Abstract
SARS-CoV-2 main protease (Mpro) cleaves the viral polypeptide 1a and 1ab in a site-specific ((L-Q|(S, A, G)) manner and produce functional enzymes for mediating viral replication. Numerous studies have reported synthetic competitive inhibitors against this target enzyme but increase in substrate concentration often reduces the effectiveness of such inhibitors. Allosteric inhibition by natural compound can provide safe and effective treatment by alleviating this limitation. Present study deals with in silico allosteric inhibition analysis of quercetin, against SARS-CoV-2-Mpro. Molecular docking of quercetin with Mpro revealed consistent binding of quercetin at a site other than active site in multiple runs, with the highest binding energy of − 8.31 kcal/mol, forming 6 H-bonds with residues Gln127, Cys128, Lys137, Asp289 and Glu290. Molecular dynamic simulation of 50 ns revealed high stability of Mpro-quercetin complex with RMSD values ranging from 0.1 to 0.25 nm. Moreover, native-Mpro and Mpro-quercetin complex conformations extracted at different time points from simulation trajectories were subjected to active site-specific docking with modelled substrate peptide (AVLQSGFR) by ZDOCK server. Results displayed site-specific cleavage of peptide when docked with native-Mpro. While substrate peptide remained intact when docked with Mpro-quercetin complex, also the binding energy of peptide reduced from 785 to 86 from 1 to 50 ns as quercetin induced alterations in the active site cavity reducing its affinity for the substrate. Further, no interactions were noticed between peptide and active site residues of Mpro-quercetin complex conformations at 40 and 50 ns. Hence, quercetin displayed effective allosteric inhibition potential against SARS-CoV-2 Mpro, and can be developed into an efficient treatment for COVID-19.
Keywords: Allosteric inhibition, Quercetin, SARS-CoV-2 main protease (Mpro), Molecular docking, Molecular dynamic simulation
Introduction
From December 2019, when SARS-CoV-2 first introduced its devastating effect to human race in China, to till date pandemic situation of COVID-19, around 1.6 million deaths and more than 72 million confirmed cases have been reported by World Health Organisation (WHO) at a global level (Coronavirus disease (COVID-19) pandemic World Health Organisation accessed on 18 December 2020). Though the current stats and highly contagious nature of the disease convey urgent need for an incredibly potent treatment, vaccines or synthetic antiviral agent developed till date have been reported to have many side effects.
Severe acute respiratory syndrome corona virus-2 (SARS-CoV-2), the novel RNA virus and the etiological agent of COVID-19 possesses about 82% identity with the sequence of SARS-CoV (Severe acute respiratory syndrome coronavirus), which victimised thousands of lives in 2003, and 50% sequence identity with Middle-east respiratory syndrome corona virus (MERS-CoV) (Ma et al. 2020). This single-stranded, positive (+) sense, enveloped RNA virus belongs to genus beta-coronavirus and family coronaviridae. The genome of SARS-CoV-2 is the largest among RNA viruses and comprises around 26–32 kilo bases (Chen et al. 2020). The large genome codes for many non-structural and structural proteins but the mutation prone nature of viral genome makes it difficult to decide the target for the development of therapies. Till date, around 116 mutations have been reported in SARS-CoV-2 (Khailany et al. 2020). The structural proteins such as the spike, nucleocapsid, membrane and glycoproteins are known to be more susceptible to mutations as compared to non-structural proteins (Wu and Yan 2005; McBride et al. 2014; Ortega et al. 2020). Thus, non-structural protein-like main protease or 3-chymo-trypsin-like protease, RNA-dependent RNA polymerase, helicase and accessary proteins can be of immense importance for therapy development. Targeting conserved regions in these non-structural counterparts can in turn increase the effectiveness of the treatment (Wu et al. 2020).
Main protease (Mpro) of SARS-CoV-2, a 33.8 kDa enzyme, plays a vital role in the cleavage of viral polyproteins (pp1a and pp1ab) and results in the release of functional replicase enzyme essential for transcription and replication of virus (Ma et al. 2020; Chen et al. 2020; Zhang et al. 2020). By autolytic cleavage, Mpro first cleaves itself from the polyproteins and subsequently by catalytic cleavage at about 11 highly conserved sites, it cleaves off the other essential proteins. Mpro has an exclusive substrate preference for amino acid Q (glutamine) at site P1 (L-Q|(S, A, G)) of polyproteins, and such substrate specificity has not yet been reported for any human protease. (Ma et al. 2020; Chen et al. 2020; Zhang et al. 2020; Zhu et al. 2011). Thus, the extremely essential contribution of Mpro in viral replication along with lack of homologous counterparts in the human host, makes it evident that Mpro can be a highly effective target for the development of enzyme inhibitor based therapeutics.
