Abstract
The stability and activity of the p53 tumor suppressor protein is tightly regulated by various posttranslational modifications, including SUMOylation. p53 can be modified by both SUMO1 and SUMO2, although how SUMOylation regulates p53 activity is still obscure. Whether p53 activity is directly regulated by deSUMOylation is unclear. Here, we show that SENP1, a SUMO-specific protease implicated in prooncogenic roles, is a p53 deSUMOylating enzyme. SENP1 interatcs with p53 and deSUMOylates p53 in cells and in vitro. Knockdown of SENP1 markedly induced p53 transactivation activity. We further show that SENP1 depletion synergizes with DNA damage-inducing agent etoposide to induce p53 activation and the expression of p21, leading to synergistic growth inhibition of cancer cells. Our results reveal that SENP1 is a critical p53 deSUMOylating enzyme and a promising therapeutic target in wild-type p53 containing cancer cells.
Keywords: p53, SUMO, SUMOylation, SENP1, SUMO protease
INTRODUCTION
The tumor suppressor protein p53 is a critical transcription factor that activates or represses the expression of a myraid of target genes to induce cell cycle arrest, apoptosis, senescence, and other anti-proliferative outcomes, thereby executing its function in maintaining genomic integrity and preventing tumorigenesis (Devine & Dai, 2013; Kruiswijk et al, 2015; Vogelstein et al, 2000). Under normal conditions, p53 is maintained at low levels mainly by the oncoprotein MDM2. MDM2 is RING-finger-containing ubiquitin (Ub) ligase that mediates p53 ubiquitination and degradation through the proteasomal system (Haupt et al, 1997; Kubbutat et al, 1997). In response to diverse cellular stress, p53 is swiftly stabilized and activated via inhibiting MDM2, evoking p53 deubiquitination, and various additional posttranslational modifications of the MDM2-p53 pathway (Kruse & Gu, 2009; Sun & Dai, 2014).
p53 can also be modified by SUMOylation, a posttranslational modificaton of proteins by small ubiquitin-like modifiers (SUMOs). SUMOylation can interfere with protein-protein interactions (Moldovan et al, 2006) or compete with other lysine-directed modifications like acetylation or ubiquitination (Desterro et al, 1998), thereby regulating protein localization, trafficking, stability and activity (Gareau & Lima, 2010; Jentsch & Psakhye, 2013). p53 can be modified by SUMO1 (Gostissa et al, 1999; Ivanschitz et al, 2015; Kwek et al, 2001; Muller et al, 2000; Rodriguez et al, 1999) and SUMO2 (Li et al, 2006; Wu & Chiang, 2009b) at lysine (K) 386 in its C terminal regulatory region. p53 SUMOylation is promoted by stress such as DNA damage and oxidative stress (Kwek et al., 2001; Li et al., 2006; Rodriguez et al., 1999). While there is a general consensus that SUMOylation does not signicantly affect p53 stability, how SUMOylation regulates p53 protein activity remains controversial. Earlier studies showed that p53 modification by SUMO1 increases its transcriptional activity (Gostissa et al., 1999; Rodriguez et al., 1999), promotes apoptosis (Muller et al., 2000), and induces p53-depedent cell senescence (Bischof et al, 2006; Li et al., 2006). Relocalization of p53 into nuclear PML bodies by SUMO1 increases p53 transactivation activity (Fogal et al, 2000). A recent study showed that PML IV-ARF interaction enhances p53 SUMO1 conjugation and activates p53 to induce cell senescence (Ivanschitz et al., 2015). Consistently, SUMOylation also targets drosophila p53 to PML body and induces p53 transactivation activity (Mauri et al, 2008). In contrast, other studies indicate that SUMOylation either does not affect p53 localization and activity (Kwek et al., 2001) or reduces p53 transactivation activity by blocking p53 acetylation by p300 (Wu & Chiang, 2009a). Likewise, SUMOylation by PIAS1 promotes p53 nuclear export (Carter et al, 2007), thus indirectly modulating p53 activity. These discrepancies may lie on that p53 SUMOylation is transient and steady-state levels of p53 SUMOylation is low presumably due to SUMO protease activity in cells. Also, SUMOylation may modulate p53 activity at selected target gene promoters in cell and context-dependent manner.
SUMOylation can be reversed via deSUMOylation by a group of SUMO proteases, including SENP1-SENP3 and SENP5-SENP7, USPL1, DESI1, DESI2 (Hickey et al, 2012; Jentsch & Psakhye, 2013; Nayak & Muller, 2014). However, whether p53 is directly regulated by deSUMOylation is still unknown. Here we report that SENP1 deSUMOylates p53 and its knockdown markedly induced p53 transactivation activity and potentiated p53 activation and cell growth inhibition in resposne to DNA damage. Together, our results reveal that SENP1 acts as a p53 SUMO protease that limits p53 activity and thus could be a therapeutic target, whose inhihition activates p53 and synergizes with genotoxic drugs in killing cancer cells.
MATERIALS AND METHODS
Cell culture, plasmids and antibodies.
