Abstract
The adaptive immune system is important for control of most viral infections. The three fundamental components of the adaptive immune system are B cells (the source of antibodies), CD4+ T cells, and CD8+ T cells. The armamentarium of B cells, CD4+ T cells, and CD8+ T cells has differing roles in different viral infections and in vaccines, and thus it is critical to directly study adaptive immunity to SARS-CoV-2 to understand COVID-19. Knowledge is now available on relationships between antigen-specific immune responses and SARS-CoV-2 infection. Although more studies are needed, a picture has begun to emerge that reveals that CD4+ T cells, CD8+ T cells, and neutralizing antibodies all contribute to control of SARS-CoV-2 in both non-hospitalized and hospitalized cases of COVID-19. The specific functions and kinetics of these adaptive immune responses are discussed, as well as their interplay with innate immunity and implications for COVID-19 vaccines and immune memory against re-infection.
The adaptive immune system is crucial for controlling viral infection, but the kinetics and magnitude of the roles of its various components differ across viral infections. Review the emerging data on the roles of B cells, CD4+ T cells, and CD8+ T cells in SARS-CoV-2 infection.
Introduction
Coronavirus disease 2019 (COVID-19), caused by the novel human pathogen severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Hu et al., 2020), is a serious disease that has resulted in widespread global morbidity and mortality. Our understanding of SARS-CoV-2 and COVID-19 has rapidly evolved during 2020. As of December 2020, the United States has experienced >300,000 deaths, winter cases are rising exceptionally fast, and the first interim phase 3 vaccine trial results have been reported. The scientific advances in understanding SARS-CoV-2 and COVID-19 have been extraordinarily rapid and broad, by any metric, which is an amazing testament to the commitment, creativity, collaboration, and expertise of the international scientific community, both in academia and industry, under extremely challenging conditions. This article will review our current understanding of the immunology of COVID-19, with a primary focus on adaptive immunity.
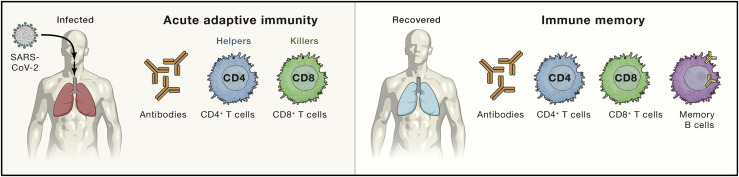
The immune system is broadly divided into the innate immune system and the adaptive immune system. Although the adaptive and innate immune systems are linked in important and powerful ways, they each consist of different cell types with different jobs. The adaptive immune system consists of three major cell types: B cells, CD4+ T cells, and CD8+ T cells (Figure 1 ). B cells produce antibodies. CD4+ T cells possess a range of helper and effector functionalities. CD8+ T cells kill infected cells. Given that adaptive immune responses are important for the control and clearance of almost all viral infections that cause disease in humans, and adaptive immune responses and immune memory are central to the success of all vaccines, it is critical to understand adaptive responses to SARS-CoV-2.
Figure 1.
The major components of adaptive immunity in viral immune responses
Virus-specific CD4+ T cells, CD8+ T cells, and antibodies (produced by B cells) constitute the three major components of acute adaptive immunity to a viral infection. Immune memory consists of memory B cells, antibodies, virus-specific CD4+ T cells, and virus-specific CD8+ T cells constitute the four major components of immune memory to a viral infection.
One integrated model of immune responses to SARS-CoV-2
This review first presents a working model of immune responses to SARS-CoV-2, to provide an overarching context, and then the review explores individual compartments and immunological facets of adaptive immunity to SARS-CoV-2 in greater detail. Importantly, this is an evolving model and should not be accepted as definitive; instead, it provides a reference point for interpreting much of the available data in the literature and to identify knowledge gaps that may provide directions for future studies.
Any virus that can cause disease in humans must have at least one immune evasion mechanism—at least one immune evasion “trick.” Without the ability to evade the immune system, a virus is usually harmless. Understanding immune evasion by a virus is frequently important for understanding the pathogenesis of the virus, as well as understanding challenges faced by the adaptive immune system and any candidate vaccine. In the case of SARS-CoV-2, the virus is clearly unusually effective at evading the triggering of early innate immune responses, such as type 1 interferons (IFNs) (see below). It is plausible that much of the nature of COVID-19 as an illness is a consequence of this one big trick of SARS-CoV-2.
In an idealized example of a generic viral infection, the innate immune system rapidly recognizes the infection and triggers the “alarm bells” of type I IFN expression and related molecules (Weaver and Murphy, 2016) (Figure 2 A). This can occur within a couple of hours of infection. The innate immune response serves three main purposes: (1) restriction of viral replication within infected cells, (2) creation of an antiviral state in the local tissue environment, including recruitment of effector cells of the innate immune system, and (3) priming the adaptive immune response. The first two activities of the innate immune system slow down the viral replication and spread. The third is a critical requirement of the innate immune system to trigger the adaptive immune response. Adaptive immune responses are slow due to the intrinsic requirement of selecting and expanding virus-specific cells from the large pools of naive B cells and T cells specific for different molecular structures and sequences (>109 cells each). Adaptive immune responses take time to generate sufficient cells to control a viral infection, ∼6–10 days after priming (Figure 2A), due to the inherent time demands for extensive proliferation and differentiation of naive cells into effector cells. Once sufficient populations of effector T cells (helper T cells and cytotoxic T cells) and effector B cells (antibody secreting cells, known as plasmablasts and plasma cells) have proliferated and differentiated, they often work together to rapidly and specifically clear infected cells and circulating virions.
Figure 2.
An integrated working model of COVID-19 immunology and disease severity
Immune response trajectories in COVID-19. Conceptual schematics of the kinetics of immune responses to SARS-CoV-2 under conditions of average COVID-19 (non-hospitalized cases) and severe or fatal COVID-19. “Innate immunity” line specifically refers to the peak kinetics of innate cytokines and chemokines detectable in blood; innate immune responses occur locally throughout the course of an infection. “T cells” refers to virus-specific CD4+ and CD8+ T cells. “Antibodies” refers to virus-specific neutralizing antibodies. Arrows indicate a time point with important differences in the presence or absence of T cell responses and the magnitude of the viral load, comparing (B) and (C).
(A) An example of a generic viral infection.
(B) Average SARS-CoV-2 infection.
(C) Severe or fatal SARS-CoV-2 infection. The period of severe COVID-19 clinical disease is shaded gray.
See also Figure S1 for additional features.
In a SARS-CoV-2 infection, the virus is particularly effective at avoiding or delaying triggering intracellular innate immune responses associated with type I and type III IFNs in vitro (Blanco-Melo et al., 2020) and in humans (Arunachalam et al., 2020; Bastard et al., 2020; Blanco-Melo et al., 2020; Laing et al., 2020). Without those responses, the virus initially replicates unabated and, equally importantly, the adaptive immune responses are not primed until the innate immune alarms occur (Figure 2B). In an average case of COVID-19, a simple model is that temporal delay in innate immune responses is enough to result in asymptomatic infection (∼40% of SARS-CoV-2 infections are asymptomatic) (Oran and Topol, 2020) or clinically mild disease (“mild” is a COVID-19 clinical definition meaning not requiring hospitalization) because the T cell and antibody responses occur relatively quickly and control the infection (Figure 2B). The presence of T cells and antibodies is associated with successful resolution of average cases of COVID-19 (Grifoni et al., 2020). Studies of acute and convalescent COVID-19 patients have observed that SARS-CoV-2-specific T cell responses are significantly associated with milder disease (Liao et al., 2020; Rydyznski Moderbacher et al., 2020; Sekine et al., 2020; Zhou et al., 2020b), suggesting that T cell responses may be important for control and resolution of a primary SARS-CoV-2 infection. These topics are discussed in detail in later sections.
Ineffective IFN innate immunity has been strongly associated with failure to control a primary SARS-CoV-2 infection and a high risk of fatal COVID-19, accompanied by innate cell immunopathology and a plasma cytokine signature of elevated CXCL10, interleukin (IL)-6, and IL-8 in many studies (Aid et al., 2020; Kuri-Cervantes et al., 2020; Li et al., 2020b; Lucas et al., 2020; Radermecker et al., 2020; Schurink et al., 2020; Del Valle et al., 2020). Impaired and delayed type I and type III IFN responses are associated with risk of severe COVID-19 (Galani et al., 2021; Hadjadj et al., 2020). The risk of a poor early innate immune response to SARS-CoV-2 is highlighted by the striking findings of very high risk of severe or fatal COVID-19 in individuals with defective type I IFN responses (Bastard et al., 2020; Zhang et al., 2020). If the innate immune response delay is too long—because of particularly efficient evasion by the virus, defective innate immunity, or a combination of both—then the virus (1) gets a large head start in replication in the upper respiratory tract (URT) and lungs, and (2) fails to prime an adaptive immune response for a long time, resulting in conditions that lead to severe enough lung disease for hospitalization (Figure 2C). These factors can be amplified by challenges of age, as elderly individuals have a smaller naive T cell pool and are therefore more likely to struggle to make a T cell response quickly that can recognize this new virus (Rydyznski Moderbacher et al., 2020), which also likely results in hampered neutralizing antibody responses, because neutralizing antibody responses are generally T cell-dependent.
If the adaptive immune response starts too late, fatal COVID-19 appears to be a situation where viral burden is high (Magleby et al., 2020) in the absence of a substantive adaptive immune response (Figure 2C). It is plausible that the innate immune system tries to fill the vacuum left by the absence of a T cell response, attempting to control the virus with an ever-expanding innate immune response. That solution ends up untenable, as a massive innate response results in excessive lung immunopathology. This conclusion is consistent with many studies finding innate cytokine/chemokine signatures of immunopathology (cited above), and particularly observation of elevated frequencies of neutrophils (the most common cell type of the innate immune system) in blood (Kuri-Cervantes et al., 2020), and massive numbers of neutrophils in lungs, associated with severe, end-stage COVID-19 disease (Li et al., 2020b; Liao et al., 2020; Radermecker et al., 2020; Schurink et al., 2020). In contrast, end-stage disease is not generally associated with preferentially elevated T cell abundance in lung tissue (Liao et al., 2020; Oja et al., 2020; Szabo et al., 2020), consistent with a working model that early adaptive immune responses are very beneficial, and late adaptive immune responses are simply too late (Figure 2C).
We describe one parsimonious working model of immune responses in COVID-19 in Figure 2, consistent with much of the available literature. However, these are still very active areas of investigation, and there are plausible alternative models as well, which are discussed below. The overall amount of data for the details of antigen-specific adaptive immune responses in acute COVID-19 infections remains limited; nevertheless, the ongoing pandemic, with well over 1 million deaths to date, requires rapid interpretation of the available data. This working model provides a useful framework and reference point to start from. Below, we provide a detailed discussion of the facets of adaptive immunity to COVID-19.
Adaptive immunity to SARS-CoV-2 infection
Humans make SARS-CoV-2-specific antibodies, CD4+ T cells, and CD8+ T cells in response to SARS-CoV-2 infection (Grifoni et al., 2020; Krammer, 2020; Stephens and McElrath, 2020) (Figure 1). Antibodies, CD4+ T cells, and CD8+ T cells can each have protective roles in controlling viral infections, but those roles and the importance of each component of adaptive immunity varies depending on the viral infection. In some infections, one of the three branches of adaptive immunity is critically important for control of the viral infection and survival of the host. For other viral infections, there is a high degree of synergy and redundancy between the branches of adaptive immunity, resulting in more paths to successful control of an infection and robust immunity. For these reasons, it is important to measure antigen-specific CD4+ T cells, CD8+ T cells, and antibodies in the same individuals, to avoid the problems illustrated in the classic blindfolded men and the elephant allegory.
CD4+ T cells
T cell responses are detected after almost all SARS-CoV-2 infections (Grifoni et al., 2020; Rydyznski Moderbacher et al., 2020; Peng et al., 2020; Sekine et al., 2020). CD4+ T cell responses to SARS-CoV-2 are more prominent than CD8+ T cell responses (Grifoni et al., 2020; Sekine et al., 2020) and have been associated with control of primary SARS-CoV-2 infection (Rydyznski Moderbacher et al., 2020). CD4+ T cells more so than CD8+ T cells were associated with control of SARS-CoV infection in animal models (Chen et al., 2010; Zhao et al., 2016). T cells specific to any viral protein can be relevant for protective immunity. Nevertheless, there is particular interest in T cell responses against SARS-CoV-2 Spike protein (“Spike”), because almost all candidate COVID-19 vaccines exclusively contain Spike (Krammer, 2020). Additionally, induction of anti-Spike antibodies depends on Spike-specific CD4+ T cells, with possible contributions of CD4+ T cells specific for other virion structural proteins (Crotty, 2015; Elsayed et al., 2018). In a study examining CD4+ T cell responses to all SARS-CoV-2 proteins in convalescent COVID-19 cases, responses were detected against almost all SARS-CoV-2 proteins (21/24) within the subject cohort, with CD4+ T cell responses only undetectable for the smallest of the proteins (Grifoni et al., 2020). The prevalence and magnitude of the SARS-CoV-2 CD4+ T cell responses correlates with the expression level of each SARS-CoV-2 protein (Grifoni et al., 2020). Spike, M, and nucleocapsid are the most prominent targets of SARS-CoV-2-specific CD4+ T cells (Grifoni et al., 2020), with substantial responses also against ORF3a and nsp3 (Le Bert et al., 2020; Grifoni et al., 2020; Nelde et al., 2021; Oja et al., 2020; Peng et al., 2020). The prominence of M as a CD4+ T cell target is intriguing, because M is a relatively small, multipass transmembrane protein without high-affinity class II restricted T cells in the naive repertoire (Mateus et al., 2020), suggesting that M is very highly expressed in vivo, or is made available to CD4+ T cells in a very immunogenic context. Although Spike is the most consistently recognized SARS-CoV-2 antigen (Braun et al., 2020; Grifoni et al., 2020; Rydyznski Moderbacher et al., 2020; Peng et al., 2020), a “megapool” of predicted class II epitopes measures ∼50% of the SARS-CoV-2-specific CD4+ T cell response outside of Spike (Grifoni et al., 2020), which substantially simplifies SARS-specific CD4+ T cell measurements. Patterns of CD4+ T cell SARS-CoV-2 antigens recognized appear to be similar during acute infection and convalescence and memory phases (Rydyznski Moderbacher et al., 2020), although ORF7/8 CD4+ T cell responses may exhibit relative selectivity for the acute phase (Tan et al., 2020a).