Several researchers have investigated and developed competitive inhibitors against SARS-CoV-2 Mpro which mechanistically inhibit the enzyme, when present in a considerable amount, by competing with the substrate polypeptide to occupy the enzyme’s active site. Comparatively high concentration or exposure of the substrate to enzyme may limit the efficiency of competitive inhibitors (Ma et al. 2020; Zhang et al. 2020; Zhu et al. 2011; Jin et al. 2020a, b; Mengist et al. 2020). In contrary, allosteric inhibitors do not compete with the substrate; thus, the high concentration of substrate does not affect their efficiency. They bind at a different site other than the active site and pose inhibition by passing perturbations at the active site which result in conformational changes in active site. These modulations reduce the affinity of the active site for the substrate (Liu et al. 2008; Strelow et al. 2012; Wenthur et al. 2014). Recent researches have reported very high sensitivity (~ 1900%) of SARS-CoV-2 Mpro in transmittance of structural alterations across the enzyme via long-range connections, compared to SARS-CoV Mpro. Such a high tendency of the SARS-CoV-2 Mpro enzyme to convey any perturbation occurring at one site to other seems to be promising and encourage investigation of potential allosteric inhibitors (Estradaa 2020; Doshi et al. 2016; Negre et al. 2018).
Quercetin, a natural flavonol, has been reported for its antiviral effect against a wide variety of influenza virus strains and provide inhibition at an early infection stage (Wu et al. 2015). It also possesses antioxidant, anti-inflammatory, anti-cancer, anti-bacterial and anti-proliferative effects and thus contributes to the treatment of numerous diseases (Anand David et al. 2016; Batiha et al. 2020). Fruits (such as apple, raspberries, cherries, and red grapes), leafy green vegetables and some herbs are major natural sources of quercetin and can be easily exploited for its extraction. Being a natural compound, it shows least toxicity in vivo compared to the till date known synthetic antiviral compounds (Panche et al. 2016; Salehi et al. 2020).
In the present study, we analysed the effective allosteric inhibition of SARS-CoV-2 Mpro by a natural well-established nutraceutical compound, quercetin. Molecular docking and molecular dynamic (MD) simulations were performed to evaluate the structural binding affinity of quercetin for Mpro. Allosteric inhibitory effect of quercetin was revealed by analysing the binding affinity of Mpro and Mpro-quercetin complexes (conformations extracted at different time points from MD simulation trajectory), for the short essential peptide sequence (AVLQSGFR containing the cleavage site of Mpro) of polypeptide 1a and 1ab at the enzyme’s active site.
Methodology
Molecular docking of SARS-CoV-2 Mpro with quercetin
The 3D coordinate structure file of SARS-CoV-2 Mpro (PDB ID: 6Y84) was procured form RCSB Protein Data Bank. The 3D coordinates of quercetin were generated using online smile translator tool (https://cactus.nci.nih.gov/translate/). AutoDockTools-1.5.6 (http://autodock.scripps.edu/resources/adt) developed by “The Scripps Research Institute” was employed for preparing Mpro protein structure file and performing blind docking analysis. The water molecules were deleted from the protein structure, all atoms were assigned to AD4 type, Kollman charge of − 135.203 was added, polar hydrogens were added and non-polar hydrogen were merged. Molecular docking of Mpro with quercetin was performed using Lamarckian Genetic Algorithm parameters: number of GA runs 10, population size 150, max number of evaluations 2,500,000, max number of generations 27,000, gene mutation rate 0.02 and crossover rate of 0.8. The lowest binding energy conformation was considered best for further analysis. The docking complex conformation and 2D interactions were studied using system-based freeware Pymol (https://pymol.org/2/) and Discovery studio 2019 software (BIOVIA, Dassault Systèmes, Discovery Studio Modeling Environment, Release 2019, San Diego: Dassault Systèmes, 2019).