Human p53-proficient osteosarcoma U2OS and p53 deficient lung non-small cell carcinoma H1299 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin, and 0.1 mg/ml streptomycin at 37 °C in a 5% CO2 humidified atmosphere as described previously (He et al, 2016; Sun et al, 2012). Flag-SENP1, its catalytically-inactive C603S mutant, V5-SENP1, GST-SENP1 (WT and the C603S mutant), His-SUMO1, His-SUMO2 plasmids as well as the p53 expressing plasmids were previously described (Sun et al., 2012; Sun et al, 2018). Anti-p53 (Do-1, Santa Cruz Biotechnology), anti-MDM2 (SMP14, Santa Cruz Biotechnology), anti-p21 (Ab-11, NeoMarkers), anti-Flag (M2, Sigma), anti-V5 (Invitrogen), and anti-SENP1 (abcam) were purchased.
Transfection, immunoblot (IB) and co-immunoprecipitation (Co-IP) analyses.
Cells were transfected with plasmids using TransIT®-LT1 reagents (Mirus Bio Corporation) following the manufacturers’ protocol. Cells were harvested at 36–48 hours posttransfection and lysed in lysis buffer consisting of 50 mM Tris-HCl (pH 8.0), 0.5% Nonidet P-40, 1 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM DTT, 1 μg/ml pepstatin A, and 1 mM leupeptin. Equal amounts of cell lysate were used for IB analysis (Sun et al., 2018; Sun et al, 2015). Co-IP was conducted as described previously (Sun et al., 2018; Sun et al., 2015). Bound proteins were detected by IB using antibodies as indicated in figure legends.
In vivo SUMOylation assay.
In vivo SUMOylation assays were performed in cells using a Ni2+-NTA pulldown method as previously described (Sun et al., 2012; Sun et al., 2015). Brifely, cells were transfected with His-SUMO1 or His-SUMO2 and the plasmids indicated in each figure. The cells were harvested at 36–48 hours after transfection and 20% of the cells were directly lysed for IB and the remaining cells were subjected to Ni2+-NTA pulldown under denaturing conditions. After wash, the bead bound proteins were analyzed by IB.
In vitro deSUMOylation assay.
Recombinant GST-SENP1 and its C603S mutant proteins were expressed in E. coli and purified using GSH beads followed by glutathione elution. SUMOylated p53 was purified from H1299 cells transfected with Flag-p53 and His-SUMO1 or His-SUMO2 using anti-Flag affinity purification (Sun et al., 2018). The SUMOylated p53 was then incubated with 0.5 μM purified GST-SENP1 (wt or the C603S mutant) or control GST alone in deSUMOylation buffer consisting of 50 mM Tris-HCl (pH 7.5), 2 mM DTT at 37°C for 2 hours. The reactions were resolved in SDS-PAGE gel followed by IB.
Glutathione S-transferase (GST)-fusion protein association assays.
His-tagged p53 protein was purified from bacteria through a Ni2+-NTA (Qiagen) column and eluted with 0.25 M imidazole as previously described (Sun et al., 2015). GST-fusion protein-protein association assays were conducted as described (Sun et al., 2012; Sun et al., 2018). Briefly, purified His-p53 proteins (200 ng) were incubated with the glutathione-Sepharose 4B beads (Sigma) containing 200 ng of GST-SENP1 and GST alone, respectively, in a final volume of 50 μl of binding buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 100mM NaCl, 10% glycerol, 0.5 mg/ml BSA and 5 mM β-mercaptoethanol) for 45 minutes at room temperature with gentle agitation. The beads were then washed five times with 500 μl of the binding buffer and bound proteins were analyzed using IB.
RNA interference (RNAi).
The 21-nucleotide siRNA duplexes with a 3’ dTdT overhang were synthesized by Dharmacon Inc (Lafayette, CO). The target sequences for SENP1 are 5’-GGACCAGCTTTCGCTTTCT-3’ (siRNA-1), 5’-GTGAACCACAACTCCGTATTC-3’ (siRNA-2). The control scramble RNA sequence was described (Sun et al., 2012). Cells were transfected with these siRNA duplexes using SilentFect Lipid Reagent (Bio-Rad) following the manufacturer’s protocol or infected with shRNA-encoding lentiviruses as described (Sun et al., 2015). The cells were analyzed 48 hours after transfection or infection.
Reverse transcriptase-Quantitative polymerase chain reaction (RT-qPCR) analysis.
Total RNA was isolated from cells using Qiagen RNeasy Mini Kits (Qiagen, Valencia, CA). Reverse transcriptions were performed as described (Sun et al., 2012). Quantitative real-time PCR was performed on an ABI StepOne™ real-time PCR system (Applied Biosystems) using SYBR Green Mix (Bio-Rad) as described previously (Sun et al., 2012). All reactions were carried out in triplicate. Relative gene expression was calculated using the ΔCτ method following the manufacturer’s instruction. The primers for p21, mdm2, and GAPDH were described (Sun et al., 2012).
Chromatin Immunoprecipitation (ChIP)-qPCR.
ChIP analysis was performed essentially as described (Dai et al, 2007; Sun et al., 2015) using anti-p53 (DO-1) antibodies or control mouse IgG. Immunoprecipitated DNA fragments were analyzed for promoter occupancy by qPCR. The primers used for p21 promoter were 5’-GTGGCTCTGATTGGCTTTCTG-3’ and 5’-CTGAAAACAGGCAGCCCAAGG-3’. The primers for mdm2 promoter were 5’-GGTTGACTCAGCTTTTCCTCTTG-3’ and 5’-GGAAAATGCATGGTTTAAATAGCC-3’.
Cell viability assay.