CD4+ T cells in acute COVID-19
Studies of SARS-CoV-2 CD4+ T cells are more common from convalescent patients than from acute cases, for logistical reasons, but it is important to understand the kinetics of the T cell responses during acute infection to be able to interpret the functional contributions of adaptive immunity to control of natural infection. One of the challenges in the literature is that intensive care unit (ICU) patients are frequently the most accessible acute patients for T cell studies, but antigen-specific T cell measurements in ICU patients is the study of late stage disease, the sequela of which might render results challenging to interpret. The situation earlier during acute disease is of particular interest. SARS-CoV-2-specific CD4+ T cells can be detected as early as days 2–4 post-symptom onset (PSO) (Rydyznski Moderbacher et al., 2020; Tan et al., 2020a; Weiskopf et al., 2020). Notably, SARS-CoV-2-specific CD4+ T cells had the strongest association with lessened COVID-19 disease severity, compared with antibodies and CD8+ T cells (Rydyznski Moderbacher et al., 2020). Rapid induction of SARS-CoV-2-specific CD4+ T cells in acute COVID-19 was associated with mild disease and accelerated viral clearance (Tan et al., 2020a). In contrast, the strikingly extended absence of SARS-CoV-2-specific CD4+ T cells was associated with severe or fatal COVID-19 (>day 22 PSO in some cases) (Braun et al., 2020; Rydyznski Moderbacher et al., 2020; Tan et al., 2020a).
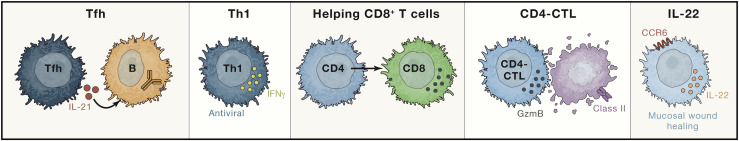
CD4+ T cell functions in SARS-CoV-2 infection
CD4+ T cells have the ability to differentiate into a range of helper and effector cell types, with capacity to instruct B cells, help CD8+ T cells, recruit innate cells, have direct antiviral activities, and facilitate tissue repair (Figure 3 ). Virus-specific CD4+ T cells commonly differentiate into Th1 cells and T follicular helper cells (Tfh). Th1 cells have antiviral activities via production of IFNγ and related cytokines. Tfh cells are the specialized providers of B cell help and are critical for the development of most neutralizing antibody responses, as well as memory B cells and long-term humoral immunity (Crotty, 2019). Circulating Tfh cells (cTfh) specific for SARS-CoV-2 are generated during acute SARS-CoV-2 infection (Meckiff et al., 2020; Rydyznski Moderbacher et al., 2020). SARS-CoV-2 memory cTfh cells are also generated (Juno et al., 2020; Rydyznski Moderbacher et al., 2020; Neidleman et al., 2020; Sekine et al., 2020). Although neutralizing antibody titers have not correlated with reduced COVID-19 severity (Dan et al., 2021; Piccoli et al., 2020; Robbiani et al., 2020), SARS-CoV-2 cTfh cell frequencies have been associated with reduced disease severity (Rydyznski Moderbacher et al., 2020). Of note, a substantial fraction of SARS-CoV-2 cTfh are CCR6+, potentially indicative of mucosal airway homing, which has also been observed for the common cold coronavirus HKU1 (Juno et al., 2020).
Figure 3.
CD4+ T cell functions observed in COVID-19
Virus-specific CD4+ T cells may differentiate into multiple distinct cell types in response to SARS-CoV-2, and exhibit a range of helper and effector functions. These include Tfh cells, which provide help to B cells for affinity maturation and antibody production; Th1 cells, which can have direct antiviral functions through cytokine secretion plus recruitment of innate cells; CD4 T cells, that help CD8 T cells to proliferate and differentiate; CD4-CTL, which can have direct cytotoxic activity against virally infected cells in a class II antigen presentation restricted manner; and CD4 T cells that produce IL-22, which has roles in wound healing.
In addition to helping antibody responses, CD4+ T cells also help CD8+ T cell responses. Although the exact CD4+ T cells providing the CD8+ help remains unclear, IL-21 can be important for CD4+ T cell help to CD8+ T cells (Buchholz and Busch, 2019; Zander et al., 2019), and IL-21 is a canonical cytokine of TFH cells.
Although helping B cells and CD8+ T cells are important functions of CD4+ T cells, CD4+ T cells also differentiate into effector cells with more direct anti-pathogen activities, such as Th1 cells. IFNγ+ CD4+ T cells protect mice from lethal SARS-CoV infection (Zhao et al., 2016). The dominant cytokine produced by SARS-CoV-2-specific CD4+ T cells from COVID-19 patients is IFNγ (Braun et al., 2020; Grifoni et al., 2020; Rydyznski Moderbacher et al., 2020; Weiskopf et al., 2020), with a clear IFNγ, tumor necrosis factor (TNF), IL-2 protein signature of canonical Th1 cells (Braun et al., 2020; Rydyznski Moderbacher et al., 2020; Peng et al., 2020; Sekine et al., 2020). CD4-CTLs are related cell types with direct cytotoxic activity, which are associated with protective immunity against multiple severe viral infections (Weiskopf et al., 2015). A CD4-CTL transcriptional signature has been seen (Meckiff et al., 2020), although the cytotoxicity degranulation marker CD107a (Buggert et al., 2018; Weiskopf et al., 2015) has been minimally observed on SARS-CoV-2-specific CD4+ T cells (Peng et al., 2020; Sekine et al., 2020). One function of CD4+ T cells is to recruit other effector cells to a site of viral antigen, and gene expression of CCL3/4/5 (MIP-1 s) and XCL1 chemokines by SARS-CoV-2-specific CD4+ T cells may contribute to effector cell recruitment (Meckiff et al., 2020).
CCR6 is a chemokine receptor associated with migration to mucosal tissues. Expression of CCR6 by a subset of SARS-CoV-2-specific CD4+ T cells may reflect underlying Th17 attributes of those cells; however, undetectable or low amounts of IL-17α protein expression have been reported (Braun et al., 2020; Grifoni et al., 2020; Juno et al., 2020; Rydyznski Moderbacher et al., 2020; Oja et al., 2020; Sekine et al., 2020; Weiskopf et al., 2020). In contrast, IL-22, which can also be made by mucosal CD4+ T cells, appears to be robustly expressed by SARS-CoV-2-specific CD4+ T cells (Grifoni et al., 2020; Rydyznski Moderbacher et al., 2020; Weiskopf et al., 2020). CCR6+ IL-22+ and CCR6+ IL-17+ cells can be largely independent cell types in other infections (Morou et al., 2019). Notably, IL-22 is strongly associated with tissue repair, particularly of lung and gut epithelial cells (Dudakov et al., 2015), suggesting that the SARS-CoV-2 CD4+ T cell response may actively participate in lung tissue repair during COVID-19. SARS-CoV-2 memory CD4+ T cells appear to retain the capacity to make IL-22 (Rydyznski Moderbacher et al., 2020). Last, there were initially great concerns about potential immune deviation to Th2 and immunopathogenesis (Peeples, 2020), but SARS-CoV-2-specific CD4+ T cells from patients consistently do not have Th2 characteristics (Grifoni et al., 2020; Rydyznski Moderbacher et al., 2020; Sekine et al., 2020).
CD8+ T cells
CD8+ T cells are critical for clearance of many viral infections, due to their ability to kill infected cells. In SARS-CoV-2 infections, the presence of virus-specific CD8+ T cells has been associated with better COVID-19 outcomes (Rydyznski Moderbacher et al., 2020; Peng et al., 2020). Overall, circulating SARS-CoV-2-specific CD8+ T cells are less consistently observed than CD4+ T cells (Grifoni et al., 2020; Rydyznski Moderbacher et al., 2020; Sekine et al., 2020). Predicted HLA class I epitope peptides are able to identify a substantial fraction of the SARS-COV-2-specific CD8 T cell response (Grifoni et al., 2020). SARS-CoV-2 CD8+ T cells are specific for a range of SARS-CoV-2 antigens, with Spike, nucleocapsid, M, and ORF3a well represented (Le Bert et al., 2020; Grifoni et al., 2020; Nelde et al., 2021; Peng et al., 2020; Sekine et al., 2020). Like SARS-CoV-2-specific CD4+ T cell responses, SARS-CoV-2-specific CD8+ T cell responses can develop rapidly during acute COVID-19 (Rydyznski Moderbacher et al., 2020), with a report of virus specific CD8+ T cells as early as day 1 PSO (Schulien et al., 2020). In acute COVID-19, SARS-CoV-2-specific CD8+ T cells exhibit high levels of molecules associated with potent cytotoxic effector functions, such as IFNγ, granzyme B, perforin, and CD107a (Rydyznski Moderbacher et al., 2020; Schulien et al., 2020; Sekine et al., 2020). Memory SARS-CoV-2 CD8+ T cells have similar expression profiles (Grifoni et al., 2020; Rydyznski Moderbacher et al., 2020; Peng et al., 2020; Sekine et al., 2020; Zuo et al., 2020a).
Antibodies and B cells
The vast majority of SARS-CoV-2 infected individuals seroconvert within 5–15 days PSO, with ∼90% seroconversion by day 10 PSO (Long et al., 2020; Rydyznski Moderbacher et al., 2020; Premkumar et al., 2020; Ripperger et al., 2020; Suthar et al., 2020). The primary antigens examined for seroconversion are Spike and nucleocapsid. Nucleocapsid and Spike immunoglobulin G (IgG) titers are highly correlated (Piccoli et al., 2020). Spike is the target of SARS-CoV-2 neutralizing antibodies, and the receptor binding domain (RBD) of Spike is the target of >90% of neutralizing antibodies in COVID-19 cases (Brouwer et al., 2020; Piccoli et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Suthar et al., 2020), with some neutralizing antibodies instead targeting the N-terminal domain (NTD) (Liu et al., 2020). The estimates of seroconversion to SARS-CoV-2 Spike range from 91%–99% in large studies (Gudbjartsson et al., 2020; Wajnberg et al., 2020). Spike IgG, IgA, and IgM develop simultaneously in infected individuals (Isho et al., 2020; Premkumar et al., 2020; Suthar et al., 2020).
Neutralizing antibodies develop rapidly in most SARS-CoV-2-infected people, on the same time frame as seroconversion. The neutralizing antibodies are produced by B cells with a wide range of heavy chain and light chain V genes (Robbiani et al., 2020; Rogers et al., 2020). SARS-CoV-2 neutralizing antibodies also exhibit little to no somatic hypermutation (Gaebler et al., 2020; Robbiani et al., 2020; Rogers et al., 2020). Altogether, these data indicate that development of neutralizing antibodies against SARS-CoV-2 is relatively easy, because it can be accomplished by many B cells with little or no affinity maturation required. The data also indicate that SARS-CoV-2 neutralizing antibody responses generally develop from naive B cells, not from pre-existing cross-reactive memory B cells (Anderson et al., 2020; Ng et al., 2020; Nguyen-Contant et al., 2020; Shrock et al., 2020; Song et al., 2020). Thus, neutralizing epitopes on the SARS-CoV-2 RBD domain, particularly those corresponding to the ACE2 receptor binding footprint, appear to be highly immunogenic and easily recognized by antibodies. However, it should also be noted that circulating SARS-CoV-2 neutralizing antibody titers are relatively low in a substantial fraction of recovered COVID-19 cases (Dan et al., 2021; Robbiani et al., 2020; Wajnberg et al., 2020), indicating that either the neutralizing antibody potency or the serum concentration is suboptimal in this subset.
Although it was noted above that antibodies stop viruses outside of cells, antibodies can also kill virally infected cells (Lu et al., 2018), and this can be an important mechanism of action in vivo (Benhnia et al., 2009; Lu et al., 2018). Currently there is no direct evidence of this in human SARS-CoV-2 infections, but Fc receptor-associated functions in serum were correlated with protective immunity in a SARS-CoV-2 NHP vaccine model (Yu et al., 2020), neutralizing antibodies with Fc-receptor binding capacity were more protective in mice (Schäfer et al., 2021), and humans who did not survive COVID-19 infection had antibody responses with reduced Fc-dependent antibody effector activity (Zohar et al., 2020).
