Abstract
In response to physiological demand, the pituitary gland generates new hormone-secreting cells from committed progenitor cells throughout life. It remains unclear to what extent pituitary stem cells (PSCs), which uniquely express SOX2, contribute to pituitary growth and renewal. Moreover, neither the signals that drive proliferation nor their sources have been elucidated. We have used genetic approaches in the mouse, showing that the WNT pathway is essential for proliferation of all lineages in the gland. We reveal that SOX2+ stem cells are a key source of WNT ligands. By blocking secretion of WNTs from SOX2+ PSCs in vivo, we demonstrate that proliferation of neighbouring committed progenitor cells declines, demonstrating that progenitor multiplication depends on the paracrine WNT secretion from SOX2+ PSCs. Our results indicate that stem cells can hold additional roles in tissue expansion and homeostasis, acting as paracrine signalling centres to coordinate the proliferation of neighbouring cells.
Research organism: Mouse
Introduction
How stem cells interact with their surrounding tissue has been a topic of investigation since the concept of the stem cell niche was first proposed (Schofield, 1978). Secreted from supporting cells, factors such as WNTs, FGFs, SHH, EGF, and cytokines regulate the activity of stem cells (Nabhan et al., 2018; Palma et al., 2005; Tan and Barker, 2014). Furthermore, communication is known to take place in a bidirectional manner (Doupé et al., 2018; Tata and Rajagopal, 2016).
The anterior pituitary (AP) is a major primary endocrine organ that controls key physiological functions including growth, metabolism, reproduction, and the stress response and exhibits tremendous capability to remodel its constituent hormone populations throughout life, in response to physiological demand. It contains a population of Sox2 expressing stem cells that self-renew and give rise to lineage-committed progenitors and functional endocrine cells (Andoniadou et al., 2013; Rizzoti et al., 2013). During embryonic development, SOX2+ undifferentiated precursor cells of Rathke’s pouch, the pituitary anlage (Arnold et al., 2011; Castinetti et al., 2011; Fauquier et al., 2008; Pevny and Rao, 2003), generate all committed endocrine progenitor lineages, defined by the absence of SOX2 and expression of either POU1F1 (PIT1), TBX19 (TPIT), or NR5A1 (SF1) (Bilodeau et al., 2009; Davis et al., 2011). These committed progenitors are proliferative and give rise to the hormone-secreting cells. Demand for hormone secretion rises after birth, resulting in dramatic organ growth and expansion of all populations by the second postnatal week (Carbajo-Pérez and Watanabe, 1990; Taniguchi et al., 2002). SOX2+ pituitary stem cells (PSCs) are most active during this period, but the bulk of proliferation and organ expansion during postnatal stages derives from SOX2− committed progenitors. The activity of SOX2+ PSCs gradually decreases and during adulthood is minimally activated even following physiological challenge (Andoniadou et al., 2013; Gaston-Massuet et al., 2011; Gremeaux et al., 2012; Zhu et al., 2015). By adulthood, progenitors carry out most of the homeostatic functions, yet SOX2+ PSCs persist throughout life in both mice and humans (Gonzalez-Meljem et al., 2017; Xekouki et al., 2019). The signals driving proliferation of committed progenitor cells are not known, and neither is it known if SOX2+ PSCs can influence this process beyond their minor contribution of new cells.
The self-renewal and proliferation of numerous stem cell populations rely on WNT signals (Basham et al., 2019; Lim et al., 2013; Takase and Nusse, 2016; Wang et al., 2015; Yan et al., 2017). WNTs are necessary for the initial expansion of Rathke’s pouch as well as for PIT1 lineage specification (Osmundsen et al., 2017; Potok et al., 2008). In the postnatal pituitary, the expression of WNT pathway components is upregulated during periods of expansion and remodelling. Gene expression comparisons between neonatal and adult pituitaries or in GH-cell ablation experiments (Gremeaux et al., 2012; Willems et al., 2016) show that the WNT pathway is upregulated during growth and regeneration.
Our previous work revealed that during disease, the paradigm of supporting cells signalling to the stem cells may be reversed; mutant stem cells expressing a degradation-resistant β-catenin in the pituitary promote cell non-autonomous development of tumours through their paracrine actions (Andoniadou et al., 2013; Gonzalez-Meljem et al., 2017). Similarly, degradation-resistant β-catenin expression in hair follicle stem cells led to cell non-autonomous WNT activation in neighbouring cells promoting new growth (Deschene et al., 2014). In the context of normal homeostasis, stem cells have been shown to influence daughter cell fate in the mammalian airway epithelium and the Drosophila gut via ‘forward regulation’ models, where the fate of a daughter cell is directed by a stem cell via juxtacrine Notch signalling (Ohlstein and Spradling, 2007; Pardo-Saganta et al., 2015). It remains unknown if paracrine stem cell action can also promote local proliferation in normal tissues.
Here, we used genetic approaches to determine if paracrine stem cell action takes place in the AP and to discern the function of WNTs in pituitary growth. Our results demonstrate that postnatal pituitary expansion, largely driven by committed progenitor cells, depends on WNT activation. Importantly, we show that SOX2+ PSCs are the key regulators of this process, acting through secretion of WNT ligands acting in a paracrine manner on neighbouring progenitors. Identification of this forward-regulatory model elucidates a previously unidentified function for stem cells during tissue expansion.
Results
WNT-responsive cells in the pituitary include progenitors driving major postnatal expansion
To clarify which cells respond to WNT signals in the postnatal AP, we first characterised the AP cell types activating the WNT pathway at P14, a peak time for organ expansion and a time point when a subpopulation of SOX2+ stem cells are proliferative. The Axin2-CreERT2 mouse line (van Amerongen et al., 2012) has been shown to efficiently label cells with activated WNT signalling in the liver, lung, breast, skin, testes, and endometrium among other tissues (Lim et al., 2013; Moiseenko et al., 2017; Syed et al., 2020; van Amerongen et al., 2012; Wang et al., 2015). Axin2 positive cells were labelled by GFP following tamoxifen induction in Axin2CreERT2/+;ROSA26mTmG/+ mice and pituitaries were analysed 2 days post-induction. We carried out double immunofluorescence staining using antibodies against uncommitted (SOX2), lineage committed (PIT1, TPIT, SF1), and hormone-expressing endocrine cells (GH, PRL, TSH, ACTH, or FSH/LH) together with antibodies against GFP labelling the WNT-activated cells. We detected WNT-responsive cells among all the different cell types of the AP including SOX2+ PSCs, the three committed populations and all hormone-secreting cells (Figure 1A, Figure 1—figure supplement 1A).
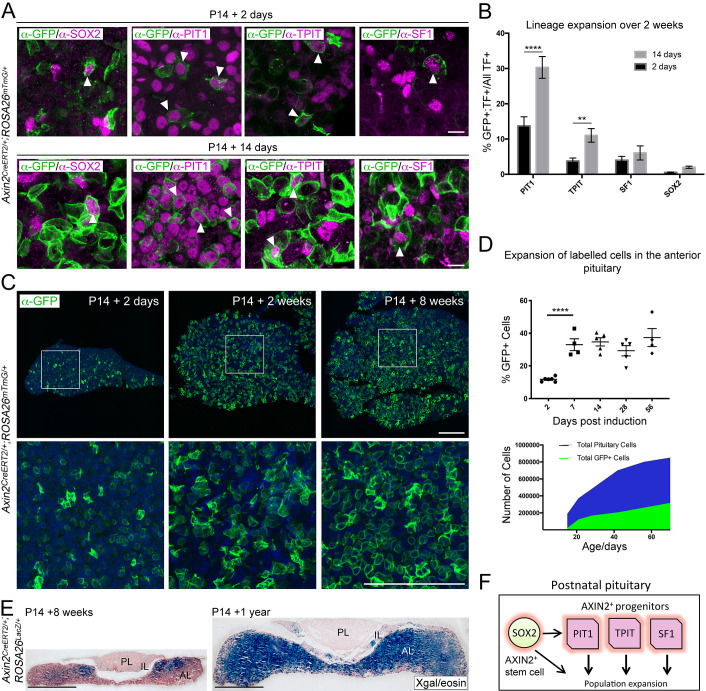
Figure 1. Axin2 expressing cells contribute to pituitary growth and expansion of all lineages.
(A) Immunofluorescence staining against GFP (green) with markers of pituitary stem cells (PSCs) or lineage commitment (magenta) in Axin2CreERT2/+; ROSA26mTmG/+ pituitaries harvested from mice induced at P14 and lineage traced for 2 days (top panel) and 14 days (bottom panel). Scale bar: 10 μm. (B) Quantification of lineage expansion between 2 and 14 days following induction at P14. Graph shows that the proportion of lineage committed cells (either PIT1+, TPIT+, or SF1+) and PSCs (SOX2+), that is, that are transcription factor (TF)+ cells that are GFP+ increases between 2 days (black bars) and 14 days (grey bars) post-induction. PIT1 p=0.000004, TPIT p=0.008 multiple t-tests. n = 4 animals per time point. (C) Immunofluorescence staining against GFP (green) in pituitaries harvested from Axin2CreERT2/+;ROSA26mTmG/+ mice induced at P14 and lineage traced for 2 days, 2 weeks, and 8 weeks. Bottom panel shows magnified fields of view of regions of interest indicated by white boxes in panels above. Scale bars: 50 μm. (D) Top panel showing the quantification of the proportion of all cells in Axin2CreERT2/+;ROSA26mTmG/+ pituitaries that are GFP+ at 2, 7, 14, 28, and 56 days post-induction as analysed by flow cytometry. Days 2–7 p<0.0001 unpaired t-test. Data points show individual measurements from biological replicates, n = 4–8 pituitaries per time point. (Bottom) Graph of the absolute number of GFP+ cells (green) and as a proportion of total cells (blue) at the time points indicated. (E) X-gal staining in Axin2CreERT2/+;ROSA26LacZ/+ pituitaries harvested from mice induced at P14 and lineage traced for 8 weeks (left) and 1 year (right). Scale bars: 500 μm. (F) Model summarising the major contribution of WNT-responsive progenitors of all lineages to pituitary growth, in addition to that of SOX2+ PSCs.
Figure 1—figure supplement 1. Axin2 expressing cells contribute to pituitary growth and expansion of all lineages.
Figure 1—figure supplement 2. Axin2 expressing cells contribute to pituitary growth and expansion of all lineages.