U2OS cells were seeded in 96 well plates (1000 cells per well) followed by siRNA transfection and treatment with etoposide. Cell viability were measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays (Promega) following the manufacturer’s instructions. Cells were incubated with 0.5mg/ml MTT in medium for 3 hours. After incubation, MTT medium was removed and DMSO (100ul per well) was added for fully dissolving the purple formazan. The absorbance was measured at OD560nm and OD690nm. The reduced Abs (Abs560nm –Abs690nm) represents the relative number of viable cells per well.
Statistical analysis.
Standard two-tailed Student’s t-test was used to analyze statistical differences between two groups from at least three independent experiments. For comparison of multiple independent groups, one-way ANOVA (Analysis of Variance) with post-hoc Tukey HSD test was used for multiple comparisons between groups using R v3.6.1. p < 0.05 was considered statistically significant.
RESULTS
SENP1 depletion activates p53 and induces p21 levels.
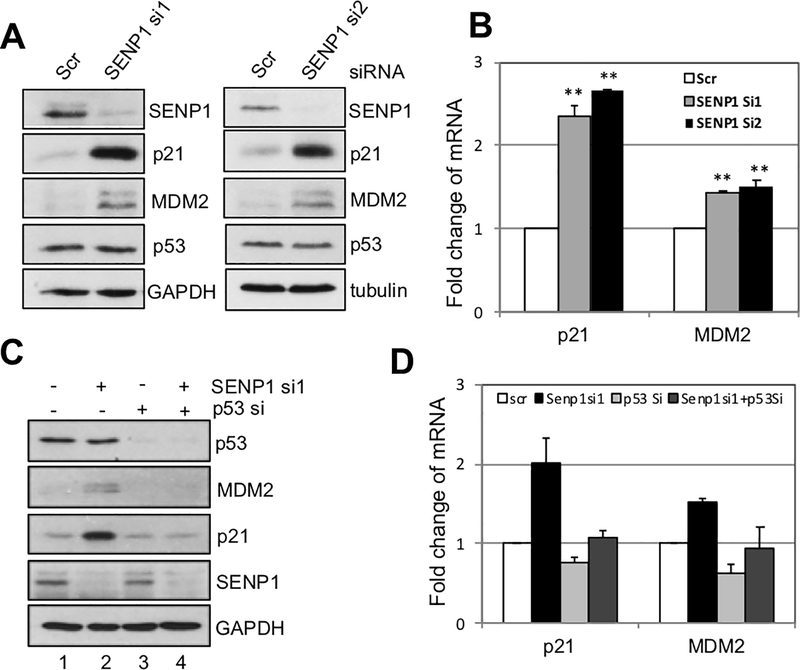
To understand whether SENP1 regulates p53 levels and activity, we first performed siRNA-mediated knockdown experiments. As shown in Fig. 1A, knockdown of SENP1 in U2OS cells using two different siRNAs markedy induced the levels of p21 and MDM2, two of the p53 target genes, whereas the levels of p53 protein were not significantly altered. Consistent with the p53 activation, RT-qPCR analysis showed that the levels of p21 and mdm2 mRNA were significantly induced by SENP1 knockdown (Fig. 1B). To test whether the induction of p21 and MDM2 is due to p53 activation, we co-depleted SENP1 and p53 in cells. As shown in Fig. 1C, knockdown of p53 completely abolished the induction of p21 and MDM2 proteins by SENP1 knockdown (compare lane 4 to lane 2). Again, this occurs at transcriptional levels as knockdown of p53 also abolished the induction of p21 and mdm2 mRNA levels by SENP1 depletion (Fig. 1D). These results demonstrate that depletion of SENP1 activates p53 and induces its target gene expression without affecting p53 levels.
Figure 1. Knockdown of SENP1 activates p53.
(A). Knockdown of SENP1 induces p53 activity, but not its levels. U2OS cells were transfected with scrambled (scr) and two individual SENP1 siRNA for 48 hours. Cell lysates were assayed for expression of SENP1, p53, p21, and MDM2 by IB. (B). Knockdown of endogenous SENP1 increases the mRNA expression of p53 targets p21 and mdm2. Total RNAs were extracted from U2OS cells transfected with siRNAs as in (A) and subjected to RT-qPCR assays.
Relative expression of p21 and mdm2 mRNA was normalized against the expression of GAPDH. **P<0.01, compared to scrambled RNA control. (C) (D). U2OS cells transfected with SENP1 siRNA and p53 siRNA alone or together were assayed by IB (C) and RT-qPCR (D) to detect the expression of p21 and MDM2 proteins and mRNA. IB, immunoblot; mRNA, messenger RNA; RT-qPCR, reverse transcriptase-quantitative polymerase chain reaction; siRNA, small interfering RNA.
SENP1 interacts with p53.