Molecular dynamic simulation
The best conformation of Mpro-quercetin docked complex having the lowest binding energy was considered for MD simulation study using GROMACS 2020 (http://www.gromacs.org/) (Pronk et al. 2013; Abraham et al. 2015). MD simulations of Mpro-quercetin complex, Mpro and quercetin were performed individually for 50 ns each at a temperature of 300 K. The topology of quercetin was prepared using online server CgenFF (https://cgenff.umaryland.edu/) and the topology of protein was made using GROMACS (Vanommeslaeghe et al. 2010). The CHARMM36 force field was used for all three simulations. Solvation was performed in a rhombic dodecahedron box using TIP3P three-point water model followed by ions addition to neutralise the system. Energy minimization was performed using steepest descent minimization algorithm for 50,000 steps. NVT and NPT equilibration simulations were done for 100 ps with position restrain at 300 K. Finally, equilibrated system was subjected to MD production run of 50000 ps at pressure and temperature of 1 bar and 300 K, respectively. During MD production run, trajectory coordinates were recorded at every 0.02 ps. The root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), and number of hydrogen bonds (h-bond) formed with respect to time were evaluated and plots were prepared using Origin software [version 2020 (https://www.originlab.com)]. The MD simulation trajectories were analysed by Visual Molecular Dynamic (VMD) software (https://www.ks.uiuc.edu/Research/vmd/) developed by University of Illinois at Urbana-Champaign (Humphrey et al. 1996). The 3D coordinates at every 100 ps were extracted from the trajectory files into a single PDB file for further evaluation by Pymol freeware (https://pymol.org/2/). The 3D images of Mpro-quercetin complex were generated at different time points using Pymol software. Eventually, individual 3D coordinates depicting Mpro-quercetin complex conformations and Mpro conformations at different time points (1 ns, 10 ns, 20 ns, 30 ns, 40 ns and 50 ns) during MD run were extracted from the full 3D coordinates PDB files made from simulation trajectories for further docking analysis to study allosteric inhibition effect of quercetin.
Modelling of the substrate peptide
The substrate peptide (AVLQSGFR), which is a part of substrate polypeptide 1a and 1ab and contains the cleavage site of SARS-CoV-2 Mpro, was modelled using PEPFOLD3 online tool (https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/) that works on Hidden Markov Model and was energy minimised using GROMACS minimizer (https://mobyle.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py#forms::Gromacs_Minimizer) associated with the server (Lamiable et al. 2016; Lindhal et al. 2001).
Allosteric inhibition analysis
To study the allosteric inhibition effect of quercetin against SARS-CoV-2 Mpro, the binding affinities of SARS-CoV-2 Mpro conformations for its substrate peptide at its active site were analysed when quercetin was bound to it at an independent site (in Mpro) other than the active site. The binding affinity of Mpro conformations in absence of quercetin were also analysed for reference. The 3D coordinate structures of SARS-CoV-2 Mpro and SARS-CoV-2 Mpro-quercetin complex conformations were extracted form MD simulation trajectories at 1 ns, 10 ns, 20 ns, 30 ns, 40 ns and 50 ns and the reference 3D structure of SARS-CoV-2 Mpro (PDB ID: 6Y84) was procured from RCSB Protein Data Bank. These structures were docked with the modelled peptide (AVLQSGFR) structure using ZDOCK online server (Pierce et al. 2011, 2014; Vreven et al. 2011). The active site residues (His41, Cys145, Asp187, Arg188, Gln189 and Thr190) were selected to assure specific binding of the substrate peptide at the enzyme’s active site. The ZDOCK score of the best docking cluster was considered in each docking performed for the evaluation of binding affinity of Mpro (either in complexed conformation with quercetin or in isolation) for its substrate peptide. The binding interactions were analysed using Pymol freeware (https://pymol.org/2/). The ZDOCK scores and the docking interactions of Mpro and Mpro-quercetin complex structures for the substrate peptide were recorded and interaction images were generated using Pymol software. The changes in structure, interactions and ZDOCK scores were compared between Mpro-peptide complex and Mpro-quercetin-peptide complex conformations.