To confirm if the three committed lineages as well as uncommitted SOX2+ PSCs all expand in response to WNT, we further lineage traced Axin2-expressing cells for 14 days after tamoxifen administration at P14. Double labelling revealed an increase in all four populations between 2 and 14 days (Figure 1A,B). This increase reached significance for the PIT1 (13.7% at 2 days to 30.3% at 14 days, p=0.000004) and TPIT (3.78% to 11.03%, p=0.008) populations, but not SF1 (0.5% to 4%, n.s.). As this time course ends at P28 at the commencement of puberty, we repeated the analysis for SF1 cells to P42, which spans puberty and the expansion of gonadotrophs (Figure 1—figure supplement 1B). This reveals a significant expansion in WNT-responsive SF1+ cells as a proportion of the total SF1+ population (p=0.0048, n = 3). Lineage tracing of the PIT1-derivates (GH+ somatotrophs, PRL+ lactotrophs, and TSH+ thyrotrophs) reveals that there is a preferential expansion of somatotrophs and thyrotrophs (Figure 1—figure supplement 1C). Only a minority of SOX2+ PSCs were WNT-responsive at 2 days (0.57%) and this population expanded to 2% at 14 days (n.s.), suggesting that these are self-renewing. GFP+ cells were traced for a period of 8 weeks post-induction, which revealed that WNT-responsive descendants continued to expand at the same rate as the rest of the pituitary (n = 4–8 mice per time point at P16, P21, P28, P42, and P70) (Figure 1C,D). The time period between 2 and 7 days saw the greatest increase in GFP+ cells, during which the labelled population nearly tripled in size (Figure 1D). The persistence of labelled cells was evident in longer-term traces using the ROSA26lacZ/+ reporter (Axin2CreERT2/+;ROSA26lacZ/+), up to a year following induction at P14 (Figure 1E, n = 4). Clonal analysis using the Confetti reporter demonstrated that individual Axin2-expressing cells (Axin2CreERT2/+;ROSA26Confetti/+) gave a greater contribution after 4 weeks compared to lineage tracing from Sox2-expressing cells (Sox2CreERT2/+;ROSA26Confetti/+), in support of predominant expansion from WNT-responsive lineage-committed progenitors (Figure 1—figure supplement 1D).
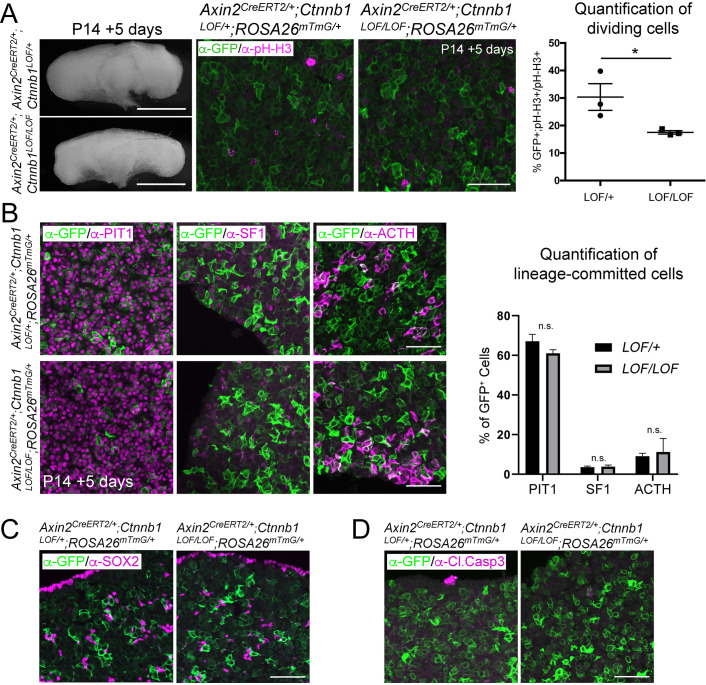
To establish if signalling mediated by β-catenin is necessary for organ expansion we carried out deletion of Ctnnb1 in the Axin2+ population from P14 during normal growth (Axin2CreERT2/+;Ctnnb1lox(ex2-6)/lox(ex2-6) hereby Axin2CreERT2/+;Ctnnb1LOF/LOF). Due to morbidity, likely due to Axin2 expression in other organs, we were limited to analysis up to 5 days post-induction. Deletion of Ctnnb1 resulted in a significant reduction in the number of dividing cells, marked by pH-H3 (40% reduction, Figure 1—figure supplement 2A, p=0.0313, n = 3), confirming that activation of the WNT pathway is necessary for expansion of the pituitary populations. This deletion did not result in significant differences in overall numbers among the three lineages, as determined by the numbers of PIT1+, SF1+, or ACTH+ cells among the targeted population (Figure 1—figure supplement 2B, n = 4 controls, two mutants). The number of SOX2+ stem cells and cells undergoing cell death also remained unaffected during the 5-day period (Figure 1—figure supplement 2C and D). Taken together, these results confirm that postnatal AP expansion depends on WNT-responsive progenitors across all lineages, in addition to SOX2+ PSCs (Figure 1F).
WNT/β-catenin signalling is required for long-term AP expansion from SOX2+ PSCs
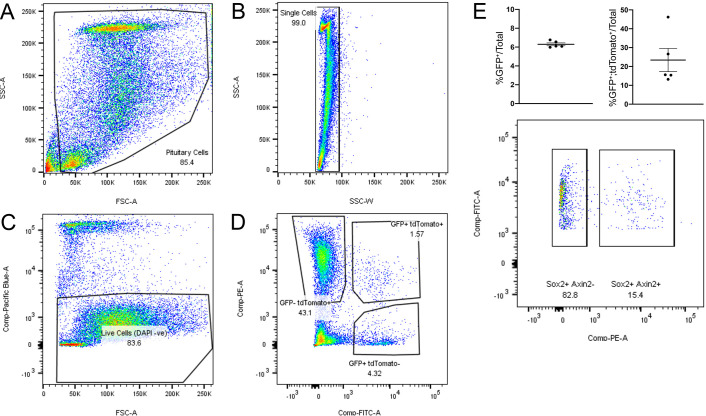
We further explored the role of WNT pathway activation in postnatal SOX2+ stem cells. To permanently mark WNT-responsive cells and their descendants whilst simultaneously marking SOX2+ PSCs, we combined the tamoxifen-inducible Axin2CreERT2/+;ROSA26tdTomato/+ with the Sox2Egfp/+ strain, where cells expressing SOX2 are labelled by enhanced green fluorescent protein (EGFP) (Axin2CreERT2/+;Sox2Egfp/+;ROSA26tdTomato/+). Following tamoxifen administration from P21, tdTomato- and EGFP-labelled cells were analysed by flow sorting after 72 hr, by which point all induced cells robustly express tdTomato (Figure 2A, Figure 2—figure supplement 1). Double-labelled cells comprised 23.4% of the SOX2+ population (n = 5 individual pituitaries) (Figure 2A, arrowheads), with the majority of tdTomato+ cells found outside of the SOX2+ compartment. It was previously shown that only around 2.5–5% of SOX2+ PSCs has clonogenic potential through in vitro assays (Andoniadou et al., 2012; Andoniadou et al., 2013; Pérez Millán et al., 2016). To determine if WNT-responsive SOX2+ cells are stem cells capable of forming colonies, we isolated double-positive tdTomato+;EGFP+ cells (i.e. Axin2+;Sox2+) as well as the single-expressing populations and plated these in equal numbers in stem cell-promoting media at clonal densities (Figure 2B). Double-positive tdTomato+;EGFP+ cells showed a significant increase in the efficiency of colony formation compared to single-labelled EGFP+ cells (average 9% compared to 5%, n = 5 pituitaries, p=0.0226, Mann–Whitney U-test [two-tailed]), demonstrating WNT-responsive SOX2+ PSCs have a greater clonogenic potential under these in vitro conditions, confirming in vivo data in Figure 1B. As expected from previous work, none of the single-labelled tdTomato+ cells (i.e. SOX2 negative) was able to form colonies (Andoniadou et al., 2012).
Figure 2. Activation of WNT signalling in SOX2+ pituitary stem cells (PSCs) and their descendants is necessary for long-term growth.
(A) Schematic of the experimental timeline used in panels A and B. Endogenous expression of tdTomato (magenta, Axin2 targeted cells) and EGFP (green, Sox2 expressing cells) in Axin2CreERT2/+;Sox2Egfp/+;ROSA26tdTomato/+ pituitaries harvested at P24 sectioned in the frontal plane. Nuclei are counterstained with Hoechst in the merged panel. Scale bar: 50 μm. (B) A representative culture plate showing colonies derived from Tomato+, EGFP+, or Tomato+;EGFP+ cells that were isolated from Axin2CreERT2/+;Sox2Egfp/+;ROSA26tdTomato/+ pituitaries by fluorescence-activated cell sorting (FACS) plated in stem cell promoting media at clonogenic densities and stained with crystal violet (left panel). The proportion of colony-forming cells in each subpopulation was quantified by counting the number of colonies per well (right panel). Each data point indicates individual wells, n = 5 separate pituitaries. p=0.0226, Mann–Whitney U-test (two-tailed). Scale bar: 10 mm. (C) Immunofluorescence staining against SOX2 (green) and Ki-67 (magenta) in Sox2+/+Ctnnb1LOF/LOF (control) and Sox2CreERT2/+Ctnnb1LOF/LOF (mutant) pituitaries from mice induced at P14 and analysed 22 weeks after induction (at P168) (bottom panel). Scale bar: 50 μm. (D) Dorsal view of whole mount pituitaries of Sox2+/+;Ctnnb1LOF/LOF (control) and Sox2CreERT2/+;Ctnnb1LOF/LOF (mutant), 22 weeks after induction (i.e. P168). Scale bars: 1 mm. (E) Model summarising the effect of Ctnnb1 deletion in SOX2+ PSCs. PL, posterior lobe; IL, intermediate lobe; AL, anterior lobe.
Figure 2—figure supplement 1. Activation of WNT signalling in SOX2+ pituitary stem cells (PSCs) and their descendants is necessary for long-term growth.
Figure 2—figure supplement 2. Activation of WNT signalling in SOX2+ pituitary stem cells (PSCs) and their descendants is necessary for long-term growth.
To confirm that PSCs with active WNT signalling through β-catenin have a greater propensity to form colonies in vitro, we analysed postnatal pituitaries from TCF/Lef:H2B-EGFP mice, reporting the activation of response to WNT signals. This response is detected through expression of an EGFP-tagged variant of histone H2B, which is incorporated into chromatin and diluted in descendants with cell division (Ferrer-Vaquer et al., 2010). Therefore, cells responding to, or having recently responded to, WNT as well as their immediate descendants will be EGFP+. At P21, EGFP+ cells were abundant in all three lobes and particularly in the marginal zone harbouring SOX2+ stem cells (Figure 2—figure supplement 2A). Through double mRNA in situ hybridisation against Egfp and Sox2 in TCF/Lef:H2B-EGFP pituitaries, we confirmed that Sox2-expressing cells activate H2B-EGFP expression at this time point (Figure 2—figure supplement 2B). Isolation by fluorescence-activated cell sorting (FACS) and in vitro culture of the postnatal EGFP+ compartment revealed an enrichment of cells with clonogenic potential in the EGFPHigh fraction compared to EGFPLow or negative cells (Figure 2—figure supplement 2C, n = 5 pituitaries). Together these results reveal that a proportion of postnatal SOX2+ stem cells respond to WNTs through downstream β-catenin/TCF/LEF signalling and that these cells have greater clonogenic capacity in vitro.