In general, and in various animal models, higher antigen load drives higher antibody titers. This appears to hold true in the case of SARS-CoV-2, where neutralizing antibody titers (and total Spike antibody titers) have positively correlated with COVID-19 disease severity in large cohort studies (Piccoli et al., 2020; Robbiani et al., 2020). Similar observations were made for SARS and Middle Eastern respiratory syndrome (MERS) (Sariol and Perlman, 2020).
The relationship between neutralizing antibodies, Tfh cells, and COVID-19 disease severity appears to be complex. High neutralizing antibody titers are associated with severe disease and potentially extrafollicular B cell responses (Piccoli et al., 2020; Woodruff et al., 2020), while SARS-CoV-2-specific Tfh cells have different associations depending on the study (Juno et al., 2020; Meckiff et al., 2020; Rydyznski Moderbacher et al., 2020; Oja et al., 2020). Disparate T and B cell responses could be due to a disconnect between B cell and T cell responses (Rydyznski Moderbacher et al., 2020; Oja et al., 2020; Tan et al., 2020a) due to the altered early innate immune response, which may result in kinetic delays or dysregulation of T cell priming (Arunachalam et al., 2020; Zhou et al., 2020b). To fill these knowledge gaps, more longitudinal studies of the kinetics of antibody and virus-specific T cell responses during acute SARS-CoV-2 infections of varying disease severity are required.
Protection and pathogenesis
SARS-CoV-2 infects epithelial cells of the URT (nasal passages and throat) and lungs (bronchi and lung alveoli). These sites are involved with different aspects of SARS-CoV-2 pathology and transmission, relevant to the adaptive immune response. Severe COVID-19 involves extensive lung infection. SARS-CoV-2 URT infection is important for transmission and is associated with milder disease symptoms. As summarized above, based on the available data, a useful working model is that COVID-19 disease severity and duration is largely a function of early evasion of innate immune recognition and the subsequent kinetics of the adaptive immune response (Figures 2A and 2B), with severe or fatal COVID-19 disease being largely the product of innate immune system lung immunopathology brought on by high viral loads and a lack of timely T cell responses (Figures 2C and S1).
Immunological mechanisms of control of SARS-CoV-2 infection
In oversimplified terms, antibodies mostly stop viruses outside of cells and T cells stop viruses inside of cells. This powerful division of labor has proven to be a successful complementary approach to control most viral infections in most individuals. In a natural infection, the adaptive immune response takes time to develop, and many cells are already infected by the time an antibody response develops. Frequently antibodies alone cannot clear an ongoing infection, it also takes T cells. This may be why SARS-CoV-2 neutralizing antibody titers have not correlated with lessened disease severity in primary COVID-19 (Baumgarth et al., 2020; Rydyznski Moderbacher et al., 2020; Tan et al., 2020a). It is always challenging to interpret immunological cause and effect in humans, due to the inherently correlative nature of the data; nevertheless, the absence of a correlation between neutralizing antibodies and recovery from COVID-19 is notable. Given that SARS-CoV-2-specific CD4+ T cells and CD8+ T cells correlated with reduced disease severity, while neutralizing antibodies in the same individuals did not (Rydyznski Moderbacher et al., 2020), one parsimonious interpretation is that T cells do the heavy lifting for control of a primary SARS-CoV-2 infection.
Injection of high doses of SARS-CoV-2 neutralizing monoclonal antibodies into SARS-CoV-2-infected humans had relatively limited effects on COVID-19 in clinical trials (Chen et al., 2020; Weinreich et al., 2020). Viral loads dropped an extra ∼0.5 log10 after 7 days in treated subjects compared to subjects who received placebo control. In contrast, individuals who seroconverted on their own exhibited 1,000- to 10,000-fold lower viral loads (Weinreich et al., 2020). Given that the monoclonal antibody injections provided SARS-CoV-2 neutralizing antibody titers ∼100 times higher than native neutralizing antibody responses in people (monoclonal antibody neutralizing titers of ∼100,000) (Weinreich et al., 2020), the results are most consistent with a dominant role for T cells in control and clearance of an ongoing SARS-CoV-2 infection. This is not to say that such therapeutic monoclonal antibody interventions are not worthwhile, but the simplest interpretation of the data is that even high concentrations of neutralizing antibodies have limited effect on the control of SARS-CoV-2 replication, and it may essentially be buying additional time for the development of an effective T cell response to clear the infection. Additionally, monoclonal antibody therapy has not, to date, shown efficacy in improving outcomes of hospitalized COVID-19 (Lundgren et al., 2020).
Human genetic and pharmacological evidence supports the possibility of control of SARS-CoV-2 in the absence of neutralizing antibodies. Two unrelated adults in Italy with agammaglobulinemia and no circulating B cells developed COVID-19 and fully recovered from infection (Soresina et al., 2020), suggesting that antibodies can be dispensable for protective immunity to SARS-CoV-2 in otherwise healthy adults. Many people have pharmaceutical depletion of B cells for unrelated conditions. Three studies, with a total of 31 COVID-19 cases, of subjects on B cell depletion therapy reported all COVID-19 cases resolved without intensive care (Montero-Escribano et al., 2020; Novi et al., 2020; Safavi et al., 2020). One report presented two fatal cases of COVID-19 in patients >65 years of age on B cell depletion therapy and other immunosuppressive drugs (Tepasse et al., 2020). However, no antigen-specific T or neutralizing antibody data are available from any of those reports. Separately, there are multiple reports of healthy individuals successfully controlling a SARS-CoV-2 infection with little to no neutralizing (or RBD IgG) antibodies detectable post-infection, while having significant SARS-CoV-2-specific T cell memory (Rydyznski Moderbacher et al., 2020; Nelde et al., 2021; Schulien et al., 2020; Sekine et al., 2020). Those observations imply the ability to control COVID-19 without substantial contribution from neutralizing antibodies, as long as a strong T cell response is present. Thus, with the currently available data, it is plausible that SARS-CoV-2 infection may be controlled by a combination of CD4+ and CD8+ T cells without neutralizing antibodies. Nevertheless, in aggregate the data support a model wherein a coordinated, early response by all three branches of adaptive immunity is likely to be most successful at controlling SARS-CoV-2 infection and limiting COVID-19 severity. More data are required to directly test these models thoroughly.
This does not mean that neutralizing antibodies are not functional or valuable. Neutralizing antibodies are associated with protective immunity against 2° infection with SARS-CoV-2 or SARS-CoV, discussed below.
Antibodies can be a useful surrogate marker of CD4+ T cell responses in many infections and vaccines, as antibody assays are much easier and more sensitive in small blood volumes than antigen-specific T cell assays. At large cohort scales it is usually impractical to measure virus-specific T cells. Thus, correlations between antibodies and T cells are of great interest. Serum antibody titers were poorly predictive of the presence of SARS-CoV-2 CD4+ T cells in a study of 188 COVID-19 cases (Dan et al., 2021). These considerations have increased interest in potentially using early T cell responses as a diagnostic in acute COVID-19 (Tan et al., 2020a). A cytokine release assay analogous to the tuberculosis IFNγ release assay is a possibility (Lewinsohn et al., 2017; Murugesan et al., 2020; Petrone et al., 2020), as are plasma CXCL10 (Rydyznski Moderbacher et al., 2020) and surrogate markers in flow cytometry for virus-specific T cells (Mathew et al., 2020; Sekine et al., 2020).
Immunopathogenesis
Sometimes the immune response to a viral infection is more damaging than the viral infection itself. Therefore, in addition to considering roles for adaptive immunity in control of COVID-19, immunopathogenesis must also be considered. While the data indicate that overactive innate immunity is the cause of severe or fatal lung pathology in COVID-19 (Figure 2, discussed above), alternative models to seriously consider are (1) T cells are the central cause of immunopathogenesis, or (2) antibodies are the central cause of immunopathogenesis. The initial emphasis on the T cell immunopathology model came from a Th2-bias hypothesis (Peeples, 2020) and that has not been borne out, as discussed above in the CD4+ T cell section. Another possibility has been T cell immunopathology in the lung by cells that are not detectable in blood. Local immunity is discussed below but, in brief, available data have not supported that model either. Although there have been reports of positive associations between T cell responses and COVID-19 immunopathology, the majority of the data points to innate immune cells in the lungs as the primary mediators of immunopathology. There are people with severe COVID-19 with high frequencies of circulating CD8+ T cells of indeterminate specificity (Mathew et al., 2020), leaving open the possibility that those cells are involved in immunopathogenesis, in contrast to individuals with severe disease and weak virus-specific T cell responses (Mathew et al., 2020; Rydyznski Moderbacher et al., 2020; Tan et al., 2020a). One of the challenges is that, late in ICU patients, there may be complex intertwined adverse events after many things have already gone haywire. Studies beginning earlier during disease are more informative, and larger longitudinal studies of virus-specific T cell responses in acute disease are sorely needed.
Regarding antibodies as the cause of immunopathogenesis, this notion primarily derives from some SARS animal model vaccine studies and an understanding of dengue. Although the term antibody-dependent enhancement (ADE) of infection is frequently used, ADE was not observed in SARS or MERS, and the instances of antibody-associated disease pathology with certain SARS immunizations were not FcR-dependent and appear more likely to be immune complex driven inflammation (Sariol and Perlman, 2020). ADE is rarely observed for viral infections in vivo. It is unclear if immune complex driven inflammation occurs in severe COVID-19. The quality of the Spike, neutralizing, and nucleocapsid antibodies is similar in both hospitalized and non-hospitalized cases (Piccoli et al., 2020). A bias toward nucleocapsid antibodies over Spike antibodies in fatal cases was observed in a pair of studies (Atyeo et al., 2020; Zohar et al., 2020), which is consistent with a role for Spike antibodies in reducing viral loads.
Autoantibodies have been reported in COVID-19 cases. It is not uncommon for transient autoreactivity to be observed in serum in response to an acute viral infection. However, autoantibodies triggered by pathogens can be pathogenic (e.g., S. pyogenes and rheumatic heart disease), and it is unclear if autoantibodies in response to SARS-CoV-2 infection are unusually prevalent or pathogenic (Zuo et al., 2020b).
Viruses have developed multitudinous ways to evade the immune system, including disrupting or diverting immune responses. Thus, although it is still possible that SARS-CoV-2 has additional important immune evasion or disruption mechanisms yet to be determined, the currently available data are encouraging for vaccine development, because adaptive immune responses to SARS-CoV-2 are associated with control of infection, and no clear causal association of adaptive immunity with COVID-19 disease severity has been observed in the literature.
Local immune responses
To control an infection, the immune system must act at the site of the infection (Figure 4 ). Most measurements of human adaptive immunity are made from blood samples, because this is by far the most convenient means of measuring immune responses. Immunological cells and antibodies in the blood do not necessarily reflect what is present in an infected tissue (Masopust and Soerens, 2019). It is therefore important to understand the relationship between the immune response in the blood and the immune response in tissues and to directly measure immune responses in affected tissues whenever possible. Currently, such data are limited for COVID-19 patients. Nevertheless, data indicate that blood measurements are largely reflective of the local immune response.
Figure 4.
Components of local immunity
Human immune responses are most often measured in blood, but immune responses at local sites of infection and/or portals of entry are important and may not be directly reflected by blood measurements. Local immunity in lungs, nasal passages, and the oral cavity and salivary glands can consist of Trm CD8+ T cells, CD4+ T cells, and IgG and IgA antibodies constitutively present in those tissues as immune memory, which can be supplemented by additional cells and antibodies upon infection.
With antibodies, both IgG and IgA are secreted at mucosal tissues. Encouragingly, most individuals are positive for Spike IgG and IgA in blood (referenced above), and Spike IgG and IgA levels in blood were indicative of levels in saliva (Isho et al., 2020). Anti-RBD secretory IgA has enhanced SARS-CoV-2 neutralization potency because secretory IgA is dimeric (Wang et al., 2020b). Data for SARS-CoV-2 antibodies in lungs are very limited, but given the tight relationship between the lungs and the circulatory system, it is expected that blood IgG and IgA levels are reflected in the lung airways. With T cells, low SARS-CoV-2 T cell frequencies in circulation are associated with severe COVID-19, but a concern has been that the SARS-CoV-2-specific T cells may not be in blood because they redistributed to lungs. However, one early study of bronchoalveolar lavage samples from COVID-19 patients found more T cells from lungs of moderate cases compared to severe cases (Liao et al., 2020), consistent with a primary association of T cells with protection, not immunopathology. Two recent studies have also found a lack of T cells in COVID-19 bronchoalveolar lavage samples (Oja et al., 2020; Szabo et al., 2020), including a lack of SARS-CoV-2-specific CD4+ T cells with a tissue-resident phenotype (Oja et al., 2020). T cells that permanently stay in non-lymphoid tissues, such as lung, are known as tissue-resident memory T cells (Trm) (Figure 4). Presence of T cells in COVID-19 bronchoalveolar lavages was associated with younger age and survival (Szabo et al., 2020). In contrast, neutrophils are highly enriched in both blood and lungs in severe or fatal COVID-19. Thus, although much more research is needed, particularly of the URT, measurements of adaptive immunity in blood appear to be a passable reflection of the lung and oral cavity in the context of SARS-CoV-2 infection.