Results and discussion
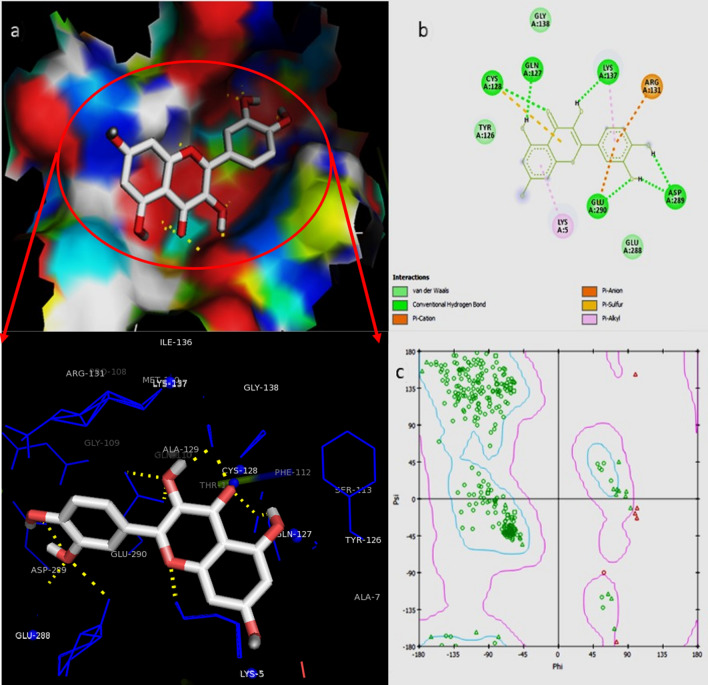
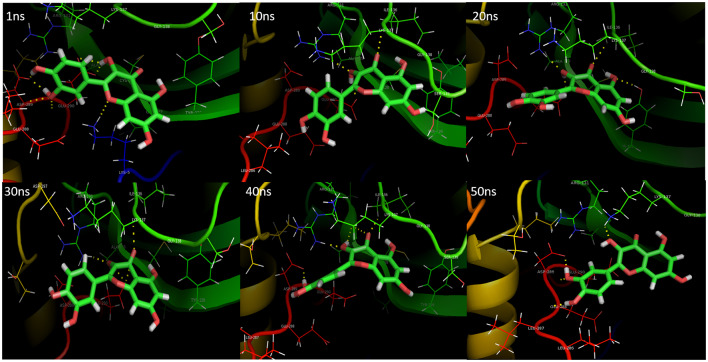
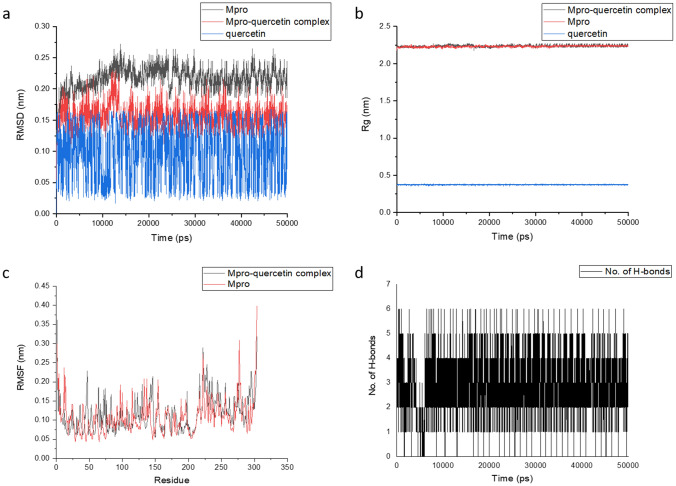
The present study deals with the in-depth analysis of allosteric inhibition effect of quercetin against the SARS-CoV-2 Mpro enzyme for the development of a natural, non-toxic, high potential inhibitor. Molecular docking of SARS-CoV-2 Mpro with quercetin resulted in binding of quercetin at an independent site other than the active site in majority of docking runs proving high binding affinity of quercetin for the site. The best docking conformation resulted in binding energy of − 8.31 kcal/mol and Ki value of 810.43 nM. The 3D binding cavity of quercetin in Mpro and the 2D interactions revealed effective interaction of quercetin resulting in formation of 6 hydrogen bonds with the conserved residues Gln127, Cys128, Lys137, Asp289 and Glu290 of Mpro protein (Fig. 1). Sencanski et al., in their recent in silico study on SARS-CoV-2 Mpro inhibitors predicted allosteric site comprising residues Arg4, Lys5, Arg131, Lys137, Trp207, Asp289, Leu287, Glu288, Gln127, Asp197, Gly138, Glu290, Tyr126, Phe291, and Leu286 which is same as depicted for Mpro-quercetin complex in Fig. 1. They reported Raltegravir, Rolitetracycline, Tolvaptan, Ciclesonide and Rescinnamine as high potential allosteric inhibitors with binding energies ranging from − 7.9 to − 7.2 kcal/mol which proves high binding potential of quercetin to the allosteric site of Mpro (Sencanski et al. 2020). El-Baba et al., in their mass spectrometry based study for the investigation of allosteric inhibitor of Mpro revealed binding of potential allosteric inhibitor x1187 at the binding site comprising residues Phe8, Met6, Gln127, Ser139, Asp295, Arg298, and Gln299. Gln127 being common to the mentioned studies and to our analysis may be considered as effective residue for allosteric inhibition thus depicting the accuracy of allosteric site selection in the present study (El-Baba et al. 2020). Iftikhar et al. in their in-silico study on inhibitors against three potential enzyme targets of SARS-CoV-2 reported Bagrosin as an allosteric inhibitor, binding with an energy of − 7.9 kcal/mol at an allosteric site lying close to the site reported in the present study, of SARS-CoV-2 proteinase with Lys5 as common interacting residue (Iftikhar et al. 2020). The Ramachandran plot of the docked Mpro-quercetin complex in turn displayed high stability of the Mpro-quercetin complex as most of the residues lie in favourable regions (Fig. 1). MD simulations each of 50 ns were performed for the best docked SARS-CoV-2 Mpro-quercetin complex (protein–ligand complex), SARS-CoV-2 Mpro (protein) and quercetin (ligand) using GROMACS software (Pronk et al. 2013; Abraham et al. 2015). The MD simulation trajectory of SARS-CoV-2 Mpro-quercetin complex was analysed and interaction images at different time points are presented