To further address the role of the canonical WNT response in the activity of SOX2+ PSCs in vivo, we expressed a loss-of-function allele of β-catenin specifically in Sox2-expressing cells (Sox2CreERT2/+;Ctnnb1lox(ex2-6)/lox(ex2-6) hereby Sox2CreERT2/+;Ctnnb1LOF/LOF) from P14. Twenty-two weeks following induction, at P168, there was a substantial drop in the number of cycling cells in the pituitary of Sox2CreERT2/+;Ctnnb1LOF/LOF mutants compared to Sox2+/+;Ctnnb1LOF/LOF controls (Figure 2C, n = 2 pituitaries per genotype). This was accompanied by AP hypoplasia following the loss of Ctnnb1 in SOX2+ PSCs (Figure 2D). Therefore, in this small sample size, the proliferative capacity of Ctnnb1-deficient SOX2+ PSCs and of their descendants was impaired long term, leading to reduced growth. In vivo genetic tracing of targeted cells over the 22-week period (Sox2CreERT2/+;Ctnnb1LOF/+;ROSA26mTmG/+ compared to Sox2CreERT2/+;Ctnnb1LOF/LOF;ROSA26mTmG/+ pituitaries) revealed that targeted (Ctnnb1-deficient) SOX2+ PSCs were capable of giving rise to the three committed lineages PIT1, TPIT, and SF1 (Figure 2—figure supplement 2D), indicating that the loss of Ctnnb1 does not prevent differentiation of SOX2+ PSCs into the three lineages. Downregulation of β-catenin was confirmed by immunofluorescence in SOX2+ (mGFP+) derivatives (Figure 2—figure supplement 2E). Although limited by a small sample size, we conclude that WNT/β-catenin signalling is likely required cell-autonomously in SOX2+ stem cells and their descendants (Figure 2E).
SOX2+ stem cells express WNT ligands
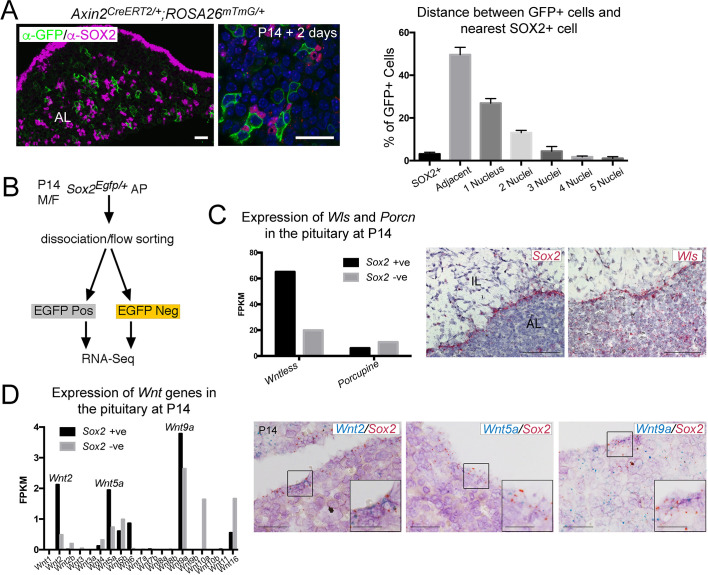
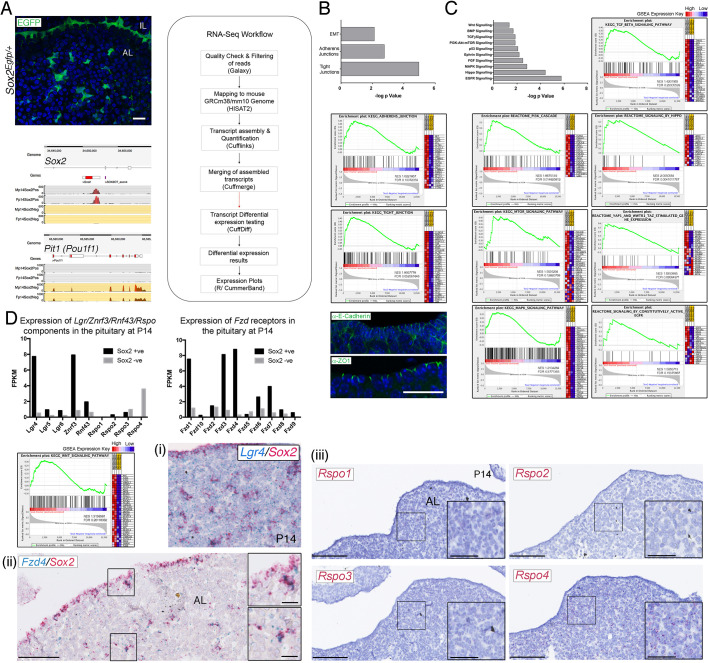
Having established that WNT activation is responsible for promoting proliferation in the AP, we next focused on identifying the source of WNT ligands. Axin2 expressing cells from Axin2CreERT2/+;ROSA26mTmG/+ mice were labelled at P14 by tamoxifen induction. Cells expressing Axin2 at the time of induction are labelled by GFP expression in the membrane. Double immunofluorescence staining for GFP together with SOX2 revealed that Axin2 expressing cells (mGFP+) are frequently located in close proximity to SOX2+ PSCs (Figure 3A). Two-dimensional quantification of the two cell types revealed that over 50% of mGFP+ cells were in direct contact with SOX2+ nuclei (n = 3 pituitaries, >500 SOX2+ cells per gland, Figure 3A). The analysis did not take into account the cellular processes of SOX2+ cells. These results led us to speculate that SOX2+ PSCs may be a source of key WNT ligands promoting proliferation of lineage-committed cells.
Figure 3. SOX2+ pituitary stem cells (PSCs) are as a source of WNT ligands in the pituitary.
(A) Immunofluorescence staining against GFP (green) and SOX2 (magenta) in Axin2CreERT2/+; ROSA26mTmG/+ pituitaries 48 hr post-induction. Graph representing a quantification of the proximity of individual GFP+ cells to the nearest SOX2+ cell as quantified by the number of nuclei separating them. Plotted data represents the proportion of GFP+ cells that fall into each category of the total GFP+ cells, taken from n = 3 separate pituitaries. Scale bars: 50 μm. (B) Experimental paradigm for RNA Seq analysis of Sox2 positive and negative cells. (C) Graphs representing the FPKM values of Wls and Porcupine in Sox2 positive and negative cells (black and grey bars, respectively). mRNA in situ hybridisation for Sox2 and for Wls on wild-type sagittal pituitaries at P14, demonstrating strong Wls expression in the marginal zone epithelium. Scale bars: 250 μm. (D) Bar chart showing the FPKM values of Wnt genes in the Sox2+ and Sox2− fractions. Double mRNA in situ hybridisation against Wnt2, Wnt5a, and Wnt9a (blue) together with Sox2 (red) validating expression in the Sox2+ population. Boxed regions through the marginal zone epithelium are magnified. Scale bars: 100 μm and 50 μm in boxed inserts.
Figure 3—figure supplement 1. SOX2+ pituitary stem cells (PSCs) are as a source of WNT ligands in the pituitary.
In order to determine if SOX2+ PSCs express WNT ligands, we carried out gene expression profiling of SOX2+ and SOX2− populations at P14, through bulk RNA-sequencing. Pure populations of Sox2-expressing cells excluding lineage-committed populations were isolated from Sox2Egfp/+ male and female pituitaries at P14 based on EGFP expression as previously shown (Andoniadou et al., 2012; Figure 3B, Figure 3—figure supplement 1A). Analysis of global gene expression signatures using ‘gene set enrichment analysis’ (GSEA) (Subramanian et al., 2005) identified a significant enrichment of molecular signatures related to epithelial-to-mesenchymal transition, adherens, and tight junctions in the EGFP+ fraction, characteristic of the SOX2+ population (Figure 3—figure supplement 1B). The SOX2+ fraction also displayed enrichment for genes associated with several signalling pathways known to be active in these cells, including epidermal growth factor receptor (EGFR) (Iwai-Liao et al., 2000), Hippo (Lodge et al., 2016; Lodge et al., 2019; Xekouki et al., 2019), MAPK (Haston et al., 2017), FGF (Higuchi et al., 2017), Ephrin (Yoshida et al., 2015; Yoshida et al., 2017), and p53 (Gonzalez-Meljem et al., 2017; Figure 3—figure supplement 1C, Supplementary file 1). Additionally, PI3K, TGFβ, and BMP pathway genes were significantly enriched in the SOX2+ population (Figure 3—figure supplement 1C, Supplementary file 1). Query of the WNT-associated genes did not suggest a global enrichment in WNT targets (e.g. enrichment of Myc and Jun, but not of Axin2 or Lef1) (Figure 3—figure supplement 1D, Supplementary file 1). Instead, SOX2+ PSCs expressed a unique transcriptomic fingerprint of key pathway genes including Lgr4, Znrf3, Rnf43 capable of regulating WNT signal intensity in SOX2+ PSCs, as well as enriched expression of the receptors Fzd1, Fzd3, Fzd4, Fzd6, and Fzd7 (Figure 3—figure supplement 1D). The predominant R-spondin gene expressed in the pituitary was Rspo4, specifically by the EGFP-negative fraction (Figure 3—figure supplement 1D). The gene profiling revealed that Wls expression encoding Gpr177/WLS, a necessary mediator of WNT ligand secretion (Carpenter et al., 2010; Takeo et al., 2013; Wang et al., 2015), is enriched in SOX2+ PSCs (Figure 3C). Analysis of Wnt gene expression confirmed enriched expression of Wnt2, Wnt5a, and Wnt9a in SOX2+ PSCs, and the expression of multiple additional Wnt genes by both fractions at lower levels (SOX2+ fraction: Wnt5b, Wnt6, Wnt16; SOX2− fraction: Wnt2, Wnt2b, Wnt3, Wnt4, Wnt5a, Wnt5b, Wnt9a, Wnt10a, Wnt16) (Figure 3D). These results reveal that SOX2+ PSCs express the essential components to regulate activation of the WNT pathway and express Wnt genes as well as the necessary molecular machinery to secrete WNT ligands.
