The value was statistically significant only within the age category of 18–29 years. For the age categories of 30–40 years and 41–55 years, the difference was statistically not significant.
Table 3.
Prevalence of Capnocytophagaspecies -in age category in three groups
| Age categorized (years) | Group | PCR | Total (%) | Chi-square test (χ2, P) | |
|---|---|---|---|---|---|
| Negative (%) | Positive (%) | ||||
| 18-29 | Gingivitis | 74 (74) | 26 (26) | 100 (100.00) | 7.6742, <0.0216 (significant) |
| Periodontitis | 78 (78) | 22 (22) | 100 (100.00) | ||
| Healthy individuals | 61 (61) | 39 (39) | 100 | ||
| Total | 213 (71) | 87 (29) | 300 (100.00) | ||
| 30-40 | Gingivitis | 70 (70) | 30 (30) | 100 (100.00) | 1.885, 0.3897 (not significant) |
| Periodontitis | 69 (69) | 31 (31) | 100 (100.00) | ||
| Healthy Individuals | 77 (77) | 23 (23) | 100 | ||
| Total | 216 (72) | 84 (28) | 300 (100.00) | ||
| 41-55 | Gingivitis | 79 (79) | 21 (21) | 100 (100.00) | 0.8159, 0.6650 (not significant) |
| Periodontitis | 80 (80) | 20 (20) | 100 (100.00) | ||
| Healthy individuals | 75 (75) | 25 (25) | 100 | ||
| Total | 234 (78) | 66 (22) | 300 (100.00) | ||
Interpretation: The prevalence of Capnocytophaga species was higher in healthy group when compared to gingivitis and periodontitis. However, the value was statistically significant only within the age categories of 18-29 years when compared by Chi-square test. For the age category of 30-40 and 41-55 years the difference was NOT statistically significant. PCR: Polymerase chain reaction
When the overall prevalence of these species was studied in 3 groups, it showed that these organisms were present in 87% in healthy individuals, 77% in gingivitis and 73% in periodontitis. This difference was statistically significant [Table 4].
Table 4.
Over all prevalence of Capnocytophaga species by polymerase chain reaction - restriction fragment length polymorphism in three groups
| Group | PCR-RFLP | Total (%) | Chi-square test | |
|---|---|---|---|---|
| Negative (%) | Positive (%) | |||
| Periodontitis | 27 (27) | 73 (73) | 100 (100.0) | 6.2692, 0.0435 (significant) |
| Healthy | 13 (13) | 87 (87) | 100 (100.0) | |
| Gingivitis | 23 (23) | 77 (77) | 100 (100.0) | |
| Total | 63 (21) | 237 (79) | 300 (100.0) | |
Interpretation: The healthy group had significantly high prevalence (87%) of Capnocytophagaspecies compared to chronic periodontitis (73%) and gingivitis (77%). The difference was statistically significant when compared by Chi-square test (P<0.05). PCR-RFLP: Polymerase chain reaction -restriction fragment length polymorphism
DISCUSSION
Standard cultural methods for identifying bacteria in human plaque samples are technically difficult because of the fastidious nature of bacteria.[7] It is now widely accepted that PCR technology provides a more sensitive means of detection for putative bacterial species as compared to conventional culture techniques. In the present study, an attempt was made to find out the prevalence of Capnocytophaga species by PCR.
300 participants, 100 each of Gingivitis, Chronic periodontitis and Healthy individuals between the age group of 18–55 years were included in the study. Among these participants, 150 (50%) were male and 150 (50%) were female. In this study, the overall prevalence of Capnocytophaga species was found to be 79%. Out of seven species, only C. gingivalis, C. ochracea and C. granulosa were detected by PCR. We could not detect C. sputigena, C. haemolytica, C. leadbetteri and C. genospecies in any of our study groups.
Capnocytophaga species are more commonly present in plaque than in saliva samples,[8] and they are established in oral cavities of children in their early years.[5] In this study, when the prevalence of these organisms was considered in different age groups from 18 to 55 years, it was found to be more in healthy individuals between the age groups 18–29 years, as compared to 30–40 years and 41–50 years. Some workers reported the prevalence of these organisms to be approximately 50% by PCR from saliva samples in children aged 2–10 years.[9] Some authors have shown the prevalence of these organisms to be more in healthy individuals between 2 and 12 years age.[5] Some have found Capnocytophaga species to be a major colonizer of the sulcus gingiva of children between the ages range of 3–10 years.[10] These findings, along with our study, suggest that these bacteria are commonly present in the oral cavity in the early years and are at risk of developing periodontitis in adolescence.