in Fig. 2. The RMSD, RMSF, Rg and hydrogen bonds plots were generated. The RMSD values of both SARS-CoV-2 Mpro and SARS-CoV-2 Mpro-quercetin complex fluctuated within a range of 0.15 nm with respect to time thus displayed high stability of protein–ligand complex during the simulation (Fig. 3a) (Kato et al. 2017). The variations in Rg of the Mpro-quercetin complex and Mpro alone, during simulation period showed similar fashion (Fig. 3b). Very low variation in Rg values with respect to time revealed consistent compactness of the protein during simulation even when present in the complexed form with quercetin, hence proved high stability of the complex (Lobanov et al. 2008). The root mean square fluctuations calculated for the protein backbone of Mpro-quercetin complex were found to be similar to that of the reference Mpro protein during simulation thus provide evidence that the integrity of protein backbone was maintained and least damaging displacements in backbone were encountered by the protein when complexed with quercetin (Fig. 3c). Along with this, the high amplitude of fluctuations at the terminal residues were due to the high degree of freedom of these residues compared to that of the intermediate region of the protein chain (Martínez 2015). Further, the fluctuations in residues of loop regions were higher compared to other forms of secondary structures. The number of hydrogen bonds formed between quercetin and Mpro protein in complex form during the simulation was plotted which proved effective interaction of quercetin with the Mpro protein during the time period of simulation (Fig. 3d). The higher number of hydrogen bond display significant interaction between the quercetin and Mpro during the simulation. All these results proved highly stable binding of quercetin with SARS-CoV-2 Mpro, thus providing great scope for its potential to modulate the structural conformation of Mpro to a greater extent.
Fig. 1.
a 3D conformation views showing interacting residues of binding site. b 2D interactions and interacting residues and c Ramachandran plot of quercetin-SARS-CoV-2 main protease docked complex
Fig. 2.
MD simulation trajectory conformations of quercetin-SARS-CoV-2 main protease complex at different time points displaying interacting residues
Fig. 3.
Plot of a RMSD, b Rg, c RMSF, and d Number of hydrogen bonds for MD simulations of SARS-CoV-2 main protease protein, Mpro-quercetin complex and quercetin
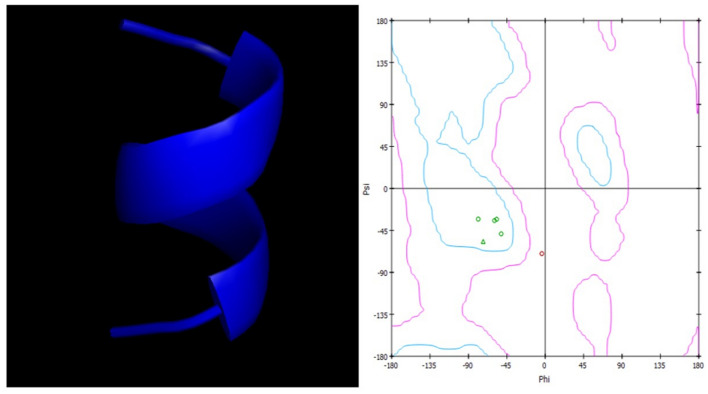
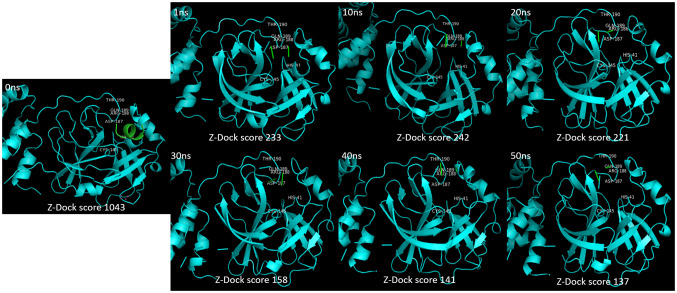
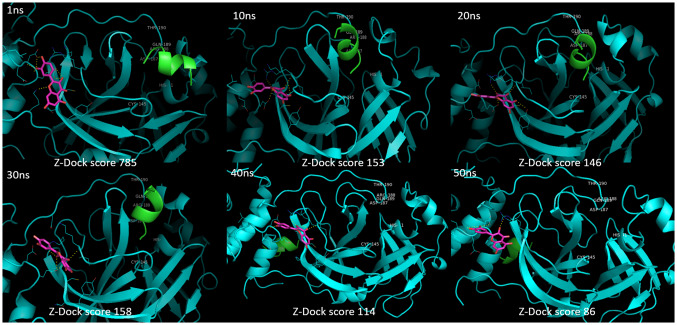