Heterogeneity in adaptive immunity in different people
Heterogeneity is a major factor in COVID-19 and immune responses to SARS-CoV-2. There is wide heterogeneity in COVID-19 disease severity, ranging from asymptomatic to fatal. There is heterogeneity in the innate immune responses to SARS-CoV-2, and heterogeneity has been observed in broad studies of adaptive immunity in acute patients (Mathew et al., 2020). There is an ∼1,000-fold range in the magnitude of antibody responses to SARS-CoV-2, as well as heterogeneity in the magnitude of SARS-CoV-2-specific CD4+ T cell and CD8+ T cell responses (Grifoni et al., 2020; Rydyznski Moderbacher et al., 2020). This person-to-person variation also persists in immune memory (see Immune Memory). For any given facet of immunity, it is important to keep the scale of the heterogeneity in perspective (i.e., does a specific example or scenario represent 10% of infections or 0.001% of infections?). It is worthwhile to consider some putative sources of COVID-19 heterogeneity as they relate to adaptive immunity. The human immune system is inherently diverse from person to person—the vast numbers of HLA alleles is the clearest example of this—and this high degree of immunological diversity is an evolutionary strategy of humans. Thus, there is no scenario where 100% of people respond to a viral infection in an identical manner. Pre-existing immunity may also contribute to the heterogeneity of COVID-19 in the population, which is discussed in a later section.
Age
Age is the largest risk factor for severe or fatal COVID-19. Compared to a 20-year-old, a 65-year-old individual in the United States has a 90× higher risk of death from COVID-19, and an individual 75 years old has a 200× higher risk of death (Centers for Disease Control and Prevention, 2020). Older individuals are less likely to make a coordinated adaptive immune response to SARS-CoV-2 (Rydyznski Moderbacher et al., 2020). A T cell response to any new viral infection depends on the repertoire of naive T cells. Notably, it is well characterized that the abundance of naive T cells declines substantially with age (Briceño et al., 2016; Qi et al., 2014; Wertheimer et al., 2014). COVID-19 disease severity was inversely correlated with the frequency of naive T cells (Mathew et al., 2020; Rydyznski Moderbacher et al., 2020). Together, data indicate that T cell responses are important for prevention of severe COVID-19 (Rydyznski Moderbacher et al., 2020), early T cell responses are better (Tan et al., 2020a), progression to a SARS-CoV-2 infection to severe COVID-19 is relatively slow, and the older the person is the more likely they possess a naive T cell pool insufficient to make a detectable SARS-CoV-2 T cell response or make a SARS-CoV-2 T cell response so slowly that the virus has too much of a head start to be overcome. This situation could be exacerbated by a general delay in priming of T cell responses due to early innate immune evasion by SARS (Arunachalam et al., 2020; Zhou et al., 2020b). In small animal models of SARS, old age is associated with greatly increased susceptibility to fatal infection, which was, at least in part, due to an insufficient T cell response connected to the presence of fewer professional antigen presenting cells (Sariol and Perlman, 2020).
The extreme shift in COVID-19 risk profile across age groups is likely due to multiple factors, at least some of which are not immunological. Nevertheless, the available data indicate that poor adaptive immunity to SARS-CoV-2 is at least one risk factor for the elderly. This observation has implications for COVID-19 vaccines. High risk groups such as the elderly are a major population target for any successful COVID-19 vaccine. If slow CD4+ T cell responses are a major risk factor for COVID-19 in the elderly, this is encouraging news for vaccine development, as one of the major strengths of immunization is that it removes the time component for generation of protective immunity (see Vaccines below).
Pediatric SARS-CoV-2 infections are not well understood. There are conflicting reports regarding the relative susceptibility of children to infection and the amount of virus shed by children. What is clear is that children <13 years of age generally have mild or no symptoms. Children make SARS-CoV-2 antibody responses distinct from adults (Weisberg et al., 2021). In rare cases, infected children develop MIS-C syndrome, which appears to be an autoimmune condition that develops post-infection (Consiglio et al., 2020; Gruber et al., 2020), conceptually similar to Kawasaki’s disease. MIS-C is successfully treated with anti-IL-6 and or IVIG (Gruber et al., 2020).
Gender, racial, and ethnicity effects
Men are at a somewhat greater risk than women for severe COVID-19. There are differences in the immune systems of men and women (Bunders and Altfeld, 2020). For SARS-CoV-2, no clear functional differences have been observed in adaptive immune responses to date (Bunders and Altfeld, 2020). Antibody titers are higher in men in some studies (Dan et al., 2021; Robbiani et al., 2020; Shrock et al., 2020). No significant differences in SARS-CoV-2-specific T cell responses have been observed (Dan et al., 2021; Peng et al., 2020). Notably, as described above, ∼10% of severe COVID-19 cases are in people with type I IFN autoantibodies, and strikingly, over 90% of those cases were men (Bastard et al., 2020). Thus, gender difference in severe COVID-19 may predominantly track back to type I IFN-related defects or other innate immune differences. Reduced expression of the type 1 IFN receptor gene IFNAR2 has been associated with severe COVID-19 in a genome-wide association study, consistent with an important role of early innate antiviral immunity in control of SARS-CoV-2 (Pairo-Castineira et al., 2020). In contrast, CCR2 expression, an important chemokine receptor of monocytes and macrophages, was positively associated with severe disease in the same study, consistent with a major myeloid cell component to COVID-19 lung inflammation.
Given the long history of misattribution of physiological factors or genetic factors and race, it is prudent to set a high bar for any conclusions of racial and ethnicity components to COVID-19 susceptibility. Socioeconomic factors and cultural behaviors clearly affect likelihood of acquisition of SARS-CoV-2 infection (Yehia et al., 2020). For example, people who continue to work during a lockdown are at higher risk than those who work from home. Geographical differences also exist in the COVID-19 pandemic in different countries around the globe, and it is currently unclear what the sources of those differences may be, although differences in living conditions are common. Circulation of other HCoVs has been speculated to play a possible role (see Pre-existing immunity section).
Seronegative SARS-CoV-2-experienced subjects
Among COVID-19 cases, almost all individuals become seropositive, and almost all individuals become positive for SARS-CoV-2 CD4+ T cells. The magnitude of the Spike-specific CD4+ T cell response correlates with Spike IgG (Grifoni et al., 2020; Rydyznski Moderbacher et al., 2020; Peng et al., 2020). However, at least some individuals are low or negative to commercial IgG assays, while retaining detectable SARS-CoV-2 CD4+ T cells. Current estimates are these individuals represent 1%–10% of PCR-confirmed SARS-CoV-2 infections (Gudbjartsson et al., 2020; Lipsitch et al., 2020; Nelde et al., 2021; Schwarzkopf et al., 2021; Sekine et al., 2020). This is not unlike what had been observed for MERS (Sariol and Perlman, 2020). Given that asymptomatic cases of SARS-CoV-2 infection tend to have lower Spike and nucleocapsid IgG titers (Piccoli et al., 2020), it is plausible that asymptomatic cases are enriched for individuals with undetectable antibodies by commercial serological assays but possess circulating SARS-CoV-2 T cells (one confounding factor is that many commercial serological assays have limited sensitivity for low level Spike IgG). Alternatively, some asymptomatic cases may have robust early T cell responses resulting in absence of clinical disease symptoms.
Heterogeneity in viral loads and tissue distribution
There are few studies that have simultaneously tracked viral loads and adaptive immune responses. This remains a crucial knowledge gap for understanding adaptive immunity to SARS-CoV-2 in humans. It appears that peak viral loads can vary >100,000-fold between cases (Magleby et al., 2020; Wölfel et al., 2020), and duration of viral replication can vary greatly, with shedding of viral RNA in stool samples reported from 0 to 42 days (Wang et al., 2020a) and evidence of virus in gut epithelium ∼90 days post-infection in some individuals (Gaebler et al., 2020). Viral replication in immunocompromised patients can last 100 days or more (Avanzato et al., 2020; Choi et al., 2020), not unlike what is observed for poliovirus, which can replicate in the gut of immunocompromised individuals for months in the absence of any reported clinical symptoms (Burns et al., 2014).
In addition to the URT and lungs, COVID-19 can involve the oral cavity, gut, and heart (Gaebler et al., 2020; To et al., 2020; Topol, 2020). Much more work needs to be done to understand adaptive immune responses in those tissues and how much variation there is from person to person. Last, it is unknown to what degree the heterogeneity of COVID-19 and adaptive immunity to SARS-CoV-2 is intrinsic to the virology and pathology of SARS-CoV-2, versus heterogeneity being predominantly intrinsic to the human population and would be observed for any new virulent, highly contagious respiratory virus unleashed on a large, non-immune population.
Long COVID
Post-acute sequelae are observed in a substantial fraction of symptomatic COVID-19 cases (Dennis et al., 2020; Sudre et al., 2020). These conditions are commonly called “long COVID” or “long haulers.” The most common post-acute sequelae are fatigue, reduced lung capacity, and inability to fully exercise or work. Rarer sequelae include vision problems, cognitive deficiencies, and a range of other outcomes, the frequencies of which are currently unclear. It is known that ICU care for any medical reason is associated with long-term recovery issues, including physical impairments, psychological problems, and cognitive impairments (Herridge et al., 2011; Pandharipande et al., 2013). Of note, COVID-19 post-acute sequelae are not restricted to severe COVID-19 cases (Dennis et al., 2020; Sudre et al., 2020). It is not known if these individuals experienced longer viral replication overall, longer viral replication in certain tissues, or whether some individuals had pre-existing medical conditions that became clinically fulminant upon SARS-CoV-2 infection (National Institute of Allergies and Infectious Diseases, 2020). It is also not known if these individuals have different adaptive immune responses to SARS-CoV-2. These are important areas for future biomedical research.
Pre-existing immunity
SARS-CoV-2 is a novel human pathogen. SARS-CoV-2 is a member of the coronavirus family that includes human coronaviruses (HCoVs) HCoV-OC43, HCoV-HKU1, HCoV-229E, and HCoV-NL63—betacoronaviruses and alphacoronaviruses that cause common colds. SARS-CoV-2 is relatively distantly related to those four endemic HCoVs, with <10% aa identity in the Spike RBD. As a result, cross-reactive circulating anti-Spike cross-neutralizing antibodies are rare (Amanat et al., 2020; Okba et al., 2020; Suthar et al., 2020; Tan et al., 2020b; Wec et al., 2020), as are cross-reactive Spike memory B cells (see above). In contrast, substantial cross-reactive T cell memory has been reported, measurable in ∼28%–50% of people, depending on the study (Le Bert et al., 2020; Braun et al., 2020; Grifoni et al., 2020). The majority of the SARS-CoV-2 cross-reactive T cells are CD4+ T cells (Grifoni et al., 2020). These have been demonstrated to be memory T cells and many are memory T cells to common cold coronaviruses with conserved epitopes (Mateus et al., 2020). Cross-reactive CD8+ T cells are observed less frequently (Grifoni et al., 2020), but may still be biologically relevant (Schulien et al., 2020).
The presence of cross-reactive memory T cells capable of recognizing SARS-CoV-2 in a fraction of the population is intriguing, because it opens the possibility of some degree of pre-existing immunity in the population (Sette and Crotty, 2020). Cross-reactive T cells can provide some degree of protective immunity against respiratory viral infections, as seen in the influenza H1N1 2009 pandemic (Sridhar et al., 2013; Wilkinson et al., 2012). The different ways in which such immunity may manifest for SARS-CoV-2 infection are discussed elsewhere (Lipsitch et al., 2020). Epidemiological evidence from a large cohort now supports the possibility of some degree of pre-existing immunity (Sagar et al., 2021). SARS-CoV-2 infected individuals with a laboratory confirmed HCoV infection within the previous 3 years were at significantly lower risk for ICU admission, after controlling for age and other factors. Firmer evidence will likely require pre- and post-infection T cell measurements with clear distinctions of individuals who have been exposed to SARS-CoV-2. Cross-reactive memory T cells are also of interest as it relates to COVID-19 vaccines, as people with cross-reactive T cells may respond differently to vaccines than people without such memory (Sette and Crotty, 2020).
Finally, although cross-reactive memory B cells are rare in adults, imprinting of antibody responses based on memory B cells to related viruses may impact protective immunity (Aydillo et al., 2020; Gostic et al., 2016). Nevertheless, given that most human antibodies against SARS-CoV-2 Spike have little somatic hypermutation (SHM), the majority of the SARS-CoV-2 Spike antibody response in adults appears to be a de novo response by naive B cells, discussed above.
Immune memory and protection from re-infection
There are four major components of immunological memory to viruses: antibodies, memory B cells, memory CD4+ T cells, and memory CD8+ T cells (Figure 1). In addition, subtypes of each of those memories, and local tissue immunological memory, may also be important. Immune memory is the source of protective immunity from a subsequent infection (Orenstein and Ahmed, 2017; Piot et al., 2019; Plotkin et al., 2018). There has been great trepidation that SARS-CoV-2 infection fails to induce immune memory and fails to induce protective immunity. There are several intertwined topics. Does SARS-CoV-2 induce immune memory? What kind of immune memory is or is not generated? Does the memory protect from SARS-CoV-2 re-infection? Does the memory protect from 2° COVID-19 disease? How durable is the immune memory?