Paracrine signalling from SOX2+ stem cells promotes WNT activation
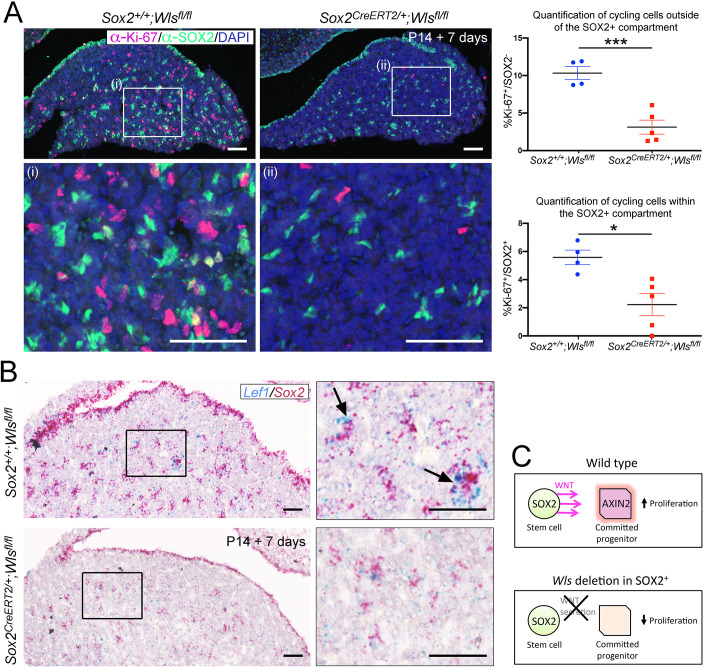
We sought to conclusively determine if WNT secretion specifically from SOX2+ PSCs drives proliferation of surrounding cells in the postnatal pituitary gland. We proceeded to delete Wls only in the Sox2-expressing population (Sox2CreERT2/+;Wlsfl/fl) from P14 by a series of tamoxifen injections. Due to morbidity, we limited analyses to 1 week following induction. Pituitaries appeared mildly hypoplastic at P21 along the medio-lateral axis (Figure 4—figure supplement 1, n = 4 controls and n = 5 mutants). To determine if this is a result of reduced proliferation, we carried out immunofluorescence using antibodies against Ki-67 and SOX2. This revealed significantly fewer cycling cells in the SOX2− population of Sox2CreERT2/+;Wlsfl/fl mutant pituitaries compared to Sox2+/+;Wlsfl/fl controls (10.326% Ki-67 in control [n = 4] compared to 3.129% in mutant [n = 5], p=0.0008, unpaired t-test) (Figure 4A). Additionally, we observed a reduction of cycling cells within the SOX2+ population (5.582% Ki-67 in control compared to 2.225% in induced Sox2CreERT2/+;Wlsfl/fl mutant pituitaries, p=0.0121, unpaired t-test) (Figure 4A), resulting in a smaller SOX2+ cell pool in mutants (23.425% SOX2+/total AP cells in Sox2+/+;Wlsfl/fl controls compared to 19.166% SOX2+/total AP cells in induced Sox2CreERT2/+;Wlsfl/fl mutant pituitaries, p=0.0238, Student’s t-test, n = 5 mutants, four controls). To determine if reduced levels of WNT activation accompanied this phenotype, we carried out double mRNA in situ hybridisation using specific probes against Lef1 and Sox2. There was an overall reduction in Lef1 expression in mutants compared to controls (n = 4 per genotype), in which we frequently observed robust expression of Lef1 transcripts in close proximity to cells expressing Sox2 (arrows, Figure 4B). Together, our data support a paracrine role for SOX2+ PSCs in driving the expansion of committed progeny through the secretion of WNT ligands (Figure 4C).
Figure 4. Paracrine secretion of WNTs from SOX2+ pituitary stem cells (PSCs) is necessary for expansion of committed cells.
(A) Immunofluorescence staining against SOX2 (green) and Ki-67 (magenta) in Sox2+/+;Wlsfl/fl (control) and Sox2CreERT2/+;Wlsfl/fl (mutant) pituitaries induced from P14 and analysed after 1 week. Nuclei were counterstained with Hoechst. (i and ii) represent magnified fields of view of regions indicated by white boxes in top panels. Scale bars: 50 μm. Graph of quantification of cycling cells marked by Ki-67 among cells negative for SOX2. Values represent mean ± SEM, p=0.0008, unpaired t-test. Graph of quantification of cycling cells marked by Ki-67 among SOX2-positive cells. Values represent mean ± SEM, p=0.0121, unpaired t-test. Each data point shows the mean of one biological replicate, n = 4 pituitaries from controls and five pituitaries from mutants. (B) Double mRNA in situ hybridisation using specific probes against Lef1 (blue) and Sox2 (red) in control and mutant pituitaries following tamoxifen induction from P14 and tracing for 7 days. Scale bars: 250 μm and 50 μm in boxed regions. (C) Model summarising paracrine WNT secretion from SOX2+ PSCs to lineage-committed progenitors and the effects of abolishing WNT secretion from SOX2+ PSCs through the deletion of Wls.
Figure 4—figure supplement 1. Paracrine secretion of WNTs from SOX2+ pituitary stem cells (PSCs) is necessary for expansion of committed cells.
Discussion
Emerging disparities between the archetypal stem cell model, exhibited by the haematopoietic system, and somatic stem cells of many organs have led to the concept that stem cell function can be executed by multiple cells not fitting a typical stem cell paradigm (Clevers and Watt, 2018). In organs with persistent populations possessing typical functional stem cell properties yet contributing minimally to turnover and repair, the necessity for such classical stem cells is questioned. Here we show that WNT signalling is required for postnatal pituitary growth by both SOX2+ PSCs and SOX2− committed progenitors. We identify an additional discreet function for SOX2+ PSCs, where these signal in a feedforward manner by secreting WNT ligands as cues to stimulate proliferation and promote tissue growth.
Consistent with previous reports, our data support that SOX2+ PSCs contribute, but do not carry out the majority of tissue expansion during the postnatal period (Zhu et al., 2015); instead, new cells primarily derive from more committed progenitors, which we show to be WNT-responsive. We demonstrate that this population of lineage-restricted WNT-responsive cells rapidly expands and contributes long-lasting clones from postnatal stages. It remains to be shown if cells among the SOX2− lineage-committed populations may also fall under the classical definition of a stem cell. Preventing secretion of WNT ligands from SOX2+ PSCs reveals that far from being dispensable, paracrine actions of the SOX2+ population that are inactive in their majority are necessary for anterior lobe expansion from lineage-committed populations. In the adrenal gland, R-spondins are necessary for cortical expansion and zonation, where deletion of Rspo3, expressed by the capsule that contains adrenocortical stem cells, results in reduced proliferation of the underlying steroidogenic cells (Vidal et al., 2016). Corroborating a model where committed pituitary progenitors depend on the paracrine actions of SOX2+ PSCs, Zhu and colleagues observed that in pituitaries with reduced numbers of PSCs, proliferation among PIT1+ cells was significantly impaired (Zhu et al., 2015). It would be intriguing to see if there is a reduction in WNT signalling in this model, or following genetic ablation of adult SOX2+ PSCs (Roose et al., 2017).
We show that a subpopulation of SOX2+ PSCs in the postnatal gland are also WNT-responsive and have greater in vitro colony-forming potential under defined conditions. This colony-forming potential is normally a property of a minority of SOX2+ PSCs at any given age and reflects their in vivo proliferative capacity (Andoniadou et al., 2012; Rizzoti et al., 2013). A role for the WNT pathway in promoting SOX2+ cell activity is supported by studies showing that pathogenic overexpression of β-catenin promotes their colony-forming ability (Sarkar et al., 2016) and their in vivo expansion (Andoniadou et al., 2012). Additionally, elevated WNT pathway activation has been described for pituitary side-population cells, enriched for SOX2+ stem cells from young, compared to old pituitaries (Gremeaux et al., 2012). This is in line with our findings that the WNT pathway has an important function in promoting the activation of SOX2+ PSCs. It remains to be shown if this response relies on autocrine WNT-signalling as for other stem cells (Lim et al., 2013); however, our results reveal reduced proliferation among SOX2+ PSCs and reduced SOX2+ cell numbers when WNT secretion from these cells is abolished, supportive of either autocrine signalling or paracrine signalling between different subsets of the SOX2+ population.
The mechanism preventing the majority of SOX2+ PSCs from responding to WNT signals remains elusive but points to heterogeneity among the population. Such regulation could occur at the level of receptor signalling; we have shown by bulk transcriptomic profiling that SOX2+ PSCs express the receptors required to respond to the WNT pathway, but also express high levels of the frizzled inhibitor Znrf3, and the R-spondin receptor Lgr4. One conceivable scenario is that high levels of Znrf3 inhibit frizzled receptors in the absence of R-spondin under normal physiological conditions, supressing a WNT response. In support of this, R-spondins have been shown to promote pituitary organoid formation (Cox et al., 2019). Whether the R-spondin/LGR/ZNRF3 module is active under physiological conditions needs to be determined. Furthermore, well-described factors expressed in PSCs are known to have inhibitory effects on β-catenin-mediated transcription, such as YAP/TAZ (Azzolin et al., 2014; Gregorieff et al., 2015) and SOX2 itself (Alatzoglou et al., 2011; Kelberman et al., 2008).