To study the allosteric inhibitory effect of quercetin against Mpro, the substrate peptide (AVLQSGFR) sequence containing the cleavage site of Mpro was modelled and Ramachandran plot was generated to check the stability of structure (Fig. 4). The structures of SARS-CoV-2 Mpro and SARS-CoV-2 Mpro-quercetin complex achieved different conformations at different time points during MD simulation. The structural conformations were extracted from MD simulation trajectory at 1 ns, 10 ns, 20 ns, 30 ns, 40 ns and 50 ns. Molecular docking of these conformations with the modelled substrate peptide at the active site of Mpro was performed. The docking analysis of Mpro conformations with substrate peptide at enzyme’s active site showed insignificant conformational change in active site at different time points (Fig. 5). In turn, cleavage of peptide was noticed when docked with 1 ns, 10 ns, 20 ns, 30 ns, 40 ns and 50 ns conformations proving significant activity of the enzyme in absence of quercetin. The ZDock scores of cleaved peptide with Mpro conformations were in the range of 233–137 due to reduce surface area of contact of cleaved peptide fragments with the enzyme. In case of docking analysis of Mpro-quercetin complex conformations with substrate peptide at Mpro active site, significant changes in the active site conformation were observed at 1 ns, 10 ns, 20 ns, 30 ns, 40 ns and 50 ns (Fig. 6). The leading active site residues (His41, Cys145, Asp187, Arg188, Gln189 and Thr190) were found to be displaced resulting in change in area of substrate binding cavity thus no peptide cleavage occurred in the presence of quercetin. No interaction of the substrate peptide was noticed with the leading active site residue Cys145 and His41 in the presence of quercetin, as was observed when Mpro was docked with substrate peptide in the absence of quercetin. This was due to significant dislocation of residues from their native positions and opening of loops containing Cys45 and His41. The peptide was found intact at the active site in 1 ns, 10 ns, 20 ns and 30 ns conformations with significant reduction in ZDock scores from 785 to 158. In 40 ns and 50 ns conformations, the severe changes in the position of the loop containing Asp187, Arg188, Gln189 and Thr190 and its penetration into the cavity along with the change in relative conformation of beta sheets caused reduction in the affinity of the substrate peptide for the active site and no bonding of substrate was found at the catalytic site resulting in low ZDock scores of 114 and 86, respectively. The significant changes in the positioning of active site residues and reduction in the exposed area due to perturbations induced by quercetin interactions at an allosteric site are apparent by the Mpro-quercetin 3D conformations, which were extracted at a different time point during the simulation period of 50 ns, docked with substrate peptide (Fig. 6).
Fig. 4.
Modelled structure of substrate peptide (AVLQSGFR) containing cleavage site of SARS-CoV-2 main protease and its Ramachandran plot
Fig. 5.
Docking conformations of substrate peptide (AVLQSGFR) with different time point conformations of SARS-CoV-2 main protease of MD simulation of 50 ns showing cleavage of substrate and the resulting ZDock scores
Fig. 6.