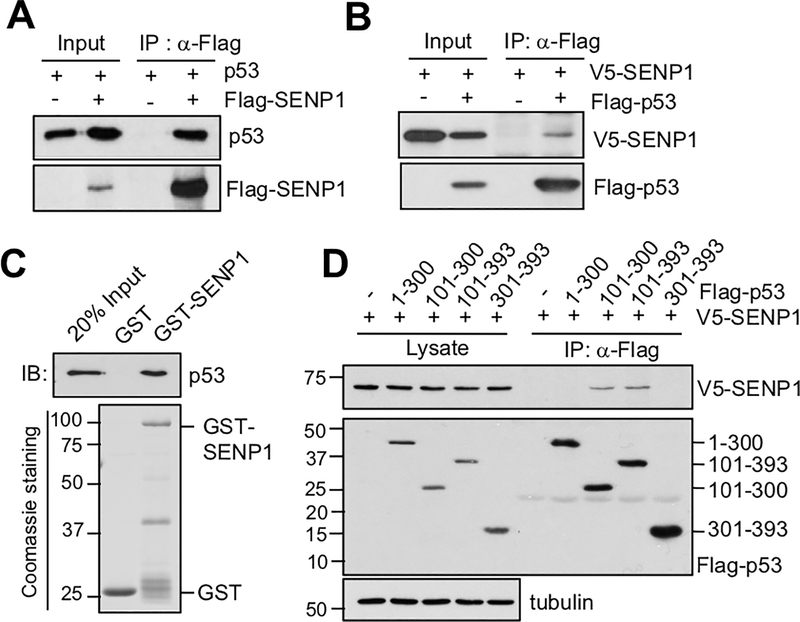
Next, we wanted to test whether SENP1 directly regulates p53 by physically interacting with p53. We performed co-immunoprecipitation (co-IP) assays in H1299 cells transfected with SENP1 and p53 alone or together. As shown in Fig. 2A, p53 was co-immunoprecipitated with Flag-SENP1 using anti-Flag antibody when both proteins are expressed. Simimlarly, V5-SENP1 was also co-immunoprecipitated with Flag-p53 using anti-Flag antibody when both proteins are expressed (Fig. 2B). To determine whether SENP1 directly binds to p53, we conducted GST-fusion protein-protein association assays. As shown in Figure 2C, purified His-p53 was bound by purified GST-SENP1 protein, but not GST alone. These results demonstrate that SENP1 directly binds to p53 in cells and in vitro. To map the domain through which SENP1 binds to p53, we performed co-IP assay in H1299 cells co-transfected with V5-SENP1 and Flag-tagged p53 deletion mutants using anti-Flag antibody. As shown in Fig. 2D, SENP1 binds to the central DNA-binding domain containing mutants (amino acids 101–300 and 101–393), but not the N-terminal transactivation domain (TAD) and the C-terminal domains. Therefore, SENP1 binds to p53 at its DNA-binding domain.
Figure 2. SENP1 interacts with p53.
(A) (B). SENP1 interacts with p53 in cells. H1299 cells transfected with p53 together with control pcDNA3-Flag vector or Flag-SENP1 (A) or with V5-SENP1 together with control Flag vector or Flag-p53 (B) were assayed by co-IP using anti-Flag antibody followed by IB. (C). SENP1 directly interacts with p53 in vitro. Purified GST or GST-SENP1 immobilized on glutathione beads was incubated with purified His-p53. Bound proteins were assayed by IB with anti-p53 antibody (top panel). Coomassie staining of GST and GST-SENP1 proteins are shown in the bottom panel. (D). SENP1 binds to the central DNA-binding domain of p53. H1299 cells were transfected with V5-SENP1 together with control Flag vector or Flag-tagged deletion mutants of p53 as indicated. The cell lysates were immunoprecipitated with anti-Flag antibodies followed by IB with anti-V5 or anti-Flag antibodies. Co-IP, co-immunoprecipitation; GST, glutathione S-transferase; IB, immunoblot
SENP1 deSUMOylates p53.
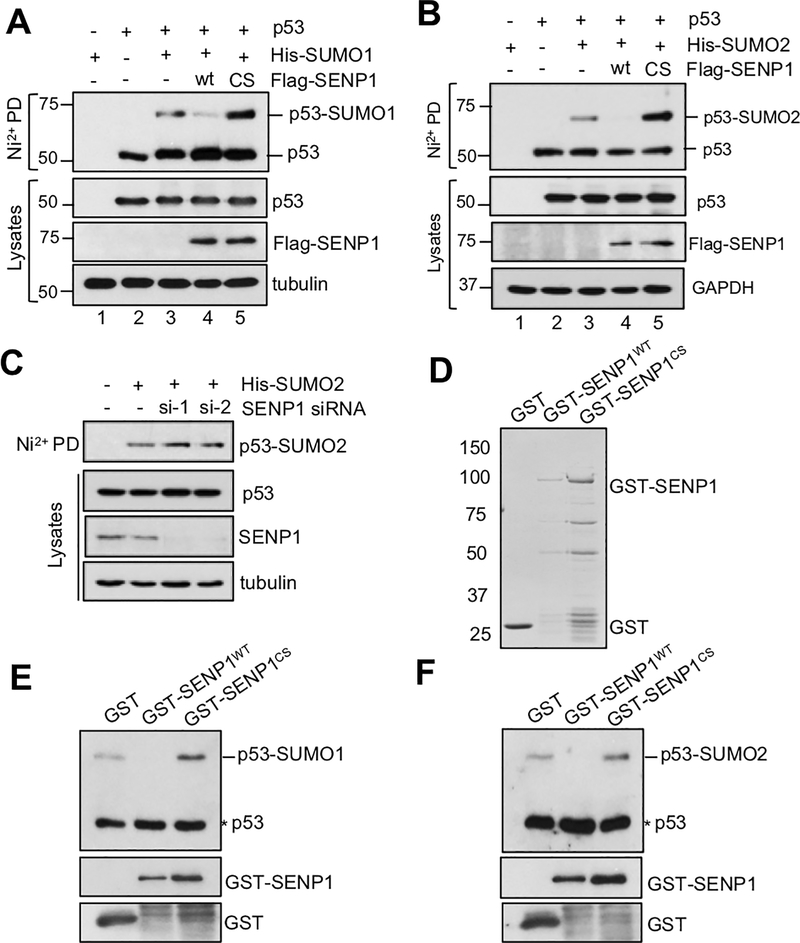