In summary, we demonstrate an alternative mechanism for stem cell contribution to homeostasis, whereby these can act as paracrine signalling hubs to promote local proliferation. Applicable to other organs, this missing link between SOX2+ PSCs and committed cell populations of the AP is key for basic physiological functions and renders stem cells integral to organ expansion.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference |
Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Axin2CreERT2/+ | Roel Nusse, Stanford University The Jackson Laboratory |
JAX:018867, RRID:IMSR_JAX:018867 | |
| Genetic reagent (Mus musculus) | Sox2CreERT2/+ | (Andoniadou et al., 2013) PMID:24094324 DOI: 10.1016/j.stem.2013.07.004 |
MGI:5512893 | |
| Genetic reagent (Mus musculus) | ROSA26mTmG/mTmG | The Jackson Laboratory | JAX:007676, RRID:IMSR_JAX:007676 | |
| Genetic reagent (Mus musculus) | ROSA26Confetti/Confetti | The Jackson Laboratory | JAX:017492, RRID:IMSR_JAX:017492 | |
| Genetic reagent (Mus musculus) | ROSA26tdTomato/tdTomato | The Jackson Laboratory | JAX:007909, RRID:IMSR_JAX:007909 | |
| Genetic reagent (Mus musculus) | Ctnnb1fl(ex2-6)/ fl(ex2-6) (CtnnbLOF/LOF) | The Jackson Laboratory | JAX:004152, RRID:IMSR_JAX:004152 | |
| Genetic reagent (Mus musculus) | Wlsfl/fl | The Jackson Laboratory | JAX:012888, RRID:IMSR_JAX:012888 | |
| Genetic reagent (Mus musculus) | Sox2eGFP/+ |
Ellis et al., 2004
PMID:15711057 DOI: 10.1159/000082134 |
MGI:3589809 | |
| Genetic reagent (Mus musculus) | TCF/Lef:H2B-GFP | The Jackson Laboratory | JAX:013752, RRID:IMSR_JAX:013752 | |
| Cell line (Mus musculus) | Primary anterior pituitary cells | This paper | N/A | Freshly isolated from Mus musculus. |
| Antibody | Anti-GFP (Chicken Polyclonal) | Abcam | ab13970, RRID:AB_300798 | IF(1:400) |
| Antibody | Anti-SOX2 (Goat Polyclonal) | Immune Systems Ltd | GT15098, RRID:AB_2195800 | IF(1:200) |
| Antibody | Anti-SOX2(Rabbit Monoclonal) | Abcam | ab92494, RRID:AB_10585428 | IF(1:100) |
| Antibody | Anti-SOX9(Rabbit Monoclonal) | Abcam | ab185230, RRID:AB_2715497 | IF(1:500) |
| Antibody | Anti-POU1F1 (PIT1) (Rabbit Monoclonal) | Gifted by Dr S. J. Rhodes (IUPUI, USA) | 422_Rhodes, RRID:AB_2722652 | IF(1:500) |
| Antibody | Anti-SF1 (NR5A1, clone N1665)(Mouse Monoclonal) | Thermo Fisher Scientific | 434200, RRID:AB_2532209 | IF(1:300) |
| Antibody | Anti-TBX19 (TPIT), (Rabbit Polyclonal) | Gifted by Dr J. Drouin (Montreal Clinical Research Institute, Canada) | Ac1250 #71, RRID:AB_2728662 | IF(1:200) |
| Antibody | Anti-Ki67 (Rabbit Monoclonal) | Abcam | ab15580, RRID:AB_443209 | IF(1:100) |
| Antibody | Anti-pH-H3 (Rabbit Polyclonal) | Abcam | ab5176, RRID:AB_304763 | IF(1:500) |
| Antibody | Anti-GH (Rabbit Polyclonal) | National Hormone and Peptide Program (NHPP) | AFP-5641801 | IF(1:1000) |
| Antibody | Anti-TSH (Rabbit Polyclonal) | National Hormone and Peptide Program (NHPP) | AFP-1274789 | IF(1:1000) |
| Antibody | Anti-PRL (Rabbit Polyclonal) | National Hormone and Peptide Program (NHPP) | AFP-4251091 | IF(1:1000) |
| Antibody | Anti-ACTH(Mouse Monoclonal) | Fitzgerald | 10C-CR1096M1, RRID:AB_1282437 | IF(1:400) |
| Antibody | Anti-LH (Rabbit Polyclonal) | National Hormone and Peptide Program (NHPP) | AFP-697071P | IF(1:300) |
| Antibody | Anti-FSH (Rabbit Polyclonal) | National Hormone and Peptide Program (NHPP) | AFP-HFS6 | IF(1:300) |
| Antibody | Anti-ZO-1 (Rat Monoclonal) | Santa Cruz | SC33725, RRID:AB_628459 | IF(1:300) |
| Antibody | Anti-E-CADHERIN(Rabbit Monoclonal) | Cell Signaling | 3195S, RRID:AB_2291471 | IF(1:300) |
| Antibody | Anti-Rabbit 488 (Goat Polyclonal) | Life Technologies | A11008, RRID:AB_143165 | IF(1:400) |
| Antibody | Anti-Rabbit 555 (Goat Polyclonal) | Life Technologies | A21426, RRID:AB_1500929 | IF(1:400) |
| Antibody | Anti-Rabbit 633 (Goat Polyclonal) | Life Technologies | A21050, RRID:AB_141431 | IF(1:400) |
| Antibody | Anti-Goat 488 (Donkey Polyclonal) | Abcam | ab150133, RRID:AB_2832252 | IF(1:400) |
| Antibody | Anti-Chicken 488 (Goat Polyclonal) | Life Technologies | A11039, RRID:AB_142924 | IF(1:400) |
| Antibody | Anti-Chicken 647 (Goat Polyclonal) | Life Technologies | A21449, RRID:AB_1500594 | IF(1:400) |
| Antibody | Anti-Rat 555 (Goat Polyclonal) | Life Technologies | A21434, RRID:AB_141733 | IF(1:400) |
| Antibody | Anti-Mouse 555 (Goat Polyclonal) | Life Technologies | A21426, RRID:AB_1500929 | IF(1:400) |
| Antibody | Anti-Rabbit Biotinylated (Donkey Polyclonal) | Abcam | ab6801, RRID:AB_954900 | IF(1:400) |
| Antibody | Anti-Rabbit Biotinylated (Goat Polyclonal) | Abcam | ab207995 | IF(1:400) |
| Antibody | Anti-Mouse Biotinylated (Goat Biotinylated) | Abcam | ab6788, RRID:AB_954885 | IF(1:400) |
| Sequence-based reagent | RNAscope probe M. musculus Axin2 | Advanced Cell Diagnostics | 400331 | |
| Sequence-based reagent | RNAscope probe M. musculus Lef1 | Advanced Cell Diagnostics | 441861 | |
| Sequence-based reagent | RNAscope probe M. musculus Wls | Advanced Cell Diagnostics | 405011 | |
| Sequence-based reagent | RNAscope probe M. musculus Rspo1 | Advanced Cell Diagnostics | 401991 | |
| Sequence-based reagent | RNAscope probe M. musculus Rspo2 | Advanced Cell Diagnostics | 402001 | |
| Sequence-based reagent | RNAscope probe M. musculus Rspo3 | Advanced Cell Diagnostics | 402011 | |
| Sequence-based reagent | RNAscope probe M. musculus Rspo4 | Advanced Cell Diagnostics | 402021 | |
| Sequence-based reagent | RNAscope probe M. musculus Lgr4 | Advanced Cell Diagnostics | 318321 | |
| Sequence-based reagent | RNAscope probe M. musculus Wnt9a | Advanced Cell Diagnostics | 405081 | |
| Sequence-based reagent | RNAscope probe M. musculus Wnt2 | Advanced Cell Diagnostics | 313601 | |
| Sequence-based reagent | RNAscope probe M. musculus Wnt5a | Advanced Cell Diagnostics | 316791 | |
| Sequence-based reagent | RNAscope probe eGFP | Advanced Cell Diagnostics | 400281 | |
| Sequence-based reagent | RNAscope probe M. musculus Jun | Advanced Cell Diagnostics | 453561 | |
| Sequence-based reagent | RNAscope probe M. musculus Axin2 (Channel 2) | Advanced Cell Diagnostics | 400331-C2 | |
| Sequence-based reagent | RNAscope probe M. musculus Sox2 (Channel 2) | Advanced Cell Diagnostics | 401041-C2 | |
| Sequence-based reagent | RNAscope probe eGFP (Channel 2) | Advanced Cell Diagnostics | 400281-C2 | |
| Sequence-based reagent | RNAscope probe M. musculus Sox2 | Advanced Cell Diagnostics | 401041 | |
| Sequence-based reagent | RNAscope probe M. musculus Pou1f1 | Advanced Cell Diagnostics | 486441 | |
| Sequence-based reagent | RNAscope probe Duplex Positive Control Ppib-C1, Polr2a-C2 | Advanced Cell Diagnostics | 321641 | |
| Sequence-based reagent | RNAscope probe Duplex Negative Control DapB (both channels) | Advanced Cell Diagnostics | 320751 | |
| Sequence-based reagent | RNAscope probe Singleplex Positive Control Ppib | Advanced Cell Diagnostics | 313911 | |
| Sequence-based reagent | RNAscope probe: Singleplex Negative Control DapB | Advanced Cell Diagnostics | 310043 | |
| Peptide, recombinant protein | Streptavidin 488 | Life Technologies | S11223 | IF(1:400) |
| Peptide, recombinant protein | Streptavidin 555 | Life Technologies | S32355 | IF(1:400) |
| Peptide, recombinant protein | Streptavidin 633 | Life Technologies | S21375 | IF(1:400) |
| Commercial assay or kit | RNAScope 2.5 HD Assay-RED | Advanced Cell Diagnostics | 322350 | |
| Commercial assay or kit | RNAScope 2.5 HD Duplex Assay | Advanced Cell Diagnostics | 322430 | |
| Commercial assay or kit | LIVE/DEAD Fixable Near IR-Dead Cell Stain Kit | Life Technologies | L34975 | |
| Commercial assay or kit | FIX and PERM Cell Permeabilization Kit | Life Technologies | GAS003 | |
| Chemical compound, drug | Tamoxifen | Sigma | T5648 | |
| Chemical compound, drug | Corn Oil | Sigma | C8267 | |
| Chemical compound, drug | Collagenase Type 2 | Worthington | 4178 | C |
| Chemical compound, drug | 10× Trypsin | Sigma | 59418C | |
| Chemical compound, drug | Deoxyribonuclease I | Worthington | LS002172 | |
| Chemical compound, drug | Fungizone | Gibco | 15290 | |
| Chemical compound, drug | Hank’s Balanced Salt Solution (HBSS) | Gibco | 14170 | |
| Chemical compound, drug | Foetal Bovine Serum | Sigma | F2442 | |
| Chemical compound, drug | HEPES | Thermo Fisher | 15630 | |
| Chemical compound, drug | bFGF | R&D Systems | 233-FB-025 | |
| Chemical compound, drug | Cholera Toxin | Sigma | C8052 | |
| Chemical compound, drug | DMEM-F12 | Thermo Fisher | 31330 | |
| Chemical compound, drug | Penicillin/Streptomycin | Gibco | 15070-063 | |
| Chemical compound, drug | Neutral Buffered Formalin | Sigma | HT501128 | |
| Chemical compound, drug | Hoechst 33342 | Thermo Fisher | H3570 | 1:1000 |
| Chemical compound, drug | Declere | Sigma | D3565 | |
| Chemical compound, drug | Neo-Clear | Sigma | 65351-M | |
| Software, algorithm | FlowJo | FlowJo, LLC |
https://www.flowjo.com/
RRID:SCR_008520 |
|
| Software, algorithm | Prism 7 | GraphPad Software | https://www.graphpad.com/ | |
| Software, algorithm | Image Lab | Bio-Rad Laboratories | http://www.bio-rad.com/ | |
| Software, algorithm | NDP View | Hamamatsu Photonics | https://www.hamamatsu.com/ | |
| Software, algorithm | HISAT v2.0.3 | Kim et al., 2015 |
https://github.com/infphilo/hisat2
RRID:SCR_015530 |
|
| Software, algorithm | DESeq2 v2.11.38 | Love et al., 2014 |
https://github.com/Bioconductor-mirror/DESeq2
RRID:SCR_015687 |
|
| Software, algorithm | featureCounts v1.4.6p5 | Liao et al., 2014 |
http://subread.sourceforge.net/
RRID:SCR_012919 |
|
| Software, algorithm | The Galaxy Platform | Afgan et al., 2016; Blankenberg et al., 2010; Goecks et al., 2010 |
https://usegalaxu.org
RRID:SCR_006281 |
|
| Software, algorithm | Gene Set Enrichment Analysis (GSEA) | Subramanian et al., 2005 |
software.broadinstitute.org/gsea/index.jsp
RRID:SCR_003199 |
|
| Software, algorithm | Cufflinks | Trapnell et al., 2012 |
https://github.com/cole-trapnell-lab/cufflinks
RRID:SCR_014597 |
|
| Other | Deposited Data, RNA-Seq | BioProject (NCBI) | PRJNA421806 |
Mice
All procedures were performed under compliance of the Animals (Scientific Procedures) Act 1986, Home Office License (P5F0A1579). KCL Biological Services Unit staff undertook daily animal husbandry. Genotyping was performed on ear biopsies taken between P11 and P15 by standard PCR using the indicated primers. These experiments were not conducted at random and the experimenters were not blind while conducting the animal handling and assessment of tissue. Images are representative of the respective genotypes. For all studies, both male and female animals were used and results combined.