Docking images of substrate peptide (AVLQSGFR) with different time point conformations of SARS-CoV-2 main protease-quercetin complex of MD simulation of 50 ns showing intact peptide with reducing ZDock scores
Many researches have reported active site inhibitors for SARS-CoV-2 Mpro which compete with the substrate polypeptide to occupy the active site (Ma et al. 2020; Zhang et al. 2020; Zhu et al. 2011; Jin et al. 2020a, b; Mengist et al. 2020). An adequate concentration of such inhibitors, greater than the substrate concentration, is often required so that the inhibitor can compete with substrate and can effectively bind to the enzyme’s active site thereby blocking the binding of the substrate (Strelow et al. 2012). Ma et al. in their in vitro study, reported GC376, boceprevir and calpain inhibitors as potent active site inhibitors of SARS-CoV-2 Mpro. These inhibitors provided 50–60% inhibition of enzyme with IC50 (Inhibitory constant) values ranging from 3 to 13 μM which are higher compared to the Ki (inhibitory constant) value of 810.43 nM predicted for allosteric inhibitor quercetin in our analysis (Ma et al. 2020). Jin et al. in their in silico and in vitro studies reported N3 as an effective inhibitor of Mpro with half-maximal cytotoxic concentration of more than 133 μM. Further, via high throughput screening, they determined 7 inhibitors having IC50 values ranging from 0.67 to 21.4 μM (Jin et al. 2020a). In another study, an antineoplastic agent carmofur was reported to inhibit viral replication by targeting the Mpro at its active site with an EC50 value of 24.30 μM and IC50 value of 1.82 μM (Jin et al. 2020b). The fact that effectiveness of competitive inhibitors or active site-binding inhibitors depends on substrate concentration and decreases with the increase in the concentration of substrate, put forward a demand for a concentration-independent inhibitor (Ramsay and Tipton 2017). In our analysis, we revealed quercetin as an effective allosteric inhibitor which can bind to the Mpro even when the active site is occupied by the substrate and will not be affected by any change in substrate concentration (Ramsay and Tipton 2017; Robinson 2015). Although very few studies have recently reported effective allosteric inhibitors against SARS-CoV-2 Mpro, a topological analysis of the Mpro revealed its very high sensitivity against any perturbation which it encounters and displayed excessively high tendency of the enzyme in transmitting it across the structure (Estradaa 2020; Doshi et al. 2016; Negre et al. 2018). These evidences can provide immense support to our present analysis of allosteric inhibition of Mpro by quercetin where binding of the compound in consideration, at a different site modulates the active site to reduce its affinity for the substrate. Hence, all the above results and facts prove that quercetin by binding at an allosteric site in SARS-CoV-2 Mpro can effectively inhibit the binding and thus the catalytic cleavage of substrate polypeptides 1a and 1ab. Such exclusive allosteric inhibition by a natural flavonol can massively hamper the replication of SARS-CoV-2 virus and can be developed into an exceedingly efficient and safe treatment against the current pandemic of COVID-19.
Conclusion
In the present in silico study, we analysed a natural flavonol quercetin for its effective allosteric inhibition against the SARS-CoV-2 Mpro. This flavonol allosteric inhibitor can be effective in both free and substrate conjugated form of Mpro. Binding energy of -8.31 kcal/mol for Mpro-quercetin complex in molecular docking analysis provided evidence for effective binding of the compound with Mpro. MD simulation of Mpro-quercetin complex proved stable interactions of quercetin with Mpro for a duration of 50 ns. In further molecular docking analysis of substrate peptide with Mpro (in both quercetin conjugated and free form) proved a significant reduction in the binding affinity of Mpro, when complexed with quercetin, for its substrate polypeptide. The docking ZDock scores of substrate peptide to Mpro binding reduced from 1043 for native state (0 ns) Mpro in the absence of quercetin to 86 for 50 ns conformation in the presence of quercetin. Henceforth, quercetin being a natural compound can be an efficient allosteric inhibitor against the effective target SARS-CoV-2 Mpro. Furthermore, exhaustive research in this concern is highly demanded to develop a natural, safe and effective treatment for the extremely dreaded respiratory syndrome of COVID-19.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
Both the authors have contributed equally in designing of work, conducting research as well as in manuscript preparation.
Compliance with ethical standards
Conflict of interest
There is no conflict of interest among the authors.
Footnotes
Shalja Verma and Anand Kumar Pandey have contributed equally.