As SENP1 is a nuclear SUMO protease, we next tested whether it regulates p53 activity by deSUMOylating p53. H1299 cells transfected with p53, His-SUMO1 or His- SUMO2 together with wt SENP1 or its catalytically inactive C603S mutant were subjected to Ni2+-NTA bead pulldown under denaturing conditions. As shown in Fig. 3A and consistent with other reports (Gostissa et al., 1999; Ivanschitz et al., 2015; Kwek et al., 2001; Muller et al., 2000; Rodriguez et al., 1999), p53 is mono-SUMOylated by SUMO1 (lane 3). The p53 SUMOylation was markedly reduced when wild type SENP1 (lane 4), but not the C603S mutant (lane 5), was co-expressed. Interestingly, co-expression of the C603S mutant resulted in an increased level of SUMOylated p53 (lane 5), indicating a dominant-negative effect of the mutant on p53 SUMOylation. We confirmed that p53 is mainly SUMOylated at lysine (K) 386 (data not shown). Similarly, wt SENP1, but not the C603S mutant, also abolished p53 modification by SUMO2 (Fig. 3B). To understand whether endogenous SENP1 can regulate p53 SUMOylation, we performed siRNA-mediated knockdown of endogenous SENP1. As shown in Fig. 3C, knockdown of SENP1 by two different siRNAs increased the levels of p53 SUMOylation. Thus, SENP1 deSUMOylates p53 in cells. To examine wheather SENP1 directly deSUMOylates p53, in-vitro deSUMOylation assays were performed using purified GST-SENP1 (wt and the C603S mutant) or GST alone (Fig. 3D). SUMOylated p53 was purified from U2OS cells co-transfected with Flag-p53 and His-SUMO1 or His-SUNO2 using affinity purification with anti-Flag (M2) agarose beads followed by Flag-peptide elution (14). As shown in Figs. 3E and 3F, purified recombinant wt SENP1, but not the C603S mutant, efficiently removed SUMO1 and SUMO2 from the SUMOylated p53, respectively. Thus, SENP1 directly deSUMOylates p53 in vitro. Together, these results reveal that SENP1 is a novel p53 deSUMOylating enzyme.
Figure 3. SENP1 deSUMOylates p53 in cells and in vitro.
(A) (B). SENP1 deSUMOylates p53 in cells. H1299 cells were transfected with His-SUMO1 (A) or His-SUMO2 (B), p53 together with or without Flag-SENP1 (wild type or the C603S mutant) plasmids for 48 hours. The cells were subjected to pulldown (PD) using Ni2+-NTA bead under denaturing conditions, followed by IB. (C). Knockdown of SENP1 increases p53 SUMOylation. H1299 cells co-transfected with His-SUMO2 with scrambled or SENP1 siRNA. The cells were assayed by Ni2+-NTA PD under denaturing conditions followed by IB. (D). Coomassie staining of purified recombinant GST-SENP1 (wt or the C603S mutant) and GST alone protein. (E) (F). SENP1 deSUMOylates p53 in vitro. In vitro deSUMOylation assays were performed by incubating purified SUMOylated p53 with recombinant GST-SENP1 (wild type and the C603S mutant) or GST alone, followed by IB using anti-p53 to detect the levels of p53 SUMOylation. GST, glutathione S-transferase; IB, immunoblot; siRNA, small interfering RNA.
SENP1 depletion potentiates p53 activation in cells in response to DNA damage.