The Sox2CreERT2/+ and Sox2Egfp/+ strains were kept on a CD-1 background. Axin2CreERT2/+ animals were kept on a mixed background of C57BL/6 backcrossed onto CD-1 for five generations and were viable and fertile, with offspring obtained at the expected Mendelian ratios. ROSA26mTmG/mTmG, ROSA26Confetti/Confetti, ROSA26tdTomato/tdTomato, Wlsfl/fl, Ctnnb1fl(ex2-6)/ fl(ex2-6), and TCF/LEF:H2B-EGFP mice were kept on a mixed background of 129/Sv backcrossed onto CD-1 for at least three generations. For lineage tracing studies, male Axin2CreERT2/+ or Sox2CreERT2/+ mice were bred with homozygous ROSA26mTmG/mTmG or ROSA26Confetti/Confetti dams to produce the appropriate allele combinations on the reporter background. Pups were induced at P14 or P15 with a single dose of tamoxifen (resuspended to 20 mg/ml in Corn Oil with 10% ethanol) by intraperitoneal injection, at a concentration of 0.15 mg/g of body weight. Pituitaries were harvested at the indicated time points post-induction and processed for further analysis as described below. Mice were harvested from different litters for each time point at random. For litters in which there was a surplus of experimental mice, multiple samples were harvested for each required time point.
For Wntless deletion studies, Sox2CreERT2/+;Wlsfl/+;ROSA26mTmG/mTmG males were bred with Wlsfl/fl;ROSA26mTmG/mTmG dams, to produce Sox2CreERT2/+;Wlsfl/+;ROSA26mTmG/mTmG, Sox2CreERT2/+;Wlsfl/fl;ROSA26mTmG/mTmG, and Wlsfl/fl;ROSA26mTmG/mTmG offspring. Pups of the indicated genotypes received intraperitoneal injections of 0.15 mg of tamoxifen per gram body weight on four consecutive days, beginning at P14, and harvested 3 days after the final injection.
For the β-catenin loss-of-function experiments, either Sox2CreERT2/+;Ctnnb1fl(ex2-6)/+;ROSA26mTmG/mTmG or Axin2CreERT2/+;Ctnnb1fl(ex2-6)/+;ROSA26mTmG/mTmG males were crossed with Ctnnb1fl(ex2-6)/fl(ex2-6);ROSA26mTmG/mTmG dams. Axin2CreERT2/+;Ctnnb1fl(ex2-6)/fl(ex2-6);ROSA26mTmG/mTmG and Axin2CreERT2/+;Ctnnb1fl(ex2-6)/+;ROSA26mTmG/mTmG pups were induced with a single dose of tamoxifen, at a concentration of 0.15 mg/g of body weight and kept alive for 7 days before harvesting. Sox2CreERT2/+;Ctnnb1fl(ex2-6)/+;ROSA26mTmG/mTmG and Sox2CreERT2/+;Ctnnb1fl(ex2-6)/fl(ex2-6);ROSA26mTmG/mTmG pups received two intraperitoneal injections of tamoxifen, at a concentration of 0.15 mg/g of body weight, on two consecutive days and were kept alive for the indicated length of time before harvesting.
TCF/LEF:H2B-EGFP mice were culled and the pituitaries harvested at the indicated ages for the respective experiments. For FACS experiments, mice were harvested at 21 days of age. Axin2CreERT2/+;Sox2eGFP/+ males were crossed with ROSA26tdTomat/tdTomato dams to produce Axin2CreERT2/+;Sox2eGFP/+;ROSA26tdTomato/+ that were induced with single doses of tamoxifen at 21 and 22 days of age and harvested 3 days after the first injection for FACS experiments.
Flow cytometry analysis of lineage traced pituitaries
For the quantification of cells by flow cytometry, anterior lobes of Axin2CreERT2/+;ROSA26mTmG/+ mice dissected at the indicated time points. The posterior and intermediate lobes were dissected from the anterior lobes under a dissection microscope. Untreated ROSA26mTmG/+ and wild-type pituitaries from age-matched litters were used as tdTomato only and negative controls, respectively. Dissected pituitaries were incubated in Enzyme Mix (0.5% w/v collagenase type 2 [Lorne Laboratories], 0.1× Trypsin [Gibco], 50 μg/ml DNase I [Worthington], and 2.5 μg/ml Fungizone [Gibco] in Hank’s Balanced Salt Solution [HBSS] [Gibco]) in a cell culture incubator for up to 3 hr; 850 ml of HBSS was added to each Eppendorf in order to quench the reaction. Pituitaries were dissociated by agitation, pipetting up and down 100× at first with a 1 ml pipette, followed by 100× with a 200 μl pipette. Cells were transferred to a 15 ml Falcon tube and resuspended in 9 ml of HBSS and spun down at 200 g for 5 min. The supernatant was aspirated, leaving behind the cell pellet that was resuspended in PBS and spun down at 1000 rpm for 5 min before being resuspended in a Live/Dead cell stain (Life Technologies, L34975) prepared to manufacturer’s instructions, for 30 min in the dark. Cells were washed in PBS as above. The pellet was resuspended in FIX and PERM Cell Permeabilization Kit (Life Technologies, GAS003) prepared as per manufacturer’s instructions for 10 min at room temperature. Cells were washed as above, and the pellet was resuspended in 500 µl of FACS buffer (1% foetal calf serum [Sigma], 25 mM HEPES in PBS) and filtered through 70 μm filters (BD Falcon), into 5 ml round bottom polypropylene tubes (BD Falcon). One minute prior to analysis, 1 µl of Hoechst was added to the suspended cells and incubated. Samples were analysed on a BD Fortessa and gated according to negative and single fluorophore controls. Single cells were gated according to SSC-A and SSC-W. Dead cells were excluded according to DAPI (2 ng/ml, incubated for 2 min prior to sorting). GFP+, tdTomato+, and GFP+;tdTomato+ cells were gated according to negative controls in the PE-A and FITC-A channels.
FACS for sequencing or colony forming assays
For FACS, the anterior lobes from Sox2eGFP/+, TCF/LEF:H2B-GFP, or Axin2CreERT2/+;Sox2eGFP/+;ROSA26tdTomato/+ and their respective controls were dissected and dissociated as above. After dissociation cells were spun down at 200 g in HBSS and the pellet was resuspended in 500 µl FACS buffer. Using an Aria III FACs machine (BD systems), samples were gated according to negative controls, and where applicable single fluorophore controls. Experimental samples were sorted according to their fluorescence, as indicated, into tubes containing either RNAlater (Qiagen) for RNA isolation or 1 ml of Pit Complete Media for culture (Pit Complete: 20 ng/ml) bFGF and 50 ng/ml of cholera toxin in ‘Pit Basic’ media (DMEM-F12 with 5% foetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin). Cells were plated in 12-well plates at clonal density, approximately 500 cells/well. Colonies were incubated for a total of 7 days before being fixed in 10% neutral buffered formalin (NBF) (Sigma) for 10 min at room temperature, washed for 5 min, three times, with PBS and stained with crystal violet in order for the number of colonies to be quantified.
RNA-sequencing
Total RNA was isolated from each sample and following poly-A selection, cDNA libraries were generated using TruSeq (Clontech, 634925). Barcoded libraries were then pooled at equal molar concentrations and sequenced on an Illumina Hiseq 4000 instrument in a 75 base pair, paired-end sequencing mode, at the Wellcome Trust Centre for Human Genetics (Oxford, United Kingdom). Raw sequencing reads were quality checked for nucleotide calling accuracy and trimmed accordingly to remove potential sequencing primer contaminants. Following QC, forward and reverse reads were mapped to GRCm38/mm10 using Hisat2 (Kim et al., 2015). Using a mouse transcriptome specific GTF as a guide, FeatureCounts (Liao et al., 2014) was used to generate gene count tables for every sample. These were utilised within the framework of the Deseq2 (Love et al., 2014) and FPKM values (generated by FPKM count Wang et al., 2012) were processed using the Cufflinks (Trapnell et al., 2012) pipelines that identified statistically significant gene expression differences between the sample groups. Following identification of differentially expressed genes (at an FDR < 0.05) we focused on identifying differentially expressed pathways using a significance threshold of FDR < 0.05 unless otherwise specified. The gene lists used for GSEA were as found on the BROAD institute GSEA MSigDBv.7 ‘molecular signatures database’. The deposited data set (BioProject, accession PRJNA421806) can be accessed through the following link: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA421806.
Immunofluorescence and microscopy
Freshly harvested pituitaries were washed in PBS for 10 min before being fixed in 10% NBF for 18 hr at room temperature. In short, embryos and whole pituitaries were washed in PBS three times, before being dehydrated through a series of 1 hr washes in 25%, 50%, 70%, 80%, 90%, 95%, and 100% ethanol. Tissues were washed in Neo-Clear (Sigma) at room temperature for 10 min, then in fresh preheated Neo-Clear at 60°C for 10 min. Subsequently, tissues were incubated in a mixture of 50% Neo-Clear:50% paraffin wax at 60°C for 15 min followed by three changes of pure wax for a minimum of 1 hr washes at 60°C, before being orientated to be sectioned in the frontal plane. Embedded samples were sectioned at 5 µm and mounted on to Super Frost+ slides.
For immunofluorescence, sections were deparaffinised in Neo-Clear by three washes of 10 min, washed in 100% ethanol for three times 5 min, and rehydrated in a series of 5-min ethanol washes up to distilled water (95%, 90%, 80%, 70%, 50%, 25%, H2O). Heat induced epitope retrieval was performed with 1× DeClear Buffer (citrate pH 6) in a Decloaking chamber NXGEN (Menarini Diagnostics) for 3 min at 110°C. Slides were left to cool to room temperature before proceeding to block for 1 hr at room temperature in blocking buffer (0.2% BSA, 0.15% glycine, 0.1% TritonX in PBS) with 10% serum (sheep or donkey, depending on secondary antibodies). Primary antibodies were diluted in blocking buffer with 1% of the appropriate serum and incubated overnight at 4°C. Slides were washed three times for 10 min with PBST. Slides were incubated with secondary antibodies diluted 1:400 in blocking buffer with 1% serum for 1 hr at room temperature. Slides were washed three times with PBST as above. Where biotinylated secondary antibodies were used, slides were incubated with streptavidin diluted 1:400 in blocking buffer with 1% serum for 1 hr at room temperature. Finally, slides were washed with PBST and mounted using Vectashield Antifade Mounting Medium (Vector Laboratories, H-1000).