When the prevalence of Capnocytophaga species was considered in three study participants, it was seen that the prevalence was significantly higher in samples from healthy group as compared to those of gingivitis and periodontitis. Some studies found these species in higher proportions in healthy subjects than in sites affected with periodontitis,[5,11] which is similar to our study. Some workers have reported these species to be frequently isolated after periodontal therapy in adult periodontitis participants,[12] while some have found these species to be associated with juvenile periodontitis.[13,14] Thus the significance of Capnocytophaga species in the pathogenesis of the periodontal disease is controversial.
When the prevalence of individual species was considered, it was observed that C. ochracea was found to be in higher proportion (36.33%), followed by C. granulosa (32.66%) and C. gingivalis (10%) in our study. Based on PCR with species-specific primers, some have found the prevalence of C. ochracea to be 100% and 96% for C. gingivalis in periodontally healthy Japanese children.[5] Using the same technique, some authors have found C. ochracea to be approximately 50% in Japanese children with primary dentition.[15] Some workers have reported C. ochracea to be 20% in plaque samples of a healthy individual.[10] Some studies have reported C. gingivalis to be associated with adult gingivitis and C. ochracea to be in higher proportion in subgingival plaques of subjects with juvenile periodontitis.[14] Some reports have shown C. gingivalis to be more prevalent than C. ochracea in adult gingivitis.[11] Some studies have shown the prevalence of C. granulosa to be 51% from subgingival plaque samples.[16] These findings, together with our present ones, suggest that these three species (C. ochracea C. gingivalis C. granulosa), especially C. ochracea are commonly present in the oral cavity and may be associated with periodontal diseases.
We were not able to detect C. sputigena, C. haemolytica, C. leadbetteri and C. genospecies in our study. Some studies have shown the prevalence of C. sputigena from 48% to 50%,[5,15] while some have shown that C. sputigena was less or moderately prevalent.[5,11,14] Some have reported very few isolates of C. sputigena, which were identified by PCR-RFLP method.[4] Some authors were unable to detect C. sputigena by PCR.[10] We were not able to detect C. sputigena in our study. There appears to be considerable variation in the detection rate of C. sputigena from several studies. This variation may be due to racial and geographical differences.
C. haemolytica was originally isolated from supragingival plaque in adults and later recovered from subgingival plaque from adults.[7] Some workers have shown the prevalence of C. haemolytica to be 10%.[16] In this study, we did not get any isolate of C. haemolytica. May be due to the low prevalence of C. haemolytica we were not able to get a single isolate of C. haemolytica.
C. leadbetteri and C. genospecies were identified from the oral cavity of children of 2–3 years of age.[3] There are no reports of isolation of these two species from adults. This may be the reason for not getting a single isolate of these two species as our entire study population consisted of adults. We tried to find out the prevalence of these organisms in both the genders in all our study groups. We found that these species were more identified in males than in females from gingivitis and healthy individuals, while they were identified more in females than males from the periodontitis group. There are no reports in the literature where any authors have made an attempt to find the prevalence of these organisms in both the genders.
CONCLUSION
We conclude that Capnocytophaga species are commonly present in healthy individuals and may be associated with periodontal disease. There is a need for further study for to check for the prevalence of other species of Capnocytophaga in health and disease.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The Principal, Maratha Mandal's NGH Institute of Dental Sciences and Research Centre, Belagavi, for encouraging to conduct the study.