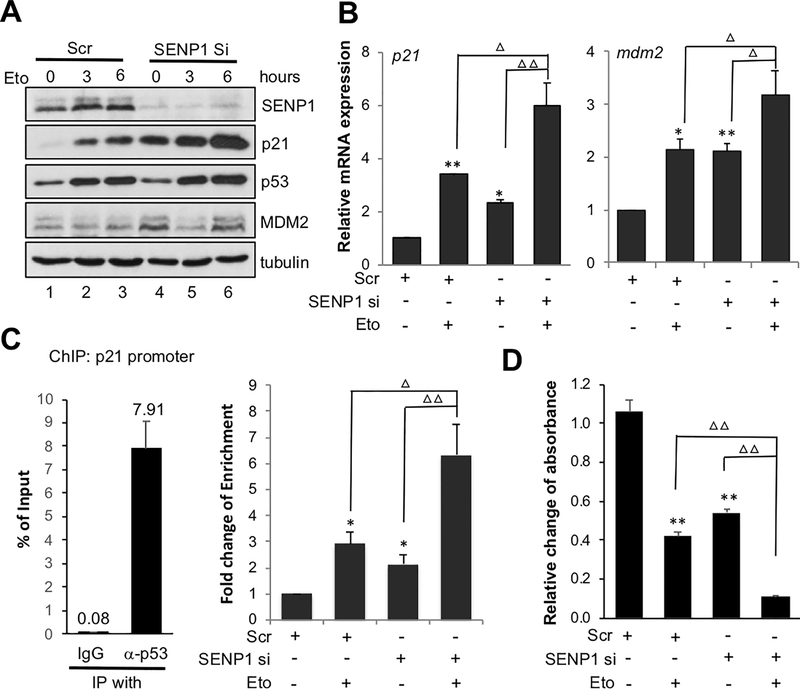
To understand whether SENP1 regulates p53 activation in response to genotoxic stress, we treated U2OS cells transfected with scrambled or SENP1 siRNA with control or DNA damage agent etoposide. Suprisingly, knockdown of SENP1 drastically increased the induction of p21 expression upon etoposide treatment (Fig. 4A, compare lane 5–6 to lanes 2–3). The expression of MDM2 was not further increased by etoposide treatment due to its destabilization upon DNA damge (Meulmeester et al, 2005). Consistently, the expression of p21 and mdm2 mRNA was synergistically increased in SENP1 knockdown cells treated with etoposide compared to SENP1 knockown and etoposide treatment alone (Fig. 4B). To test whether deSUMOylation of p53 by SENP1 affects p53 chromatin binding to target gene promoters, we performed ChIP-qPCR analysis using anti-p53 antibody and control IgG. As shown in Fig. 4C, p53 specifically binds the p21 gene promoter as shown by anti-p53 IP compared to control IgG (left panel). Either SENP1 knockdown or etoposide treament alone led to increased p53 binding to the p21 gene promoter. Treatment of SENP1 knockdown cells with etoposide further markedly increased the p53 binding to p21 promoter (right panel). This is consistent with the synergistic role of SENP1 knockdown and etoposide treatment in activating p53.
Figure 4. SENP1 depletion potentiates p53 activation and cell growth inhibition by genotoxic stress.
(A) (B). SENP1 knockdown synergizes with etoposide to induce p53 activity. U2OS cells transfected with scrambled or SENP1 siRNA for 48 hours and treated with 20 μM etoposide (Eto) for 3 and 6 hours. The cell lysates were assayed by IB to detect the expression of indicated proteins (A). The cells were also assayed by RT-qPCR (B) to detect the expression of p21 and mdm2 mRNA, normalized to the expression of GAPDH. (C). SENP1 depletion potentiates p53 promoter binding activity following genotoxic stress. U2OS cells transfected with scrambled or SENP1 siRNA for 48 hours and treated with 20 μM etoposide (Eto) for 6 hours. The cells were subjected to ChIP-qPCR assays using anti-p53 antibody or control mouse IgG. Enrichment of p53 on p21 promoter containing p53RE versus IgG in scrambled RNA transfected cells was shown on the left, with the average percentage of input from three independent experiments indicated on the top of each bar. The fold change of p53 enrichment on p21 promoter were then calculated as relative fold change compared to promoter enrichment of control scrambled RNA transfected cells (right). (D) SENP1 depletion potentiates cell growth inhibition by genotoxic agent. U2OS cells transfected with scrambled or SENP1 siRNA for 48 hours were treated with or without Eto for 12 hours. Cell viability was measured by MTT assays. *P<0.05, **P<0.01, compared to scrambled RNA control. ΔP<0.05, ΔΔP<0.01, compared to SENP1 siRNA or Eto treatment alone. ChIP, chromatin immunoprecipitation; IB, immunoblot; IgG, immunoglobulin G; mRNA, messenger RNA; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; RT-qPCR, reverse transcriptase-quantitative polymerase chain reaction; siRNA, small interfering RNA.
SENP1 depletion enhances cell growth inhibition by genotoxic agent.
Givent that SENP1 knockdown synergistically increases p53 activity induced by genotoxic agent etoposide in cells, we next tested whether knockdown of SENP1 could synergistically increase the inhibition of cell growth inhibition upon genotoxic agent. We performed cell viability assays in U2OS cells transfected with control or SENP1 siRNA for 48 hours followed by etoposide treatment. As shown in Fig. 4D, the combination of SENP1 knockdown and etoposide treatment markedly inhibited cell growth inhibition. These results suggest that inhibiting SENP1 may have potential in combinational therapy with chemotherapeutic agents in cancer, given that SENP1 is overexpressed in various human cancers.
DISCUSSION
SENP1 is a nuclear SUMO specific protease that deSUMOylates a variety of target proteins and plays critical roles in diverse cellular processes, including cell cycle, transcription, metabolism, DNA repair, immune response and hypoxia response, and is essential for animal development (Chen et al, 2013; Chen et al, 2019; Cheng et al, 2007; Cheng et al, 2005; Ji et al, 2007; Liu et al, 2017; Sun et al., 2018). Homozygous deletion of the SENP1 gene is embryonic lethal due to impaired erythropoiesis (Cheng et al., 2007). SENP1 deSUMOylation of HIF1α and MYC stabilizes both proteins and stimulates their activity via inhibiting ubiquitination-mediated proteasome degradation (Cheng et al., 2007; Sun et al., 2018). SENP1 also deSUMOylates and activates other proteins that promote cancer cell growth, migration and evasion such as c-JUN, PIN1 Gli1, etc (Chen et al., 2013; Chen et al., 2019; Cheng et al., 2007; Cheng et al., 2005; Ji et al., 2007; Liu et al., 2017; Sun et al., 2018). Collectively, SENP1 has emerged as a pro-oncogenic protein (Driscoll et al, 2010; Hoefer et al, 2012) and is overexpressed in many types of human cancers including prostate (Wang et al, 2013), breast (Chen et al., 2013; Sun et al., 2018), and thyroid cancers (Jacques et al, 2005). In this study, we show that SENP1 is a p53 deSUMOylating enzyme that deSUMOylates p53 in cells and in vitro. Consistent with previous studies supporting the role of SUMOylation in augmenting p53 transactivation activity, ablation of SENP1-mediated deSUMOylation markedly induces p53 activation without changing its levels. The p53 activation is well supported by p53-dependent induction of the downstream target gene p21. Thus, our finding adds a new mechanism by which SENP1 promotes cell proliferation by negatively regulating p53 activity.