The following antibodies, along with their dilutions and detection technique, were used: GFP (1:400, Alexa Fluor-488 or −647 secondary), SOX2 raised in goat (1:200, Alexa Fluor-488 secondary), SOX2 raised in rabbit (1:100, biotinylated secondary), SOX9 (1:500, biotinylated secondary), PIT1 (1:500, biotinylated secondary), SF1 (1:300, biotinylated secondary), TPIT (1:200, biotinylated secondary), Ki-67 (1:100, biotinylated secondary), pH-H3 (1:500, biotinylated secondary), GH (1:1000, biotinylated secondary), TSH (1:1000, biotinylated secondary), PRL (1:1000, biotinylated secondary), ACTH (1:400, Alexa Fluor-555 secondary), LH/FSH (1:300, biotinylated secondary), ZO-1 (1:300, Alexa Fuor-488), and E-Cadherin (1:300, Alexa Fluor-488). Nuclei were visualised with Hoechst (1:1000). Images were taken on a TCS SPS Confocal (Leica Microsystem) with a 20× objective for analysis.
mRNA in situ hybridisation
All mRNA in situ hybridisations were performed using the RNAscope singleplex or duplex chromogenic kits (Advanced Cell Diagnostics) on formalin fixed paraffin embedded sections processed as described in the above section. The protocol followed the manufacturer’s instructions with slight modifications. ImmEdge Hydrophobic Barrier PAP Pen (Vector Laboratories, H-4000) was used to draw a barrier around section while air-drying following the first ethanol washes. Pretreatment followed the standard length of time for pituitaries (12 min), while embryos were boiled for 10 min. For singleplex, the protocol proceeded to follow the instructions exactly. For duplex, Amplification nine was extended to 1 hr and the dilution of the Green Detection reagent was increased to 1:30. For both protocols, sections were counterstained with Mayer’s Haematoxylin (Vector Laboratories, H-3404), left to dry at 60°C for 30 min before mounting with VectaMount Permanent Mounting Medium (Vector Laboratories, H-5000). Slides were scanned using a Nanozoomer-XR Digital Slide Scanner (Hamamatsu) and processed using Nanozoomer Digital Pathology View (Hamamatsu).
Quantification of cells
Cell numbers were quantified in ImageJ using the cell counter plugin (Schindelin et al., 2012). At a minimum, three sections per pituitary were quantified, spaced no less than 100 µM apart in the tissue.
Statistics
All statistical analyses were performed in GraphPad Prism. Data points in graphs represent the mean values of recordings from a single biological replicate unless otherwise stated.
Acknowledgements
This study has been supported by the Medical Research Council (MR/L016729/1, MR/T012153/1) (CLA), The Lister Institute of Preventive Medicine (CLA), the Deutsche Forschungsgemeinschaft (DFG German Research Foundation) (Project Number 314061271 – TRR 205) (CLA), the Howard Hughes Medical Institute (RN), the Agence Nationale de la Recherche (ANR-18-CE14-0017), and Fondation pour la Recherche Médicale (DEQ20150331732) (PM). JPR was supported by a Dianna Trebble Endowment Fund Dental Institute Studentship, EJL by the King’s Bioscience Institute and the Guy’s and St Thomas' Charity Prize PhD Programme in Biomedical and Translational Science, and YK by a Project Support Grant from the British Society for Neuroendocrinology. We thank Dr AF Parlow and the National Hormone and Peptide Program (Harbor–University of California, Los Angeles Medical Center) for providing some of the antibodies used in this study and Prof. J Drouin and Prof. S Rhodes for TPIT and PIT1 antibodies respectively. We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z) for the generation of the sequencing data. For flow sorting and analysis, this research was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. We thank Marie Isabelle Garcia, Juan Pedro Martinez-Barbera, and Paul Le Tissier for useful discussions and critical comments on the manuscript.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Cynthia L Andoniadou, Email: cynthia.andoniadou@kcl.ac.uk.
Marianne E Bronner, California Institute of Technology, United States.
Marianne E Bronner, California Institute of Technology, United States.
Funding Information
This paper was supported by the following grants:
Medical Research Council MR/L016729/1 to Cynthia Lilian Andoniadou.
Medical Research Council MR/T012153/1 to Cynthia Lilian Andoniadou.
Deutsche Forschungsgemeinschaft 314061271 - TRR 205 to Cynthia Lilian Andoniadou.
Howard Hughes Medical Institute to Roel Nusse.
Agence Nationale de la Recherche ANR-18-CE14-0017 to Patrice Mollard.
Fondation pour la Recherche Médicale DEQ20150331732 to Patrice Mollard.
Lister Institute of Preventive Medicine to Cynthia Lilian Andoniadou.
Dianna Trebble Endowment Fund to John P Russell.
King’s Bioscience Institute and the Guy’s and St Thomas’ Charity Prize to Emily J Lodge.
British Society for Neuroendocrinology to Yasmine Kemkem.
Additional information
Competing interests
Reviewing editor, eLife.
No competing interests declared.
Author contributions
Conceptualization, Formal analysis, Investigation, Methodology, Writing - original draft, Writing - review and editing.
Resources, Writing - review and editing.
Formal analysis, Investigation.
Software, Formal analysis, Investigation.
Resources, Investigation.
Resources.
Resources, Investigation.
Investigation.
Resources.
Investigation.
Investigation.
Investigation.
Resources, Supervision.
Resources, Supervision, Funding acquisition, Methodology, Writing - review and editing.
Resources, Supervision, Funding acquisition, Methodology, Writing - review and editing.
Conceptualization, Supervision, Funding acquisition, Investigation, Methodology, Writing - original draft, Writing - review and editing.
Ethics
Animal experimentation: This study was performed under compliance of the Animals (Scientific Procedures) Act 1986, Home Office License (P5F0A1579) and KCL Biological Safety approval for project 'Function and Regulation of Pituitary Stem Cells in Mammals'.
Additional files
Gene lists generated from gene set enrichment analyses of bulk RNA-sequencing data comparing Sox2+ and Sox2− cells. Associated with Figure 3—figure supplement 1.
Data availability
Sequencing data can be accessed through the following link: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA421806.
The following dataset was generated:
Russell JP, Yianni V, Andoniadou CL. 2020. Pituitary stem cells produce paracrine WNT signals to control the expansion of their descendant progenitor cells. NCBI BioProject. PRJNA421806
References
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Research. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alatzoglou KS, Andoniadou CL, Kelberman D, Buchanan CR, Crolla J, Arriazu MC, Roubicek M, Moncet D, Martinez-Barbera JP, Dattani MT. SOX2 haploinsufficiency is associated with slow progressing hypothalamo-pituitary tumours. Human Mutation. 2011;32:1376–1380. doi: 10.1002/humu.21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoniadou CL, Gaston-Massuet C, Reddy R, Schneider RP, Blasco MA, Le Tissier P, Jacques TS, Pevny LH, Dattani MT, Martinez-Barbera JP. Identification of novel pathways involved in the pathogenesis of human adamantinomatous craniopharyngioma. Acta Neuropathologica. 2012;124:259–271. doi: 10.1007/s00401-012-0957-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoniadou CL, Matsushima D, Mousavy Gharavy SN, Signore M, Mackintosh AI, Schaeffer M, Gaston-Massuet C, Mollard P, Jacques TS, Le Tissier P, Dattani MT, Pevny LH, Martinez-Barbera JP. Sox2+ Stem/Progenitor Cells in the Adult Mouse Pituitary Support Organ Homeostasis and Have Tumor-Inducing Potential. Cell Stem Cell. 2013;13:433–445. doi: 10.1016/j.stem.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V, Fassina A, Cordenonsi M, Piccolo S. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Basham KJ, Rodriguez S, Turcu AF, Lerario AM, Logan CY, Rysztak MR, Gomez-Sanchez CE, Breault DT, Koo BK, Clevers H, Nusse R, Val P, Hammer GD. A ZNRF3-dependent wnt/β-catenin signaling gradient is required for adrenal homeostasis. Genes & Development. 2019;33:209–220. doi: 10.1101/gad.317412.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Roussel-Gervais A, Drouin J. Distinct developmental roles of cell cycle inhibitors p57Kip2 and p27Kip1 distinguish pituitary progenitor cell cycle exit from cell cycle reentry of differentiated cells. Molecular and Cellular Biology. 2009;29:1895–1908. doi: 10.1128/MCB.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A, Galaxy Team Manipulation of FASTQ data with galaxy. Bioinformatics. 2010;26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajo-Pérez E, Watanabe YG. Cellular proliferation in the anterior pituitary of the rat during the postnatal period. Cell and Tissue Research. 1990;261:333–338. doi: 10.1007/BF00318674. [DOI] [PubMed] [Google Scholar]
- Carpenter AC, Rao S, Wells JM, Campbell K, Lang RA. Generation of mice with a conditional null allele for wntless. Genesis. 2010;48:554–558. doi: 10.1002/dvg.20651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castinetti F, Davis SW, Brue T, Camper SA. Pituitary stem cell update and potential implications for treating hypopituitarism. Endocrine Reviews. 2011;32:453–471. doi: 10.1210/er.2010-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Watt FM. Defining adult stem cells by function, not by phenotype. Annual Review of Biochemistry. 2018;87:1015–1027. doi: 10.1146/annurev-biochem-062917-012341. [DOI] [PubMed] [Google Scholar]
- Cox B, Laporte E, Vennekens A, Kobayashi H, Nys C, Van Zundert I, Uji-i H, Vercauteren Drubbel A, Beck B, Roose H, Boretto M, Vankelecom H. Organoids from pituitary as a novel research model toward pituitary stem cell exploration. Journal of Endocrinology. 2019;240:287–308. doi: 10.1530/JOE-18-0462. [DOI] [PubMed] [Google Scholar]