In the context of SARS-CoV-2, much of the concern has come from the perspective that infection with common cold HCoVs fails to induce durable protective immunity. This is a controversial topic. Much of that conclusion derives from a particular interpretation of a human challenge study from 1990 (Callow et al., 1990). That study, by the British Common Cold Unit, was excellent work but is subject to differing interpretations. Volunteers were challenged intranasally with HCoV-229E. Ten of the 15 volunteers became infected and 8 developed clinical cold symptoms. Of note, 5 of the 15 did not become infected, and those 5 had significantly higher IgA and IgG titers. Next, the donors were recalled 11 months later and challenged intranasally again with HCoV-229E. Of the original infected group, 6 out of 9 became reinfected, thus resulting in the conclusion that immunity was short-lived. However, none of the individuals developed colds. Additionally, infection was judged based on viral shedding for at least 1 day or a change in antibody titer. Notably, the mean duration of shedding among positive subjects was 2.0 days instead of the 5.6 days seen in the first challenge. Therefore, at face value, immune memory provided sterilizing immunity in 33% of subjects, protective immunity from disease in 100% of subjects, and a 65% reduction in duration of viral shedding. Another similar, independent, study by the Common Cold Unit found 100% protection from re-infection with a homologous HCoV-229E 8–12 months later (Reed, 1984). Thus, the studies provide evidence for immune memory that at least provided protection from clinical symptoms and reduced viral shedding; this also provides support for the potential efficacy of a SARS-CoV-2 vaccine.
A 30-year longitudinal serological study provides an opportunity to estimate re-infections by HCoVs (Edridge et al., 2020). Although the study is limited to ten subjects, it is confounded by the high degree of cross-reactive nucleocapsid antibodies between common cold HCoVs, and the conclusions were that reinfections are common, the data are also open to multiple interpretations, and HCoV reinfection appears to be context-dependent. For HCoV-HKU1, a betacoronavirus like SARS-CoV-2, if one assumes that a change in nucleocapsid IgG titers should be assigned to a single viral infection with the highest titer (due to cross-reactivity), the data indicate that only a single individual experienced two HCoV-HKU1 infections, which were 19 years apart. For HCoV-OC43, the other common cold betacoronavirus, individuals appeared to experience between zero and four infections over the 20–35 years of the study. No clinical symptoms information was available, so it is unknown if the infections were symptomatic. In an extreme example, one individual appeared to have nine HCoV-NL63 infections over 21 years of study. There also remains the possibility that one or more endemic HCoVs have heterologous strains that do not cross-protect (Baumgarth et al., 2020).
Studies of immune memory to SARS-CoV, the most related coronavirus to SARS-CoV-2, are limited, but there have been notable findings. Memory T cells have been detected 17 years after SARS infection (Le Bert et al., 2020). Memory B cells were reportedly lost within 6 years (Tang et al., 2011), but neutralizing antibodies were detectable for 17 years (Tan et al., 2020b). MERS has been less well studied, but MERS Spike IgG did not persist for 2 years in mild or subclinical cases, whereas T cell memory did persist (Sariol and Perlman, 2020). T cell memory in tissues (Trm) to these viruses has not been studied in humans, but such Trms may be key players in restricting re-infection in URT and lungs (Figure 4) (Lipsitch et al., 2020; Zhao et al., 2016). Given that T cell memory has a role in protection from influenza disease severity in humans (Greenbaum et al., 2009; Sridhar et al., 2013; Wilkinson et al., 2012), and with the evidence described above for SARS-CoV-2 T cells, it seems unlikely that T cell memory would not have some protective effects against 2° COVID-19 in humans.
Immune memory to SARS-CoV-2
Does SARS-CoV-2 induce immune memory? Immunological memory is not straightforward to predict. Data from at least 6 months post-infection are needed to define the likelihood of immune memory to SARS-CoV-2 lasting for years or not. Therefore, because SARS-CoV-2 is a new virus in humans, data on immune memory to the virus are limited. The simplest feature of immune memory to measure is circulating antibodies. Data from two large (>1,000 subjects) studies—one cross-sectional and one longitudinal—have indicated that circulating SARS-CoV-2 IgG titers are well maintained for 3–4 months (Gudbjartsson et al., 2020; Wajnberg et al., 2020). A different large study has instead observed ∼25% of cases becoming seronegative over the course of 6 months (Ward et al., 2020); however, that study depended on self-administered lateral flow tests that had limited sensitivity (84%). Circulating and saliva Spike IgA titers decline faster than IgG titers early on (Isho et al., 2020). Multiple studies determined that Spike memory B cells were present at least ∼30–90 days post-infection (Juno et al., 2020; Nguyen-Contant et al., 2020; Rodda et al., 2021). Virus-specific memory B cells, antibodies, and memory T cells were detected in mild COVID-19 cases at ∼90 days post-infection (Rodda et al., 2021).
A few studies are now available that have assessed T cell and B cell memory at 6 or more months post-infection. Specifically assessing T cells at 6 months post-infection in 95 subjects, one study found ∼90% positive for memory CD4+ T cells and ∼70% positive for memory CD8+ T cells (Zuo et al., 2020a). Memory CD4+ T cells were more abundant than memory CD8+ T cells. In an independent study, using different experimental techniques, 90% of cases were positive for memory CD4+ T cells and 70% were positive for memory CD8+ T cells at 6+ months post-infection (Dan et al., 2021). The majority of the memory CD8+ T cells had a terminally differentiated effector memory cell phenotype (TEMRA) (Dan et al., 2021). The CD4+ T cell memory predominantly consists of Th1 and Tfh cells (Dan et al., 2021; Zuo et al., 2020a). Notably, the study by Zuo et al. (2020a) included a large fraction of asymptomatic subjects (44%), and still almost all of the asymptomatic cases had detectable T cell memory and only moderately lower levels than symptomatic cases. The mixed cross-sectional and longitudinal design of the study by Dan et al., 2021) estimated the durability of both CD4+ and CD8+ T cell memory to have half-lives of ∼3–5 months. Those estimates match that of the yellow fever virus vaccine (Akondy et al., 2017), which has very long-lasting protective immunity. It is possible, but not certain, that the decay of T cell memory slows over time (Akondy et al., 2017; Hammarlund et al., 2003), which would be consistent with the observation of SARS-CoV memory T cells 17 years post-infection (Le Bert et al., 2020).
Memory B cells have been assessed at 6 months post-COVID-19 in multiple studies (Dan et al., 2021; Gaebler et al., 2020). In a cohort of 188 cases, memory B cells specific for Spike, RBD, and nucleocapsid were each detectable in 100% of subjects at 6+ months post-infection (Dan et al., 2021). Almost all of the Spike-specific memory B cells were IgG, with only ∼5% IgA representation (Dan et al., 2021). IgG RBD memory B cells were also consistently found in an independent cohort (Gaebler et al., 2020). Notably, frequencies of RBD (and Spike) memory B cells increased over time, with more RBD memory B cells at 6 months PSO than at 1 month PSO (Dan et al., 2021; Gaebler et al., 2020), consistent with observations at 3 months PSO (Rodda et al., 2021). In addition to increased frequencies of RBD memory B cells at 6 months PSO, the cells had undergone affinity maturation and expressed higher potency neutralizing antibodies (Gaebler et al., 2020), consistent with extended germinal center responses after a viral infection (Cyster and Allen, 2019).
The study by (Dan et al., 2021) (by authors of this review) is notable for including memory B cells, memory CD4+ T cells, memory CD8+ T cells, and antibodies. There are very few studies comparing all four of those compartments of immune memory to a viral infection in the same individuals; this was by far the largest such study. The five measurements of RBD IgG, Spike IgA, RBD-specific memory B cells, SARS-CoV-2-specific CD4+ T cells, and SARS-CoV-2-specific CD8+ T cells were used as metrics to gauge the quality of immune memory to SARS-CoV-2, due to their potential roles in protective immunity from 2° COVID-19. The majority of COVID-19 cases were positive for all five of those immune memory compartments at ∼1 month PSO. By 5+ months PSO, ∼95% of individuals were still positive for at least three out of five SARS-CoV-2 immune memory compartments (Dan et al., 2021). Of note, heterogeneity is a defining feature of COVID-19 immune memory. Virus-specific antibody, memory B cell, memory CD4+ T cell, and memory CD8+ T cells spanned large ranges between individuals, and changed with differing patterns over time. Antibody titers were not a surrogate indicator of the magnitude of memory T cells (Dan et al., 2021), suggesting that simple antibody serodiagnostic tests will not be a robust indicator of protective immunity in people previously infected with SARS-CoV-2. Much more data on immune memory are expected in the coming months, but data so far indicate that T cell memory, B cell memory, and antibodies are all likely to persist for years in most individuals infected by SARS-CoV-2 (Dan et al., 2021; Gaebler et al., 2020; Wajnberg et al., 2020; Zuo et al., 2020a).
Protection from re-infection or 2° COVID-19
In the context of re-infection, immune memory can be present, thus the initial race between virus and priming of adaptive immunity is removed from the equation. Sterilizing immunity against viruses can only be accomplished by high-titer neutralizing antibodies, and it has been demonstrated that passive transfer of neutralizing antibodies in advance of SARS-CoV-2 infection (mimicking the conditions of 2° infection) can effectively prevent or limit URT infection, lung infection, and symptomatic disease in animal models (Baum et al., 2020; Rogers et al., 2020; Zost et al., 2020). Nevertheless, successful protection against clinical disease can be accomplished by other forms of adaptive immune memory. The relatively slow course of severe COVID-19 in humans (median 19 days PSO for fatal cases) (Zhou et al., 2020a) leaves open the reasonable possibility that protective immunity against symptomatic or severe 2° COVID-19 may very well involve memory compartments such as circulating memory T and B cells (Altmann and Boyton, 2020; Baumgarth et al., 2020). Memory B and T cells can take several days to reactivate and generate recall responses. Mechanisms of protection can vary based on the kinetics of the infection. For example, clinical hepatitis after hepatitis B virus (HBV) infection is prevented by immune memory even without vaccine-elicited circulating antibodies, because of the relatively slow course of HBV disease (Van Damme and Van Herck, 2007; Rosado et al., 2011).
Direct evidence of protection from re-infection first came from two nonhuman primate studies. Animals infected with SARS-CoV-2 were protected from a high dose re-challenge with SARS-CoV-2 1 month later, and protection was associated with neutralizing antibodies (Chandrashekar et al., 2020; Deng et al., 2020). A shortcoming of each of those reports is that the time passed between the initial infection and the re-challenge was short, thus the data do not speak to how durable protective immunity is against SARS. Additionally, although the current nonhuman primate models are quite valuable, infection requires high doses of SARS-CoV-2, and the disease kinetics are relatively short and mild. An important new nonhuman primate study has shown that depletion of CD8+ T cells reduces protection from re-infection in the URT (McMahan et al., 2020). This is the first direct evidence for a role of CD8+ T cells in protective immunity from COVID-19, and it likely reflects the presence of CD8+ Trm cells in nasal passages that developed in response to the primary infection. It is notable that the impact of the CD8+ T cells was observed in the URT by day 1 post-infection. Additionally, depletion of CD8+ T cells had essentially no effect on protection of lungs; few animals had even briefly detectable virus in lungs, even after intratracheal administration of SARS-CoV-2, indicating that other components of adaptive immune memory suffice to control virus in lungs of previously infected animals.
Direct evidence of protection of humans from 2° COVID-19 thus far comes primarily from two studies. An ∼118-day study in the United Kingdom of ∼2,800 individuals observed no symptomatic re-infections (Wyllie et al., 2020), whereas an ∼1,200 person study observed no symptomatic re-infections over 6 months (Lumley et al., 2020). Although SARS-CoV-2-specific T cells and antibodies were observed in those studies, specific correlates of protection can only be established by studies observing a significant number of re-infections over time. There are anecdotal examples of SARS-CoV-2 re-infection, but unfortunately there is currently insufficient information to tell how rare re-infections are and whether they occur in individuals with detectable immune memory. For most viral infections that elicit immunological memory, protective immunity from re-infection—or at least re-infection resulting in clinical disease—lasts for multiple years or more in most individuals.
SARS-CoV-2 variants
SARS-CoV-2 genetic variation has been a topic of intense interest. Whether SARS-CoV-2 will be able to exhibit sufficient genetic flexibility to escape humoral immune responses in the near term is unclear. Many RNA viruses, such as measles and polioviruses, exhibit antigenic stability and unchanging serotypes over periods of many years. As a result, the measles and polio vaccines remain highly effective now, 70 years after they were first introduced. Although the situation for any coronavirus is unclear, it is highly unlikely that SARS-CoV-2 mutations would escape T cell immunity, because a very broad array of SARS-CoV-2 epitopes are recognized in humans with COVID-19 (Grifoni et al., 2020) consisting of CD4+ and CD8+ T cell responses to >10 epitopes distributed throughout the SARS-CoV-2 genome, which vary from person to person (Tarke et al., 2020). For antibody responses, SARS-CoV-2 mutations exist that could affect individual neutralizing antibody epitopes. However, a key attribute of the neutralization epitopes on SARS-CoV-2 Spike is that the surface area on RBD that is targeted by neutralizing antibodies is large enough that no single viral mutation is expected to avoid neutralization by polyclonal human serum (Li et al., 2020a; Thomson et al., 2020). This is consistent with the broad range of SARS-CoV-2 neutralizing antibodies isolated from humans, indicating that SARS-CoV-2 is a relatively easy neutralization target that elicits a diverse array of antibodies in each person (see Antibodies and B cells section, above). The Spike D614G variant that is now globally common (that binds ACE2 more tightly and is more transmissible) (Hou et al., 2020; Korber et al., 2020) is neutralized by plasma from subjects infected with the original D614 virus (Korber et al., 2020). The Spike N439 variant has enhanced binding to ACE2 but has been shown to still be robustly neutralized by serum from the vast majority of COVID-19 patients (Thomson et al., 2020). Thus, although it is important to track SARS-CoV-2 evolution, it is unlikely that the virus will be able to evolve escape variants that avoid the majority of humoral and cellular immune memory in COVID-19 cases or COVID-19 vaccine recipients soon.