Regulation of p53 by SUMO proteases has been reported by several previous studies. For example, repression of SENP1 was shown to induce p53-depedent cell senescence in normal human fibroblast cells (Yates et al, 2008). Likewise, SENP2 has been shown to reduce p53 levels via deSUMOylating MDM2 (Jiang et al, 2011) and hnRNP-K (Lee et al, 2012), a p53 co-activator in response to DNA damage, whereas SENP6 suppresses p53 activity by deSUMOylating and stabilizing TRIM28 (Li et al, 2018), which cooperates with MDM2 to promote p53 ubiquitination (Wang et al, 2005). However, whether p53 is directly regulated by SUMO proteases has not been reported. Our results here reveal that SENP1 is a direct regulator for p53 by deSUMOylating p53. Particularly, inhibiting SENP1-mediated deSUMOylation promotes p53 binding to target gene promoters (Fig. 4), indicating that SUMOylated p53 could access target DNA more efficiently and recruit co-activators to promote p53-mediated transcription. It is interesting to test whether SENP1 could regulate p53 activity at target gene promotes by deSUMOylating other p53 regulators and chromatin modifiers. Of note, SENP1 has been shown to possess specificity towards SUMO1 and is required for deSUMOylating SUMO1-modified proteins during mouse development (Sharma et al, 2013). Here we show that SENP1 also directly acts on SUMO2-modified p53, consistent with the previously reported SENP1 activity towards SUMO2/3-modifications (Hickey et al., 2012; Mendes et al, 2016).
Importantly, our results also indicate that SENP1 is a therapeutic target in cancer, as its knockdown synergistically enhances p53 activity, p21 induction and cell growth inhibition in cells treated with DNA damage agent etoposide (Fig. 4). As noted above, emerging evidence suggest that SENP1 has pro-oncogenic function by postively regulating various oncogenic pathways, such as HIF1α and MYC, via deSUMOylation. Thus, in addtion to the HIF1α and MYC pathways, inhibiting SENP1 has additional therapeutic role in wild-type p53 containing cancers by inducing p53-dependent cell growth inhibition. Future studies include testing the synergistic effeccts of SENP1 inhibition with additional chemotherapeutic drugs as well as identification of small molecule inhibitors to inhibit SENP1 activity to treat cancer.
ACKNOWLEDGEMENTS
We thank members in our laboratory for active discussion. M-S. D was supported by NIH grants R01 CA160474, R01 CA186241, and R01 GM130604.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA SHARING
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Bischof O, Schwamborn K, Martin N, Werner A, Sustmann C, Grosschedl R, Dejean A. (2006). The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol Cell 22: 783–794 [DOI] [PubMed] [Google Scholar]
- Carter S, Bischof O, Dejean A, Vousden KH. (2007) C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol 9: 428–435 [DOI] [PubMed] [Google Scholar]
- Chen CH, Chang CC, Lee TH, Luo M, Huang P, Liao PH, Wei S, Li FA, Chen RH, Zhou XZ et al. (2013) SENP1 deSUMOylates and regulates Pin1 protein activity and cellular function. Cancer Res 73: 3951–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sun XX, Sears RC, Dai MS. (2019) Writing and erasing MYC ubiquitination and SUMOylation. Genes Dis 6: 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kang X, Zhang S, Yeh ET. (2007) SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 131: 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Perkins ND, Yeh ET. (2005) Differential regulation of c-Jun-dependent transcription by SUMO-specific proteases. J Biol Chem 280: 14492–14498 [DOI] [PubMed] [Google Scholar]
- Dai MS, Arnold H, Sun XX, Sears R, Lu H. (2007) Inhibition of c-Myc activity by ribosomal protein L11. EMBO J 26: 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. (1998) SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell 2: 233–239 [DOI] [PubMed] [Google Scholar]
- Devine T, Dai MS. (2013) Targeting the ubiquitin-mediated proteasome degradation of p53 for cancer therapy. Curr Pharm Des 19: 3248–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll JJ, Pelluru D, Lefkimmiatis K, Fulciniti M, Prabhala RH, Greipp PR, Barlogie B, Tai YT, Anderson KC, Shaughnessy JD Jr. et al. (2010) The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood 115: 2827–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Del Sal G. (2000) Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J 19: 6185–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. (1999) Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J 18: 6462–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. (1997) Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- He X, Li Y, Dai MS, Sun XX. (2016) Ribosomal protein L4 is a novel regulator of the MDM2-p53 loop. Oncotarget 7: 16217–16226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CM, Wilson NR, Hochstrasser M. (2012) Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13: 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefer J, Schafer G, Klocker H, Erb HH, Mills IG, Hengst L, Puhr M, Culig Z. (2012) PIAS1 is increased in human prostate cancer and enhances proliferation through inhibition of p21. Am J Pathol 180: 2097–2107 [DOI] [PubMed] [Google Scholar]