References
- Abraham MJ, Murtola T, Schulz R, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Anand David AV, Arulmoli R, Parasuraman S. Overviews of biological importance of quercetin: a bioactive flavonoid. Pharmacogn Rev. 2016;10:84–89. doi: 10.4103/0973-7847.194044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiha GE, Beshbishy AM, Ikram M, et al. The pharmacological activity, biochemical properties, and pharmacokinetics of the major natural polyphenolic flavonoid: quercetin. Foods. 2020;9:1–16. doi: 10.3390/foods9030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIOVIA . Dassault Systèmes, discovery studio modeling environment, release 2019. San Diego: Dassault Systèmes; 2019. [Google Scholar]
- Chen B, Tian E, He B, et al. Overview of lethal human coronaviruses. Sig Transduct Target Ther. 2020;5:1–16. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus disease (COVID-19) pandemic World Health Organisation. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 2 Jul 2019
- Doshi U, Holliday MJ, Eisenmesser EZ, Hamelberg D. Dynamical network of residue–residue contacts reveals coupled allosteric effects in recognition, catalysis, and mutation. Proc Natl Acad Sci USA. 2016;113:4735–4740. doi: 10.1073/pnas.1523573113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Baba TJ, Lutomski CA, Kantsadi AL, et al. Allosteric inhibition of the SARS-CoV-2 main protease: insights from mass spectrometry based assays. Angew Chem. 2020 doi: 10.1002/anie.202010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estradaa E. Topological analysis of SARS CoV-2 main protease. Chaos: an Interdisciplinary. J Nonlinear Sci. 2020;30:1–13. doi: 10.1063/5.0013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Iftikhar H, Ali HN, Farooq S, Naveed H, Shahzad-Ul-Hussan S. Identification of potential inhibitors of three key enzymes of SARS-CoV2 using computational approach. Comput Biol Med. 2020;122:1–8. doi: 10.1016/j.compbiomed.2020.103848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jin Z, Zhao Y, Sun Y, et al. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- Kato K, Nakayoshi T, Fukuyoshi S, et al. Validation of molecular dynamics simulations for prediction of three-dimensional structures of small proteins. Molecules. 2017;22:1–15. doi: 10.3390/molecules22101716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:1–6. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamiable A, Thévenet P, Rey J, et al. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44:W449–W454. doi: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Mod. 2001;7:306–317. doi: 10.1007/s008940100045. [DOI] [Google Scholar]
- Liu X, Pavlovsky AG, Viola RE. The structural basis for allosteric inhibition of a threonine-sensitive aspartokinase. J Biol Chem. 2008;283:16216–16225. doi: 10.1074/jbc.M800760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov MY, Bogatyreva NS, Galzitskaya OV. Radius of gyration as an indicator of protein structure compactness. Mol Biol. 2008;42:623–628. doi: 10.1134/S0026893308040195. [DOI] [PubMed] [Google Scholar]
- Ma C, Sacco MD, Hurst B, et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;2020:1–15. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE. 2015;10:1–10. doi: 10.1371/journal.pone.0119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengist HM, Fan X, Jin T. Designing of improved drugs for COVID-19: Crystal structure of SARS-CoV-2 main protease Mpro. Sig Transduct Target Ther. 2020;5:1–2. doi: 10.1038/s41392-020-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre CF, Morzan UN, Hendrickson HP, et al. Eigenvector centrality for characterization of protein allosteric pathways. Proc Natl Acad Sci USA. 2018;115:E12201–E12208. doi: 10.1073/pnas.1810452115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JT, Serrano ML, Pujol FH, Rangel HR. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panche AN, Diwan AD, Chandra SR. Flavonoids: an overview. J Nutr Sci. 2016;5:1–15. doi: 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE. 2011;6:1–6. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BG, Wiehe K, Hwang H, et al. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S, Páll S, Schulz R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay RR, Tipton KF. Assessment of enzyme inhibition: a review with examples from the development of monoamine oxidase and cholinesterase inhibitory drugs. Molecules. 2017;22:1–46. doi: 10.3390/molecules22071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PK. Enzymes: principles and biotechnological applications. Essays Biochem. 2015;59:1–41. doi: 10.1042/bse0590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi B, Machin L, Monzote L, et al. Therapeutic potential of quercetin: new insights and perspectives for human health. ACS Omega. 2020;5:11849–11872. doi: 10.1021/acsomega.0c01818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sencanski M, Perovic V, Pajovic SB, Adzic M, Paessler S, Glisic S. Drug repurposing for candidate SARS-CoV-2 main protease inhibitors by a novel in silico method. Molecules (Basel, Switzerland) 2020;25:1–13. doi: 10.3390/molecules25173830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelow J, Dewe W, Iversen PW, et al. et al. Mechanism of action assays for enzymes. In: Sittampalam GS, Grossman A, Brimacombe K, et al.et al., editors. Assay guidance manual. Bethesda: Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2012. [PubMed] [Google Scholar]
- Vanommeslaeghe K, Hatcher E, Acharya C, et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreven T, Hwang H, Weng Z. Integrating atom-based and residue-based scoring functions for protein-protein docking. Protein Sci. 2011;20:1576–1586. doi: 10.1002/pro.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenthur CJ, Gentry PR, Mathews TP, Lindsley CW. Drugs for allosteric sites on receptors. Annu Rev Pharmacol Toxicol. 2014;54:165–184. doi: 10.1146/annurev-pharmtox-010611-134525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Yan S. Reasoning of spike glycoproteins being more vulnerable to mutations among 158 coronavirus proteins from different species. J Mol Model. 2005;11:8–16. doi: 10.1007/s00894-004-0210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Li R, Li X, et al. Quercetin as an antiviral agent inhibits influenza A virus (IAV) entry. Viruses. 2015;8:1–6. doi: 10.3390/v8010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, George S, Schmidt MF, et al. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Res. 2011;92:204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]