Vaccine-elicited immunity
Immune memory is the source of protection by almost all vaccines, and thus COVID-19 vaccine development is closely tied to the topic of immunological memory. Immune memory and immunological features of protective immunity to SARS-CoV-2 are discussed above. Recent excellent reviews summarized the status of the major COVID-19 candidate vaccines and vaccine trials around the world (Klasse et al., 2020; Krammer, 2020), and interim results from the Pfizer/BioNtech RNA COVID-19 and Moderna vaccine phase 3 clinical trials have subsequently been released. An ideal COVID-19 vaccine would elicit long-lasting high titer neutralizing antibody titers and would provide sterilizing immunity to prevent disease and onward transmission. Even if that is not accomplished, a vaccine could still be highly effective at preventing serious COVID-19 disease. If the neutralizing antibody titers are sufficient to blunt the size of the viral inoculum, the presence of memory T cells may control the infection. The working model described above (Figure 2), with severe COVID-19 cases being associated with a failure to make a T cell response fast enough during natural infection with SARS-CoV-2, is good news for vaccines, because vaccines overcome/bypass the speed problem of adaptive immunity and T cell responses. Priming of the immune system by a vaccine happens well in advance of virus exposure. Additionally, the findings that Spike is a good target for CD4+ T cell responses, Tfh cell responses, and CD8+ T cell responses in SARS-CoV-2-infected people is good news for vaccine development (Grifoni et al., 2020; Juno et al., 2020; Rydyznski Moderbacher et al., 2020; Peng et al., 2020), because almost all COVID-19 vaccines target Spike only.
Although lung infection is a major component of severe COVID-19 (and relatively slow), URT infection is important for transmission. Notably, a vaccine that can prevent severe disease, or even most URT symptomatic diseases, would not necessarily prevent transmission of virus. For example, the current pertussis vaccine prevents clinical disease but not infection, and probably not transmission (Warfel et al., 2014), and much SARS-CoV-2 transmission occurs early, during the pre-symptomatic phase (He et al., 2020). Several non-human primate COVID-19 vaccine studies are consistent with the possibility of COVDI-19 vaccines preventing severe disease in humans but possibly not preventing URT infection (Corbett et al., 2020; van Doremalen et al., 2020; Gao et al., 2020; Guebre-Xabier et al., 2020; Mercado et al., 2020; Tostanoski et al., 2020; Vogel et al., 2020; Yu et al., 2020). It is plausible that SARS-CoV-2 infection may elicit better protective immunity in the URT than any of the major current COVID-19 vaccine candidates, because infection occurs at that site and is therefore more likely to elicit tissue-resident memory. Tissue-resident T cells were relevant for protective immunity in a SARS mouse model (Zhao et al., 2016) and B. pertussis infection versus pertussis immunization (Kapil and Merkel, 2019), but more needs to be learned about local immunity to SARS-CoV-2. A number of human vaccines against respiratory pathogens do not depend on local T cell memory, or are very unlikely to elicit URT T cell memory, such as measles, smallpox, and flu vaccines, as well as RSV vaccines in clinical trials.
The elderly present particular and important challenges for COVID-19 vaccines. Older individuals are at much higher risk for severe COVID-19. This risk is surely multifactorial, but it appears that one component of the risk is poor T cell responses due in part to a more limited naive T cell repertoire (Rydyznski Moderbacher et al., 2020). COVID-19 vaccines may overcome this problem. One key feature of vaccines is that immunization occurs well in advance of infection, giving the adaptive immune system time to respond, expand, and mature. These factors highlight the likely importance of measuring T cell responses to COVID-19 vaccines in the elderly.
Interim phase 3 vaccine trial results
The recent interim results from the Pfizer/BioNtech phase 3 trial were extraordinary news (November 9th). Although it was an interim analysis, the vaccine had 95% efficacy at preventing disease, with 170 COVID-19 cases for the U.S. Food and Drug Administration (FDA) to evaluate (94 cases in the initial release) (U.S. Food and Drug Administration, 2020b). The vaccine is also safe, having been injected into over 20,000 people, with two doses (>40,000 immunizations) and no serious safety concerns (U.S. Food and Drug Administration, 2020b). That is consistent with the safety of the phase 1 vaccine trial (Walsh et al., 2020). The vaccine (BNT162b2) is an RNA vaccine, delivered in a lipid nanoparticle, with the RNA encoding a full-length (membrane anchored) SARS-CoV-2 Spike (Walsh et al., 2020) with trimer stabilization mutations (P-P) (Pallesen et al., 2017; Wrapp et al., 2020). The Moderna RNA vaccine mRNA-1273 is also a full-length (membrane-anchored) SARS-CoV-2 Spike with P-P trimer stabilization mutations.
BNT162b2 vaccine protection was reported based on symptomatic cases, not number of infections. This endpoint was decided on because preventing systemic disease is the primary goal of COVID-19 vaccines, and it would be extraordinarily challenging to check 40,000 people for SARS2 infections continuously for months. This clinical trial (and others) tracked self-reported symptoms, and subjects with symptoms were then tested to confirm SARS-CoV-2 infection. Some observers were surprised that >90% vaccine efficacy was possible, but many vaccines are close to 100% effective, including vaccines against respiratory pathogens such as smallpox and measles.
In phase 1 trials, the Pfizer BNT162b2 COVID-19 vaccine elicited neutralizing antibody titers somewhat better than SARS-CoV-2 natural infection, over the short term (Walsh et al., 2020). T cell data were not reported for BNT162b2, but phase 1 trial data for the related vaccine BNT162b1 (encoding only the Spike RBD domain instead of full-length Spike) showed robust CD4+ T cell and CD8+ T cell responses, with the CD4+ T cell response largely consisting of Th1 cells (Sahin et al., 2020). Immune responses in the elderly are another major topic of interest. The Pfizer BNT162b2 vaccine has been tested for immunogenicity in individuals >65 years old, and the results were limited but encouraging. Antibody responses were similar in both age groups (T cell data were not reported) (Walsh et al., 2020). Interim vaccine efficacy phase 3 trials results indicated equivalent protection in older and younger age groups (94% and 95%) (U.S. Food and Drug Administration, 2020b).
Interim results from the Moderna RNA vaccine (mRNA-1273, developed in collaboration with the NIH Vaccine Research Center) phase 3 trial were released 1 week after Pfizer (November 16th). 94% efficacy at protection from COVID-19 was reported, with 196 total cases to evaluate in the FDA review (Baden et al., 2020). There were 30 severe cases of COVID-19 in the trial, all of which were in the placebo group, indicated the Moderna vaccine works very well at preventing severe COVID-19 (Baden et al., 2020). Overall, the interim results from the two COVID-19 RNA vaccine trials were virtually identical, with 94% and 95% efficacy and similar other outcomes. The safety profile of the two vaccines is also excellent, with a combined >70,000 doses administered and no serious adverse events. Both vaccines received Emergency Use Authorization from the FDA in December.
The interim phase 3 trial results were very encouraging, and the biggest unknown now is probably the durability of the vaccine-induced immunity. Because there is no licensed RNA vaccine, no clear reference point exists for how durable immunity will be for this vaccine. Are the antibodies durable? Is the T cell memory durable? Is the B cell memory durable? Those are all important questions, and it will take time to answer them. Two RNA vaccine studies encoding other antigens did not see durable antibody titers in humans or non-human primates (Feldman et al., 2019; Monslow et al., 2020). However, those studies were with other antigens and different vaccine formulations. A first look at antibody titers 90 days after the 2nd immunization with Moderna mRNA-1273 indicated a robust maintenance of RBD IgG titers, which is an encouraging early sign (Widge et al., 2021).
Prevention of transmission is also an important topic. The Pfizer and Moderna clinical trials were not designed to test this, but in the Moderna trial there were swab tests on the day of the second immunization. There were substantially fewer asymptomatic infections detected in the vaccinated group after a single immunization (67% reduced) (U.S. Food and Drug Administration, 2020a). It will be important to gather much more extensive data, but the early observation is encouraging. In both the Pfizer and Moderna clinical trials, a subset of trial participants will be tested for antibodies against Nucleocapsid, which will generally reveal whether study subjects have become infected with SARS-CoV-2 after immunization.
Interim phase 3 study results for the AstraZeneca/Oxford ChAdOx1 were also released in November. Per protocol, efficacy against COVID-19 was 62% and overall efficacy was 70% (131 total cases) (Voysey et al., 2021). Efficacy against asymptomatic SARS-CoV-2 infection was 27%. The vaccine also had only moderate efficacy in a nonhuman primate protection study (van Doremalen et al., 2020), and this vaccine does not incorporate a stabilized Spike. Interim efficacy was 90% in a subgroup of the clinical trial that inadvertently received a lower priming dose, which requires future investigation (Voysey et al., 2021).
Finally, the interim phase 3 vaccine trial results are also encouraging for other major COVID-19 candidate vaccines. Multiple candidate vaccines had similar immunogenicity profiles to the COVID-19 RNA vaccines in humans, with equivalent or better neutralizing antibody titers or CD4+ T cell responses (Folegatti et al., 2020; Jackson et al., 2020; Keech et al., 2020; Zhu et al., 2020). Th1 and Tfh cell response were elicited to the Moderna/VRC RNA vaccine (Corbett et al., 2020). CD8+ T cell responses to candidate COVID-19 vaccines in humans or non-human primates are either largely absent or not measured in the current literature (e.g., the CanSino Ad5 vector-based vaccine did not distinguish between CD4+ and CD8+ T cells by ELISPOT) (Zhu et al., 2020). Multiple major candidate vaccines had efficacy at short-term protection in pre-clinical models, including the J&J Ad26 vector-based vaccine (Corbett et al., 2020; van Doremalen et al., 2020; Guebre-Xabier et al., 2020; Mercado et al., 2020). In sum, there is no specific reason not to expect multiple other candidate COVID-19 vaccines to do well in humans now; although, of course, vaccine phase 3 clinical trial outcomes are notoriously difficult to predict. For the moment, it is a phenomenal accomplishment for the world to go from nothing to multiple vaccines with ∼95% efficacy signal within a calendar year. That is a first-ever in human history.
Conclusion
Although there is much more to be learned about adaptive immune responses to SARS-CoV-2 and their relationships to disease severity, immune memory, protection, and vaccines, an extraordinary amount has been accomplished during 2020. Studies of antigen-specific CD4+ T cells, CD8+ T cells, and antibodies together in larger cohorts of acute patients, representing a range of disease severity, are needed to further understand mechanisms of protective adaptive immune responses to COVID-19. Big unknowns remain about variable kinetics of viral loads and duration of infection and the connections between those parameters and adaptive immunity and immune memory. Understanding heterogeneous disease manifestations of COVID-19 remains a major knowledge gap, and exploring relationships between those phenomena and adaptive immunity is a priority. Additionally, duration of immune memory and protective immunity to SARS-CoV-2 after COVID-19 and in response to COVID-19 vaccines will be a high priority for years to come.
Acknowledgments
We thank Dennis Burton, Ananda Goldrath, Matthew Pipkin, Mitch Kronenberg, and other colleagues for feedback on a draft manuscript. We thank Christina Corbaci for graphic arts. This work was funded by the NIH NIAID under award AI142742 (Cooperative Centers for Human Immunology [CCHI]) and was additionally supported in part by La Jolla Institute for Immunology Institutional Funds and an anonymous donor.
Declaration of interests
A.S. is a consultant for Gritstone, Flow Pharma, Merck, Epitogenesis, Gilead, and Avalia. S.C. is a consultant for Avalia. LJI has filed for patent protection for various aspects of T cell epitope and vaccine design work.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.01.007.
Supplemental information
(A) Conceptual schematics of viral loads, elaborated to represent both URT and lungs.
(B) Innate immune responses, elaborated to represent both blood (serum/plasma) cytokines/chemokines and local tissue innate immune responses, in an average case of COVID-19.
(C) T cell responses, elaborated to individually show virus-specific CD4+ T cells and CD8+ T cells, in an average case of COVID-19.
(D) Antibody responses, elaborated to show binding antibodies and neutralizing antibodies, in an average case of COVID-19.