- Ivanschitz L, Takahashi Y, Jollivet F, Ayrault O, Le Bras M, de The H. (2015) PML IV/ARF interaction enhances p53 SUMO-1 conjugation, activation, and senescence. Proc Natl Acad Sci U S A 112: 14278–14283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques C, Baris O, Prunier-Mirebeau D, Savagner F, Rodien P, Rohmer V, Franc B, Guyetant S, Malthiery Y, Reynier P. (2005) Two-step differential expression analysis reveals a new set of genes involved in thyroid oncocytic tumors. J Clin Endocrinol Metab 90: 2314–2320 [DOI] [PubMed] [Google Scholar]
- Jentsch S, Psakhye I. (2013) Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet 47: 167–186 [DOI] [PubMed] [Google Scholar]
- Ji Z, Degerny C, Vintonenko N, Deheuninck J, Foveau B, Leroy C, Coll J, Tulasne D, Baert JL, Fafeur V. (2007) Regulation of the Ets-1 transcription factor by sumoylation and ubiquitinylation. Oncogene 26: 395–406 [DOI] [PubMed] [Google Scholar]
- Jiang M, Chiu SY, Hsu W. (2011) SUMO-specific protease 2 in Mdm2-mediated regulation of p53. Cell Death Differ 18: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk F, Labuschagne CF, Vousden KH. (2015) p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol 16: 393–405 [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. (2009) Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. (1997) Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Kwek SS, Derry J, Tyner AL, Shen Z, Gudkov AV. (2001) Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene 20: 2587–2599 [DOI] [PubMed] [Google Scholar]
- Lee SW, Lee MH, Park JH, Kang SH, Yoo HM, Ka SH, Oh YM, Jeon YJ, Chung CH. (2012) SUMOylation of hnRNP-K is required for p53-mediated cell-cycle arrest in response to DNA damage. EMBO J 31: 4441–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lu D, Dou H, Liu H, Weaver K, Wang W, Li J, Yeh ETH, Williams BO, Zheng L et al. (2018) Desumoylase SENP6 maintains osteochondroprogenitor homeostasis by suppressing the p53 pathway. Nat Commun 9: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Santockyte R, Shen RF, Tekle E, Wang G, Yang DC, Chock PB. (2006) Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. J Biol Chem 281: 36221–36227 [DOI] [PubMed] [Google Scholar]
- Liu H, Yan S, Ding J, Yu TT, Cheng SY. (2017) DeSUMOylation of Gli1 by SENP1 Attenuates Sonic Hedgehog Signaling. Mol Cell Biol 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri F, McNamee LM, Lunardi A, Chiacchiera F, Del Sal G, Brodsky MH, Collavin L. (2008) Modification of Drosophila p53 by SUMO modulates its transactivation and pro-apoptotic functions. J Biol Chem 283: 20848–20856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes AV, Grou CP, Azevedo JE, Pinto MP. (2016) Evaluation of the activity and substrate specificity of the human SENP family of SUMO proteases. Biochim Biophys Acta 1863: 139–147 [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. (2005) Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell 18: 565–576 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. (2006) PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell 23: 723–732 [DOI] [PubMed] [Google Scholar]
- Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. (2000) c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem 275: 13321–13329 [DOI] [PubMed] [Google Scholar]
- Nayak A, Muller S. (2014) SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol 15: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT. (1999) SUMO-1 modification activates the transcriptional response of p53. EMBO J 18: 6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Yamada S, Lualdi M, Dasso M, Kuehn MR. (2013) Senp1 is essential for desumoylating Sumo1-modified proteins but dispensable for Sumo2 and Sumo3 deconjugation in the mouse embryo. Cell Rep 3: 1640–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Challagundla KB, Dai MS. (2012) Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J 31: 576–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Chen Y, Su Y, Wang X, Chauhan KM, Liang J, Daniel CJ, Sears RC, Dai MS. (2018) SUMO protease SENP1 deSUMOylates and stabilizes c-Myc. Proc Natl Acad Sci U S A 115: 10983–10988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Dai MS. (2014) Deubiquitinating enzyme regulation of the p53 pathway: A lesson from Otub1. World J Biol Chem 5: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS. (2015) The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci U S A 112: 3734–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. (2000) Surfing the p53 network. Nature 408: 307–310 [DOI] [PubMed] [Google Scholar]
- Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ 3rd, Chen J. (2005) MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J 24: 3279–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Xia N, Li T, Xu Y, Zou Y, Zuo Y, Fan Q, Bawa-Khalfe T, Yeh ET, Cheng J. (2013) SUMO-specific protease 1 promotes prostate cancer progression and metastasis. Oncogene 32: 2493–2498 [DOI] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. (2009a) Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J 28: 1246–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. (2009b) p53 sumoylation: mechanistic insights from reconstitution studies. Epigenetics 4: 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates KE, Korbel GA, Shtutman M, Roninson IB, DiMaio D. (2008) Repression of the SUMO-specific protease Senp1 induces p53-dependent premature senescence in normal human fibroblasts. Aging Cell 7: 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]