REFERENCES
- 1.Tsai CY, Tang CY, Tan TS, Chen KH, Liao KH, Liou ML. Subgingival microbiota in individuals with severe chronic periodontitis. J Microbiol Immunol Infect. 2018;51:226–34. doi: 10.1016/j.jmii.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Urbán E, Terhes G, Radnai M, Gorzó I, Nagy E. Detection of periodontopathogenic bacteria in pregnant women by traditional anaerobic culture method and by a commercial molecular genetic method. Anaerobe. 2010;16:283–8. doi: 10.1016/j.anaerobe.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Frandsen EV, Poulsen K, Könönen E, Kilian M. Diversity of Capnocytophaga species in children and description of Capnocytophaga leadbetteri sp nov and Capnocytophaga genospecies AHN8471. Int J Syst Evol Microbiol. 2008;58:324–36. doi: 10.1099/ijs.0.65373-0. [DOI] [PubMed] [Google Scholar]
- 4.Ciantar M, Newman HN, Wilson M, Spratt DA. Molecular identification of Capnocytophaga spp. via 16S rRNA PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 2005;43:1894–901. doi: 10.1128/JCM.43.4.1894-1901.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi F, Okada M, Zhong X, Miura K. PCR detection of Capnocytophaga species in dental plaque samples from children aged 2 to 12 years. Microbiol Immunol. 2001;45:17–22. doi: 10.1111/j.1348-0421.2001.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 6.Kugaji MS, Bhat KG, Joshi VM, Pujar M, Mavani PT. Simplified method of detection of Dialister invisus and Olsenella uli in oral cavity samples by polymerase chain reaction. J Adv Oral Res. 2017;8:47–52. [Google Scholar]
- 7.Yamamoto T, Kajiura S, Hirai Y, Watanabe T. Capnocytophaga haemolytica sp nov and Capnocytophaga granulosa sp nov, from human dental plaque. Int J Syst Bacteriol. 1994;44:324–9. doi: 10.1099/00207713-44-2-324. [DOI] [PubMed] [Google Scholar]
- 8.Frisken KW, Higgins T, Palmer JM. The incidence of periodontopathic microorganisms in young children. Oral Microbiol Immunol. 1990;5:43–5. doi: 10.1111/j.1399-302x.1990.tb00225.x. [DOI] [PubMed] [Google Scholar]
- 9.Amano A, Kishima T, Kimura S, Takiguchi M, Ooshima T, Hamada S, et al. Periodontopathic bacteria in children with Down syndrome. J Periodontol. 2000;71:249–55. doi: 10.1902/jop.2000.71.2.249. [DOI] [PubMed] [Google Scholar]
- 10.Conrads G, Mutters R, Fischer J, Brauner A, Lütticken R, Lampert F. PCR reaction and dot-blot hybridization to monitor the distribution of oral pathogens within plaque samples of periodontally healthy individuals. J Periodontol. 1996;67:994–1003. doi: 10.1902/jop.1996.67.10.994. [DOI] [PubMed] [Google Scholar]
- 11.Holdeman LV, Moore WE, Cato EP, Burmeister JA, Palcanis KG, Ranney RR. Distribution of capnocytophaga in periodontal microfloras. J Periodontal Res. 1985;20:475–83. doi: 10.1111/j.1600-0765.1985.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 12.Mombelli A, Nyman S, Brägger U, Wennström J, Lang NP. Clinical and microbiological changes associated with an altered subgingival environment induced by periodontal pocket reduction. J Clin Periodontol. 1995;22:780–7. doi: 10.1111/j.1600-051x.1995.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 13.Tanner A. Periodontology Today. Zurich: Karger Publishers; 1988. Is the specific plaque hypothesis still tenable? pp. 123–31. [Google Scholar]
- 14.Savitt ED, Socransky SS. Distribution of certain subgingival microbial species in selected periodontal conditions. J Periodontal Res. 1984;19:111–23. doi: 10.1111/j.1600-0765.1984.tb00800.x. [DOI] [PubMed] [Google Scholar]
- 15.Kimura S, Ooshima T, Takiguchi M, Sasaki Y, Amano A, Morisaki I, et al. Periodontopathic bacterial infection in childhood. J Periodontol. 2002;73:20–6. doi: 10.1902/jop.2002.73.1.20. [DOI] [PubMed] [Google Scholar]
- 16.Ciantar M, Spratt DA, Newman HN, Wilson M. Capnocytophaga granulosa and Capnocytophaga haemolytica: Novel species in subgingival plaque. J Clin Periodontol. 2001;28:701–5. doi: 10.1034/j.1600-051x.2001.028007701.x. [DOI] [PubMed] [Google Scholar]