References
- Aid M., Busman-Sahay K., Vidal S.J., Maliga Z., Bondoc S., Starke C., Terry M., Jacobson C.A., Wrijil L., Ducat S., et al. Vascular Disease and Thrombosis in SARS-CoV-2-Infected Rhesus Macaques. Cell. 2020;183:1354–1366.e13. doi: 10.1016/j.cell.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy R.S., Fitch M., Edupuganti S., Yang S., Kissick H.T., Li K.W., Youngblood B.A., Abdelsamed H.A., McGuire D.J., Cohen K.W., et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 2017;552:362–367. doi: 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann D.M., Boyton R.J. SARS-CoV-2 T cell immunity: Specificity, function, durability, and role in protection. Sci. Immunol. 2020;5:eabd6160. doi: 10.1126/sciimmunol.abd6160. [DOI] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. medRxiv. 2020 doi: 10.1101/2020.11.06.20227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J., et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell. 2020;183:1901–1912.e9. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydillo T., Rombauts A., Stadlbauer D., Aslam S., Abelenda-Alonso G., Escalera A., Amanat F., Jiang K., Krammer F., Carratala J., et al. Antibody Immunological Imprinting on COVID-19 Patients. medRxiv. 2020 doi: 10.1101/2020.10.14.20212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., Sahly H.M.E., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2035389. Published online December 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. HGID Lab. NIAID-USUHS Immune Response to COVID Group. COVID Clinicians. COVID-STORM Clinicians. Imagine COVID Group. French COVID Cohort Study Group. Milieu Intérieur Consortium. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N., Nikolich-Žugich J., Lee F.E.-H., Bhattacharya D. Antibody Responses to SARS-CoV-2: Let’s Stick to Known Knowns. J. Immunol. 2020;205:2342–2350. doi: 10.4049/jimmunol.2000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhnia M.R.-E.-I., McCausland M.M., Moyron J., Laudenslager J., Granger S., Rickert S., Koriazova L., Kubo R., Kato S., Crotty S. Vaccinia virus extracellular enveloped virion neutralization in vitro and protection in vivo depend on complement. J. Virol. 2009;83:1201–1215. doi: 10.1128/JVI.01797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Briceño O., Lissina A., Wanke K., Afonso G., von Braun A., Ragon K., Miquel T., Gostick E., Papagno L., Stiasny K., et al. Reduced naïve CD8(+) T-cell priming efficacy in elderly adults. Aging Cell. 2016;15:14–21. doi: 10.1111/acel.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz V.R., Busch D.H. Back to the Future: Effector Fate during T Cell Exhaustion. Immunity. 2019;51:970–972. doi: 10.1016/j.immuni.2019.11.007. [DOI] [PubMed] [Google Scholar]
- Buggert M., Nguyen S., McLane L.M., Steblyanko M., Anikeeva N., Paquin-Proulx D., Del Rio Estrada P.M., Ablanedo-Terrazas Y., Noyan K., Reuter M.A., et al. Limited immune surveillance in lymphoid tissue by cytolytic CD4+ T cells during health and HIV disease. PLoS Pathog. 2018;14:e1006973. doi: 10.1371/journal.ppat.1006973. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bunders M.J., Altfeld M. Implications of sex differences in immunity for SARS-CoV-2 pathogenesis and design of therapeutic interventions. Immunity. 2020;53:487–495. doi: 10.1016/j.immuni.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C.C., Diop O.M., Sutter R.W., Kew O.M. Vaccine-derived polioviruses. J. Infect. Dis. 2014;210(Suppl 1):S283–S293. doi: 10.1093/infdis/jiu295. [DOI] [PubMed] [Google Scholar]
- Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A.J. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2020. Cases, Data, and Surveillance | CDC.https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/index.html [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lau Y.F., Lamirande E.W., Paddock C.D., Bartlett J.H., Zaki S.R., Subbarao K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J. Virol. 2010;84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2029849. Published online October 28, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.-H., Boucau J., Bowman K., et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consiglio C.R., Cotugno N., Sardh F., Pou C., Amodio D., Rodriguez L., Tan Z., Zicari S., Ruggiero A., Pascucci G.R., et al. CACTUS Study Team The Immunology of Multisystem Inflammatory Syndrome in Children with COVID-19. Cell. 2020;183:968–981.e7. doi: 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. A brief history of T cell help to B cells. Nat. Rev. Immunol. 2015;15:185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster J.G., Allen C.D.C. B Cell Responses: Cell Interaction Dynamics and Decisions. Cell. 2019;177:524–540. doi: 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Faliti C.E., Ramirez S.I., Frazier A., Yu E.D., Grifoni A., Rawlings S.A., et al. Immunological memory to SARS-CoV-2 assessed for up to eight months after infection. Science. 2021:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.-H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Bao L., Liu J., Xiao C., Liu J., Xue J., Lv Q., Qi F., Gao H., Yu P., et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis A., Wamil M., Kapur S., Alberts J., Badley A.D., Decker G.A., Rizza S.A., Banerjee R., Banerjee A. Multi-organ impairment in low-risk individuals with long COVID. medRxiv. 2020 doi: 10.1101/2020.10.14.20212555. [DOI] [Google Scholar]
- Dudakov J.A., Hanash A.M., van den Brink M.R.M. Interleukin-22: immunobiology and pathology. Annu. Rev. Immunol. 2015;33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Elsayed H., Nabi G., McKinstry W.J., Khoo K.K., Mak J., Salazar A.M., Tenbusch M., Temchura V., Überla K. Intrastructural Help: Harnessing T Helper Cells Induced by Licensed Vaccines for Improvement of HIV Env Antibody Responses to Virus-Like Particle Vaccines. J. Virol. 2018;92:e00141-18. doi: 10.1128/JVI.00141-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman R.A., Fuhr R., Smolenov I., Mick Ribeiro A., Panther L., Watson M., Senn J.J., Smith M., Almarsson Ӧ., Pujar H.S., et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., et al. Oxford COVID Vaccine Trial Group Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Ladinsky M., Cho A., Jankovic M., Schaefer-Babajew D., et al. Evolution of Antibody Immunity to SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.11.03.367391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I.E., Rovina N., Lampropoulou V., Triantafyllia V., Manioudaki M., Pavlos E., Koukaki E., Fragkou P.C., Panou V., Rapti V., et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostic K.M., Ambrose M., Worobey M., Lloyd-Smith J.O. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 2016;354:722–726. doi: 10.1126/science.aag1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J.A., Kotturi M.F., Kim Y., Oseroff C., Vaughan K., Salimi N., Vita R., Ponomarenko J., Scheuermann R.H., Sette A., Peters B. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc. Natl. Acad. Sci. USA. 2009;106:20365–20370. doi: 10.1073/pnas.0911580106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber C.N., Patel R.S., Trachtman R., Lepow L., Amanat F., Krammer F., Wilson K.M., Onel K., Geanon D., Tuballes K., et al. Mapping Systemic Inflammation and Antibody Responses in Multisystem Inflammatory Syndrome in Children (MIS-C) Cell. 2020;183:982–995.e14. doi: 10.1016/j.cell.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., Arnthorsson A.O., Helgason D., Bjarnadottir K., Ingvarsson R.F., et al. Humoral Immune Response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guebre-Xabier M., Patel N., Tian J.-H., Zhou B., Maciejewski S., Lam K., Portnoff A.D., Massare M.J., Frieman M.B., Piedra P.A., et al. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. bioRxiv. 2020 doi: 10.1101/2020.08.18.256578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund E., Lewis M.W., Hansen S.G., Strelow L.I., Nelson J.A., Sexton G.J., Hanifin J.M., Slifka M.K. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Herridge M.S., Tansey C.M., Matté A., Tomlinson G., Diaz-Granados N., Cooper A., Guest C.B., Mazer C.D., Mehta S., Stewart T.E., et al. Canadian Critical Care Trials Group Functional disability 5 years after acute respiratory distress syndrome. N. Engl. J. Med. 2011;364:1293–1304. doi: 10.1056/NEJMoa1011802. [DOI] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020:1–14. doi: 10.1038/s41579-020-00459-7. Published online October 6, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5:eabe5511. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. mRNA-1273 Study Group An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juno J.A., Tan H.-X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- Kapil P., Merkel T.J. Pertussis vaccines and protective immunity. Curr. Opin. Immunol. 2019;59:72–78. doi: 10.1016/j.coi.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse P.J., Nixon D., Moore J.P. Immunogenicity of clinically relevant SARS-CoV-2 vaccines in non-human primates and humans. Preprints 2020. 2020 doi: 10.1126/sciadv.abe8065. 2020090166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Lewinsohn D.M., Leonard M.K., LoBue P.A., Cohn D.L., Daley C.L., Desmond E., Keane J., Lewinsohn D.A., Loeffler A.M., Mazurek G.H., et al. Official American Thoracic Society/Infectious Diseases Society of America/Centers for Disease Control and Prevention Clinical Practice Guidelines: Diagnosis of Tuberculosis in Adults and Children. Clin. Infect. Dis. 2017;64:111–115. doi: 10.1093/cid/ciw778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Jiang L., Li X., Lin F., Wang Y., Li B., Jiang T., An W., Liu S., Liu H., et al. Clinical and pathological investigation of patients with severe COVID-19. JCI Insight. 2020;5:e138070. doi: 10.1172/jci.insight.138070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Lipsitch M., Grad Y.H., Sette A., Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.-W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies directed to multiple epitopes on SARS-CoV-2 spike. bioRxiv. 2020 doi: 10.1101/2020.06.17.153486. [DOI] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Lu L.L., Suscovich T.J., Fortune S.M., Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumley S.F., O’Donnell D., Stoesser N.E., Matthews P.C., Howarth A., Hatch S.B., Marsden B.D., Cox S., James T., Warren F., et al. Antibodies to SARS-CoV-2 are associated with protection against reinfection. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2034545. [DOI] [Google Scholar]
- Lundgren J.D., Grund B., Barkauskas C.E., Holland T.L., Gottlieb R.L., Sandkovsky U., Brown S.M., Knowlton K.U., Self W.H., Files D.C., et al. ACTIV-3/TICO LY-CoV555 Study Group A Neutralizing Monoclonal Antibody for Hospitalized Patients with Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2033130. Published online December 22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magleby R., Westblade L.F., Trzebucki A., Simon M.S., Rajan M., Park J., Goyal P., Safford M.M., Satlin M.J. Impact of SARS-CoV-2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients with Coronavirus Disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa851. Published online June 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D., Soerens A.G. Tissue-Resident T Cells and Other Resident Leukocytes. Annu. Rev. Immunol. 2019;37:521–546. doi: 10.1146/annurev-immunol-042617-053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2020 doi: 10.1038/s41586-020-03041-6. Published online December 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell. 2020;183:1340–1353.e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monslow M.A., Elbashir S., Sullivan N.L., Thiriot D.S., Ahl P., Smith J., Miller E., Cook J., Cosmi S., Thoryk E., et al. Immunogenicity generated by mRNA vaccine encoding VZV gE antigen is comparable to adjuvanted subunit vaccine and better than live attenuated vaccine in nonhuman primates. Vaccine. 2020;38:5793–5802. doi: 10.1016/j.vaccine.2020.06.062. [DOI] [PubMed] [Google Scholar]
- Montero-Escribano P., Matías-Guiu J., Gómez-Iglesias P., Porta-Etessam J., Pytel V., Matias-Guiu J.A. Anti-CD20 and COVID-19 in multiple sclerosis and related disorders: A case series of 60 patients from Madrid, Spain. Mult. Scler. Relat. Disord. 2020;42:102185. doi: 10.1016/j.msard.2020.102185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morou A., Brunet-Ratnasingham E., Dubé M., Charlebois R., Mercier E., Darko S., Brassard N., Nganou-Makamdop K., Arumugam S., Gendron-Lepage G., et al. Altered differentiation is central to HIV-specific CD4+ T cell dysfunction in progressive disease. Nat. Immunol. 2019;20:1059–1070. doi: 10.1038/s41590-019-0418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugesan K., Jagannathan P., Pham T.D., Pandey S., Bonilla H.F., Jacobson K., Parsonnet J., Andrews J.R., Weiskopf D., Sette A., et al. Interferon-gamma release assay for accurate detection of SARS-CoV-2 T cell response. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1537. Published online October 9, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Allergies and Infectious Diseases . 2020. Workshop on Post-Acute Sequelae of COVID-19.https://web.cvent.com/event/cf41e3b5-04e7-4e09-b25d-f33dfcd16fed/summary [Google Scholar]
- Neidleman J., Luo X., Frouard J., Xie G., Gill G., Stein E.S., McGregor M., Ma T., George A.F., Kosters A., et al. SARS-CoV-2-Specific T Cells Exhibit Phenotypic Features of Helper Function, Lack of Terminal Differentiation, and High Proliferation Potential. Cell Rep Med. 2020;1:100081. doi: 10.1016/j.xcrm.2020.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. MBio. 2020;11:e01991-20. doi: 10.1128/mBio.01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novi G., Mikulska M., Briano F., Toscanini F., Tazza F., Uccelli A., Inglese M. COVID-19 in a MS patient treated with ocrelizumab: does immunosuppression have a protective role? Mult. Scler. Relat. Disord. 2020;42:102120. doi: 10.1016/j.msard.2020.102120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oja A.E., Saris A., Ghandour C.A., Kragten N.A.M., Hogema B.M., Nossent E.J., Heunks L.M.A., Cuvalay S., Slot E., Linty F., et al. Divergent SARS-CoV-2-specific T- and B-cell responses in severe but not mild COVID-19 patients. Eur. J. Immunol. 2020;50:1998–2012. doi: 10.1002/eji.202048908. [DOI] [PubMed] [Google Scholar]
- Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., et al. Severe Acute Respiratory Syndrome Coronavirus 2−Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oran D.P., Topol E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection : A Narrative Review. Ann. Intern. Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein W.A., Ahmed R. Simply put: Vaccination saves lives. Proc. Natl. Acad. Sci. USA. 2017;114:4031–4033. doi: 10.1073/pnas.1704507114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. GenOMICC Investigators. ISARICC Investigators. COVID-19 Human Genetics Initiative. 23andMe Investigators. BRACOVID Investigators. Gen-COVID Investigators Genetic mechanisms of critical illness in Covid-19. Nature. 2020 doi: 10.1038/s41586-020-03065-y. Published online December 11, 2020. [DOI] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandharipande P.P., Girard T.D., Jackson J.C., Morandi A., Thompson J.L., Pun B.T., Brummel N.E., Hughes C.G., Vasilevskis E.E., Shintani A.K., et al. BRAIN-ICU Study Investigators Long-term cognitive impairment after critical illness. N. Engl. J. Med. 2013;369:1306–1316. doi: 10.1056/NEJMoa1301372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeples L. News Feature: Avoiding pitfalls in the pursuit of a COVID-19 vaccine. Proc. Natl. Acad. Sci. USA. 2020;117:8218–8221. doi: 10.1073/pnas.2005456117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Oxford Immunology Network Covid-19 Response T cell Consortium. ISARIC4C Investigators Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone L., Petruccioli E., Vanini V., Cuzzi G., Fard S.N., Alonzi T., Castilletti C., Palmieri F., Gualano G., Vittozzi P., et al. A whole blood test to measure SARS-CoV-2 specific response in COVID-19 patients. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.09.051. Published online October 10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piot P., Larson H.J., O’Brien K.L., N’kengasong J., Ng E., Sow S., Kampmann B. Immunization: vital progress, unfinished agenda. Nature. 2019;575:119–129. doi: 10.1038/s41586-019-1656-7. [DOI] [PubMed] [Google Scholar]
- Plotkin S., Orenstein W., Offit P. Seventh Edition. Elsevier; 2018. Plotkin’s Vaccines. [Google Scholar]
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q., Liu Y., Cheng Y., Glanville J., Zhang D., Lee J.-Y., Olshen R.A., Weyand C.M., Boyd S.D., Goronzy J.J. Diversity and clonal selection in the human T-cell repertoire. Proc. Natl. Acad. Sci. USA. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radermecker C., Detrembleur N., Guiot J., Cavalier E., Henket M., d’Emal C., Vanwinge C., Cataldo D., Oury C., Delvenne P., Marichal T. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J. Exp. Med. 2020;217:e20201012. doi: 10.1084/jem.20201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S.E. The behaviour of recent isolates of human respiratory coronavirus in vitro and in volunteers: evidence of heterogeneity among 229E-related strains. J. Med. Virol. 1984;13:179–192. doi: 10.1002/jmv.1890130208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger T.J., Uhrlaub J.L., Watanabe M., Wong R., Castaneda Y., Pizzato H.A., Thompson M.R., Bradshaw C., Weinkauf C.C., Bime C., et al. Orthogonal SARS-CoV-2 Serological Assays Enable Surveillance of Low-Prevalence Communities and Reveal Durable Humoral Immunity. Immunity. 2020;53:925–933.e4. doi: 10.1016/j.immuni.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado M.M., Scarsella M., Pandolfi E., Cascioli S., Giorda E., Chionne P., Madonne E., Gesualdo F., Romano M., Ausiello C.M., et al. Switched memory B cells maintain specific memory independently of serum antibodies: the hepatitis B example. Eur. J. Immunol. 2011;41:1800–1808. doi: 10.1002/eji.201041187. [DOI] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi F., Nourbakhsh B., Azimi A.R. B-cell depleting therapies may affect susceptibility to acute respiratory illness among patients with Multiple Sclerosis during the early COVID-19 epidemic in Iran. Mult. Scler. Relat. Disord. 2020;43:102195. doi: 10.1016/j.msard.2020.102195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L., Mizgerd J.P. Recent endemic coronavirus infection is associated with less severe COVID-19. J. Clin. Invest. 2021;131:e143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R., et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Salvat Lago M., Decker A., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2020 doi: 10.1038/s41591-020-01143-2. Published online November 12, 2020. [DOI] [PubMed] [Google Scholar]
- Schurink B., Roos E., Radonic T., Barbe E., Bouman C.S.C., de Boer H.H., de Bree G.J., Bulle E.B., Aronica E.M., Florquin S., et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. 2020;1:e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzkopf S., Krawczyk A., Knop D., Klump H., Heinold A., Heinemann F.M., Thümmler L., Temme C., Breyer M., Witzke O., et al. Cellular Immunity in COVID-19 Convalescents with PCR-Confirmed Infection but with Undetectable SARS-CoV-2-Specific IgG. Emerg. Infect. Dis. 2021;27 doi: 10.3201/2701.203772. [DOI] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Karolinska COVID-19 Study Group Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.-H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. MGH COVID-19 Collection & Processing Team Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., He W., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. Cross-reactive serum and memory B cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. bioRxiv. 2020 doi: 10.1101/2020.09.22.308965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soresina A., Moratto D., Chiarini M., Paolillo C., Baresi G., Focà E., Bezzi M., Baronio B., Giacomelli M., Badolato R. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr. Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar S., Begom S., Bermingham A., Hoschler K., Adamson W., Carman W., Bean T., Barclay W., Deeks J.J., Lalvani A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- Stephens D.S., McElrath M.J. COVID-19 and the Path to Immunity. JAMA. 2020;324:1279–1281. doi: 10.1001/jama.2020.16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudre C.H., Murray B., Varsavsky T., Graham M.S., Penfold R.S., Bowyer R.C.E., Pujol J.C., Klaser K., Antonelli M., Canas L.S., et al. Attributes and predictors of Long-COVID: analysis of COVID cases and their symptoms collected by the Covid Symptoms Study App. medRxiv. 2020 doi: 10.1101/2020.10.19.20214494. [DOI] [Google Scholar]
- Suthar M.S., Zimmerman M.G., Kauffman R.C., Mantus G., Linderman S.L., Hudson W.H., Vanderheiden A., Nyhoff L., Davis C.W., Adekunle O., et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med. 2020;1:100040. doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.A., Dogra P., Gray J.I., Wells S.B., Connors T.J., Weisberg S.P., Krupska I., Matsumoto R., Poon M.M.L., Idzikowski E., et al. Analysis of respiratory and systemic immune responses in COVID-19 reveals mechanisms of disease pathogenesis. medRxiv. 2020 doi: 10.1101/2020.10.15.20208041. [DOI] [Google Scholar]
- Tan A.T., Linster M., Tan C.W., Bert N.L., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J., et al. Early induction of SARS-CoV-2 specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.10.15.341958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.-C., Tiu C., Hu Z., Chen V.C.-W., Young B.E., Sia W.R., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.-T., Wrammert J., Ma M.-J., Lv H., Wang T.-B., Yang H., Richardus J.H., Liu W., et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., Antunes R. da S., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. bioRxiv. 2020 doi: 10.1101/2020.12.08.416750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepasse P.R., Hafezi W., Lutz M., Kühn J., Wilms C., Wiewrodt R., Sackarnd J., Keller M., Schmidt H.H., Vollenberg R. Persisting SARS-CoV-2 viraemia after rituximab therapy: two cases with fatal outcome and a review of the literature. Br. J. Haematol. 2020;190:185–188. doi: 10.1111/bjh.16896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., Filipe A. da S., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. The circulating SARS-CoV-2 spike variant N439K maintains fitness while evading antibody-mediated immunity. bioRxiv. 2020 doi: 10.1101/2020.11.04.355842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol E.J. COVID-19 can affect the heart. Science. 2020;370:408–409. doi: 10.1126/science.abe2813. [DOI] [PubMed] [Google Scholar]
- Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., Yu J., Chan C.N., Bondoc S., Starke C.E., et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration . 2020. Vaccines and Related Biological Products Advisory Committee December 17, 2020 Meeting Briefing. Moderna COVID-19 Vaccine.https://www.fda.gov/media/144434/download [Google Scholar]
- U.S. Food and Drug Administration . 2020. Vaccines and Related Biological Products Advisory Committee December 10, 2020 Meeting Briefing. Pfizer.https://www.fda.gov/media/144245/download [Google Scholar]
- Van Damme P., Van Herck K. A review of the long-term protection after hepatitis A and B vaccination. Travel Med. Infect. Dis. 2007;5:79–84. doi: 10.1016/j.tmaid.2006.04.004. [DOI] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., et al. A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates. bioRxiv. 2020 doi: 10.1101/2020.09.08.280818. [DOI] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., McMahon M., Meade P., Mendu D.R., Muellers K., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Jr., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang L., Sang L., Ye F., Ruan S., Zhong B., Song T., Alshukairi A.N., Chen R., Zhang Z., et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J. Clin. Invest. 2020;130:5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffman H.-H., Oliveira T.Y., Oren D.A., et al. Enhanced SARS-CoV-2 Neutralization by Secretory IgA in vitro. bioRxiv. 2020 doi: 10.1101/2020.09.09.288555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H., Cooke G., Atchison C., Whitaker M., Elliott J., Moshe M., Brown J.C., Flower B., Daunt A., Ainslie K., et al. Declining prevalence of antibody positivity to SARS-CoV-2: a community study of 365,000 adults. medRxiv. 2020 doi: 10.1101/2020.10.26.20219725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfel J.M., Zimmerman L.I., Merkel T.J. Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proc. Natl. Acad. Sci. USA. 2014;111:787–792. doi: 10.1073/pnas.1314688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver C., Murphy K. W.W. Norton; 2016. Janeway’s Immunobiology. [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2035002. Published online December 17, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S.P., Connors T.J., Zhu Y., Baldwin M.R., Lin W.-H., Wontakal S., Szabo P.A., Wells S.B., Dogra P., Gray J., et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 2021;22:25–31. doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Bangs D.J., Sidney J., Kolla R.V., De Silva A.D., de Silva A.M., Crotty S., Peters B., Sette A. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc. Natl. Acad. Sci. USA. 2015;112:E4256–E4263. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheimer A.M., Bennett M.S., Park B., Uhrlaub J.L., Martinez C., Pulko V., Currier N.L., Nikolich-Žugich D., Kaye J., Nikolich-Žugich J. Aging and cytomegalovirus infection differentially and jointly affect distinct circulating T cell subsets in humans. J. Immunol. 2014;192:2143–2155. doi: 10.4049/jimmunol.1301721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2021;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson T.M., Li C.K.F., Chui C.S.C., Huang A.K.Y., Perkins M., Liebner J.C., Lambkin-Williams R., Gilbert A., Oxford J., Nicholas B., et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie D., Mulchandani R., Jones H.E., Taylor-Phillips S., Brooks T., Charlett A., Ades E.-H., Makin A., Oliver I., Moore P., et al. EDSAB-HOME Investigators SARS-CoV-2 responsive T cell numbers are associated with protection from COVID-19: A prospective cohort study in keyworkers. medRxiv. 2020 doi: 10.1101/2020.11.02.20222778. [DOI] [Google Scholar]
- Yehia B.R., Winegar A., Fogel R., Fakih M., Ottenbacher A., Jesser C., Bufalino A., Huang R.-H., Cacchione J. Association of Race With Mortality Among Patients Hospitalized With Coronavirus Disease 2019 (COVID-19) at 92 US Hospitals. JAMA Netw. Open. 2020;3:e2018039. doi: 10.1001/jamanetworkopen.2020.18039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander R., Schauder D., Xin G., Nguyen C., Wu X., Zajac A., Cui W. CD4+ T Cell Help Is Required for the Formation of a Cytolytic CD8+ T Cell Subset that Protects against Chronic Infection and Cancer. Immunity. 2019;51:1028–1042.e4. doi: 10.1016/j.immuni.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. COVID-STORM Clinicians. COVID Clinicians. Imagine COVID Group. French COVID Cohort Study Group. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort. NIAID-USUHS/TAGC COVID Immunity Group Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., To K.K.-W., Wong Y.-C., Liu L., Zhou B., Li X., Huang H., Mo Y., Luk T.-Y., Lau T.T.-K., et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.-C., Guan X.-H., Li Y.-H., Huang J.-Y., Jiang T., Hou L.-H., Li J.-X., Yang B.-F., Wang L., Wang W.-J., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M., et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519.e12. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Dowell A., Pearce H., Verma K., Long H., Begum J., Aiano F., Amin-Chowdhury Z., Hallis B., Stapley L., et al. Robust SARS-CoV-2-specific T-cell immunity is maintained at 6 months following primary infection. bioRxiv. 2020 doi: 10.1101/2020.11.01.362319. [DOI] [Google Scholar]
- Zuo Y., Estes S.K., Ali R.A., Gandhi A.A., Yalavarthi S., Shi H., Sule G., Gockman K., Madison J.A., Zuo M., et al. Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci. Transl. Med. 2020;12:eabd3876. doi: 10.1126/scitranslmed.abd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Conceptual schematics of viral loads, elaborated to represent both URT and lungs.
(B) Innate immune responses, elaborated to represent both blood (serum/plasma) cytokines/chemokines and local tissue innate immune responses, in an average case of COVID-19.
(C) T cell responses, elaborated to individually show virus-specific CD4+ T cells and CD8+ T cells, in an average case of COVID-19.
(D) Antibody responses, elaborated to show binding antibodies and neutralizing antibodies, in an average case of COVID-19.