- Davis SW, Mortensen AH, Camper SA. Birthdating studies reshape models for pituitary gland cell specification. Developmental Biology. 2011;352:215–227. doi: 10.1016/j.ydbio.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschene ER, Myung P, Rompolas P, Zito G, Sun TY, Taketo MM, Saotome I, Greco V. β-Catenin activation regulates tissue growth non-cell autonomously in the hair stem cell niche. Science. 2014;343:1353–1356. doi: 10.1126/science.1248373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupé DP, Marshall OJ, Dayton H, Brand AH, Perrimon N. Drosophila intestinal stem and progenitor cells are major sources and regulators of homeostatic niche signals. PNAS. 2018;115:12218–12223. doi: 10.1073/pnas.1719169115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Developmental Neuroscience. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- Fauquier T, Rizzoti K, Dattani M, Lovell-Badge R, Robinson IC. SOX2-expressing progenitor cells generate all of the major cell types in the adult mouse pituitary gland. PNAS. 2008;105:2907–2912. doi: 10.1073/pnas.0707886105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Vaquer A, Piliszek A, Tian G, Aho RJ, Dufort D, Hadjantonakis AK. A sensitive and bright single-cell resolution live imaging reporter of wnt/ß-catenin signaling in the mouse. BMC Developmental Biology. 2010;10:121. doi: 10.1186/1471-213X-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston-Massuet C, Andoniadou CL, Signore M, Jayakody SA, Charolidi N, Kyeyune R, Vernay B, Jacques TS, Taketo MM, Le Tissier P, Dattani MT, Martinez-Barbera JP. Increased wingless (Wnt) signaling in pituitary progenitor/stem cells gives rise to pituitary tumors in mice and humans. PNAS. 2011;108:11482–11487. doi: 10.1073/pnas.1101553108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biology. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Meljem JM, Haston S, Carreno G, Apps JR, Pozzi S, Stache C, Kaushal G, Virasami A, Panousopoulos L, Mousavy-Gharavy SN, Guerrero A, Rashid M, Jani N, Goding CR, Jacques TS, Adams DJ, Gil J, Andoniadou CL, Martinez-Barbera JP. Stem cell senescence drives age-attenuated induction of pituitary tumours in mouse models of paediatric craniopharyngioma. Nature Communications. 2017;8:1819. doi: 10.1038/s41467-017-01992-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and Cancer. Nature. 2015;526:715–718. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- Gremeaux L, Fu Q, Chen J, Vankelecom H. Activated phenotype of the pituitary stem/progenitor cell compartment during the early-postnatal maturation phase of the gland. Stem Cells and Development. 2012;21:801–813. doi: 10.1089/scd.2011.0496. [DOI] [PubMed] [Google Scholar]
- Haston S, Pozzi S, Carreno G, Manshaei S, Panousopoulos L, Gonzalez-Meljem JM, Apps JR, Virasami A, Thavaraj S, Gutteridge A, Forshew T, Marais R, Brandner S, Jacques TS, Andoniadou CL, Martinez-Barbera JP. MAPK pathway control of stem cell proliferation and differentiation in the embryonic pituitary provides insights into the pathogenesis of papillary craniopharyngioma. Development. 2017;144:2141–2152. doi: 10.1242/dev.150490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Yoshida S, Kanno N, Mitsuishi H, Ueharu H, Chen M, Nishimura N, Kato T, Kato Y. Clump formation in mouse pituitary-derived non-endocrine cell line tpit/F1 promotes differentiation into growth-hormone-producing cells. Cell and Tissue Research. 2017;369:353–368. doi: 10.1007/s00441-017-2603-2. [DOI] [PubMed] [Google Scholar]
- Iwai-Liao Y, Kumabe S, Takeuchi M, Higashi Y. Immunohistochemical localisation of epidermal growth factor, transforming growth factor alpha and EGF receptor during organogenesis of the murine hypophysis in vivo. Okajimas Folia Anatomica Japonica. 2000;76:291–301. doi: 10.2535/ofaj1936.76.6_291. [DOI] [PubMed] [Google Scholar]
- Kelberman D, de Castro SC, Huang S, Crolla JA, Palmer R, Gregory JW, Taylor D, Cavallo L, Faienza MF, Fischetto R, Achermann JC, Martinez-Barbera JP, Rizzoti K, Lovell-Badge R, Robinson IC, Gerrelli D, Dattani MT. SOX2 plays a critical role in the pituitary, forebrain, and eye during human embryonic development. The Journal of Clinical Endocrinology & Metabolism. 2008;93:1865–1873. doi: 10.1210/jc.2007-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lim X, Tan SH, Koh WL, Chau RM, Yan KS, Kuo CJ, van Amerongen R, Klein AM, Nusse R. Interfollicular epidermal stem cells self-renew via autocrine wnt signaling. Science. 2013;342:1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge EJ, Russell JP, Patist AL, Francis-West P, Andoniadou CL. Expression analysis of the hippo cascade indicates a role in pituitary stem cell development. Frontiers in Physiology. 2016;7:114. doi: 10.3389/fphys.2016.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge EJ, Santambrogio A, Russell JP, Xekouki P, Jacques TS, Johnson RL, Thavaraj S, Bornstein SR, Andoniadou CL. Homeostatic and tumourigenic activity of SOX2+ pituitary stem cells is controlled by the LATS/YAP/TAZ cascade. eLife. 2019;8:e43996. doi: 10.7554/eLife.43996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseenko A, Kheirollahi V, Chao CM, Ahmadvand N, Quantius J, Wilhelm J, Herold S, Ahlbrecht K, Morty RE, Rizvanov AA, Minoo P, El Agha E, Bellusci S. Origin and characterization of alpha smooth muscle actin-positive cells during murine lung development. Stem Cells. 2017;35:1566–1578. doi: 10.1002/stem.2615. [DOI] [PubMed] [Google Scholar]
- Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359:1118–1123. doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Osmundsen AM, Keisler JL, Taketo MM, Davis SW. Canonical WNT signaling regulates the pituitary organizer and pituitary gland formation. Endocrinology. 2017;158:3339–3353. doi: 10.1210/en.2017-00581. [DOI] [PubMed] [Google Scholar]
- Palma V, Lim DA, Dahmane N, Sánchez P, Brionne TC, Herzberg CD, Gitton Y, Carleton A, Alvarez-Buylla A, Ruiz i Altaba A. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005;132:335–344. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Saganta A, Tata PR, Law BM, Saez B, Chow RD, Prabhu M, Gridley T, Rajagopal J. Parent stem cells can serve as niches for their daughter cells. Nature. 2015;523:597–601. doi: 10.1038/nature14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Millán MI, Brinkmeier ML, Mortensen AH, Camper SA. PROP1 triggers epithelial-mesenchymal transition-like process in pituitary stem cells. eLife. 2016;5:e14470. doi: 10.7554/eLife.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Rao MS. The stem-cell menagerie. Trends in Neurosciences. 2003;26:351–359. doi: 10.1016/S0166-2236(03)00169-3. [DOI] [PubMed] [Google Scholar]
- Potok MA, Cha KB, Hunt A, Brinkmeier ML, Leitges M, Kispert A, Camper SA. WNT signaling affects gene expression in the ventral diencephalon and pituitary gland growth. Developmental Dynamics. 2008;237:1006–1020. doi: 10.1002/dvdy.21511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzoti K, Akiyama H, Lovell-Badge R. Mobilized adult pituitary stem cells contribute to endocrine regeneration in response to physiological demand. Cell Stem Cell. 2013;13:419–432. doi: 10.1016/j.stem.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose H, Cox B, Boretto M, Gysemans C, Vennekens A, Vankelecom H. Major depletion of SOX2+ stem cells in the adult pituitary is not restored which does not affect hormonal cell homeostasis and remodelling. Scientific Reports. 2017;7:16940. doi: 10.1038/s41598-017-16796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Huebner AJ, Sulahian R, Anselmo A, Xu X, Flattery K, Desai N, Sebastian C, Yram MA, Arnold K, Rivera M, Mostoslavsky R, Bronson R, Bass AJ, Sadreyev R, Shivdasani RA, Hochedlinger K. Sox2 suppresses gastric tumorigenesis in mice. Cell Reports. 2016;16:1929–1941. doi: 10.1016/j.celrep.2016.07.034. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed SM, Kumar M, Ghosh A, Tomasetig F, Ali A, Whan RM, Alterman D, Tanwar PS. Endometrial Axin2+ cells drive epithelial homeostasis, regeneration, and Cancer following oncogenic transformation. Cell Stem Cell. 2020;26:64–80. doi: 10.1016/j.stem.2019.11.012. [DOI] [PubMed] [Google Scholar]
- Takase HM, Nusse R. Paracrine wnt/β-catenin signaling mediates proliferation of undifferentiated spermatogonia in the adult mouse testis. PNAS. 2016;113:E1489–E1497. doi: 10.1073/pnas.1601461113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo M, Chou WC, Sun Q, Lee W, Rabbani P, Loomis C, Taketo MM, Ito M. Wnt activation in nail epithelium couples nail growth to digit regeneration. Nature. 2013;499:228–232. doi: 10.1038/nature12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DW, Barker N. Intestinal stem cells and their defining niche. Current Topics in Developmental Biology. 2014;107:77–107. doi: 10.1016/B978-0-12-416022-4.00003-2. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Yasutaka S, Kominami R, Shinohara H. Mitoses of thyrotrophs contribute to the proliferation of the rat pituitary gland during the early postnatal period. Anatomy and Embryology. 2002;206:67–72. doi: 10.1007/s00429-002-0283-4. [DOI] [PubMed] [Google Scholar]
- Tata PR, Rajagopal J. Regulatory circuits and Bi-directional signaling between stem cells and their progeny. Cell Stem Cell. 2016;19:686–689. doi: 10.1016/j.stem.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nature Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Vidal V, Sacco S, Rocha AS, da Silva F, Panzolini C, Dumontet T, Doan TM, Shan J, Rak-Raszewska A, Bird T, Vainio S, Martinez A, Schedl A. The adrenal capsule is a signaling center controlling cell renewal and zonation through Rspo3. Genes & Development. 2016;30:1389–1394. doi: 10.1101/gad.277756.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems C, Fu Q, Roose H, Mertens F, Cox B, Chen J, Vankelecom H. Regeneration in the pituitary after Cell-Ablation injury: time-related aspects and molecular analysis. Endocrinology. 2016;157:705–721. doi: 10.1210/en.2015-1741. [DOI] [PubMed] [Google Scholar]
- Xekouki P, Lodge EJ, Matschke J, Santambrogio A, Apps JR, Sharif A, Jacques TS, Aylwin S, Prevot V, Li R, Flitsch J, Bornstein SR, Theodoropoulou M, Andoniadou CL. Non-secreting pituitary tumours characterised by enhanced expression of YAP/TAZ. Endocrine-Related Cancer. 2019;26:215–225. doi: 10.1530/ERC-18-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Janda CY, Chang J, Zheng GXY, Larkin KA, Luca VC, Chia LA, Mah AT, Han A, Terry JM, Ootani A, Roelf K, Lee M, Yuan J, Li X, Bolen CR, Wilhelmy J, Davies PS, Ueno H, von Furstenberg RJ, Belgrader P, Ziraldo SB, Ordonez H, Henning SJ, Wong MH, Snyder MP, Weissman IL, Hsueh AJ, Mikkelsen TS, Garcia KC, Kuo CJ. Non-equivalence of wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Kato T, Higuchi M, Chen M, Ueharu H, Nishimura N, Kato Y. Localization of juxtacrine factor ephrin-B2 in pituitary stem/progenitor cell niches throughout life. Cell and Tissue Research. 2015;359:755–766. doi: 10.1007/s00441-014-2054-y. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kato T, Kanno N, Nishimura N, Nishihara H, Horiguchi K, Kato Y. Cell type-specific localization of ephs pairing with ephrin-B2 in the rat postnatal pituitary gland. Cell and Tissue Research. 2017;370:99–112. doi: 10.1007/s00441-017-2646-4. [DOI] [PubMed] [Google Scholar]
- Zhu X, Tollkuhn J, Taylor H, Rosenfeld MG. Notch-Dependent pituitary SOX2(+) Stem cells exhibit a timed functional extinction in regulation of the postnatal gland. Stem Cell Reports. 2015;5:1196–1209. doi: 10.1016/j.stemcr.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]