Abstract
Tuberculosis is the world's most deadly infectious disease, with 10 million people falling ill and 1.5 million people dying from the disease every year. With the increasing number of drug-resistant Mycobacterium tuberculosis (MTB) strains and prevalence of coinfection of MTB with human immunodeficiency virus, many challenges remain in the prevention and treatment of tuberculosis. Therefore, the development of safe and effective tuberculosis vaccines is an urgent issue. In this study, we identified cytotoxic T lymphocyte epitopes on drug resistance-associated membrane protein efflux pumps of MTB, the ATP-binding cassette and the major facilitator superfamilies. First, three online software were used to predict HLA-A2-restricted epitopes. Then, the candidate epitopes were confirmed with the T2A2 cell binding affinity and peptide/MHC (pMHC) complex stability assays and in vitro immune activity experiments. Two drug-resistant T lymphocyte epitopes, designated Rv1218c-p24 and Rv2477c-p182, were selected, and their immunogenic activities studied in vivo in genetically engineered mice. The immune activities of these two epitopes were improved with the help of complete Freund's adjuvant (CFA). The epitopes identified here provide a foundation for the diagnosis and treatment of patients infected with drug resistant and the future development of a multiepitope vaccine.
1. Introduction
Despite the extensive use of the Mycobacterium bovis (Mb) bacilli Calmette Guérin (BCG) vaccine, tuberculosis (TB) is still the world's most deadly infectious disease. The World Health Organization (WHO) estimated that 10 million cases occurred worldwide in 2018, and 1.5 million people died from this disease, including 251,000 people coinfected with human immunodeficiency virus. Two-thirds of the total number of new cases of TB occurred in eight countries, including India, China, and Indonesia [1, 2]. Although antibiotics such as rifampicin, isoniazid, and ofloxacin are recommended by the WHO for the treatment of tuberculosis, and these antibiotics have significantly prevented the spread of MTB in past decades, they are associated with toxicities and may hasten the development of drug resistance in MTB, so the number of new cases remains huge [3]. Drug-resistant TB (DR-TB) can be divided into multidrug-resistant TB (MDR-TB), extensively drug-resistant TB (XDR-TB), and totally drug-resistant TB (TDR-TB). MDR-TB causes a disease that does not respond to the two most powerful first-line anti-TB drugs, isoniazid and rifampicin, where XDR-TB is a more serious form of MDR-TB, which does not respond to the most effective second-line anti-TB drugs and often leaves patients with no further appropriate medicines [3–5]. BCG is the only vaccine currently available that effectively prevents MTB, but BCG only induces a good protective immune response in childhood [6]. Therefore, the development of new strategies for the treatment and/or prevention of MDR-TB and XDR-TB is urgently required.
In recent years, immunotherapy has developed rapidly and demonstrated its unique advantages in the treatment of many diseases, including cancer and infectious diseases, and the CD8+ T cell-mediated cellular immune response plays an important role in these therapies. Several studies have reported that reactive CD8+ T cells can be detected in the circulation of purified protein derivative-positive (PPD+) healthy donors and patients with active TB, which are responsive to the bacterial proteins ESAT-6, CFP-10, CFP21, and so on [7–10]. Several groups of MHC I-restricted cytotoxic T lymphocyte (CTL) epitopes have been identified in MTB, and a key research strategy is to select ideal antigens of MTB to identify the CTL epitopes.
The main reason for the emergence of DR-TB is that MTB uses many mechanisms to eliminate antibiotics, including drug efflux pumps and simulating targets [11]. Antibiotic efflux is a ubiquitous mechanism underlying both innate and acquired drug resistance. So far, five efflux pump families have been identified: the ATP-binding cassette (ABC) superfamily [12], the major facilitator superfamily (MFS) [13], the multidrug and toxic compound extrusion family (MATE) [14], small multidrug-resistant family (SMR) [15], and the resistance nodulation division family (RND) [16]. ABC transporters and MFS are the largest superfamilies of efflux pumps in M. tuberculosis. The overexpression of some ABC transporter proteins is reported to be an important cause of drug resistance, ultimately leading to chemotherapy failure. Rv0194, Rv0933 (PstB), Rv1218c, Rv1819c, Rv2209, Rv2477c, Rv2686c, Rv2687c, Rv1273C, and Rv2937 have been identified as members of the ABC transporter superfamily that are associated with MDR-TB and XDR-TB [17–19]. The increased expression of Rv0933 (PstB) in clinical drug-resistant tuberculosis isolates may contribute to drug resistance in MTB [20], and the significantly elevated expression of Rv1218c at the transcriptional level has been observed in MDR-TB strains [21]. Overexpression of two of the proteins (Rv2477 and Rv2209) has also been observed with ofloxacin stress in MTB [22]. The deletion of the Rv1877 homolog increased bacterial susceptibility to ethidium bromide, acriflavine, and erythromycin [23]. The MFS transporter family is the largest class of proteins present in all living organisms. In general, MFS transporters are efflux pumps that transport small molecular solutes along electrochemical gradients [24]. Some MFS transporters can also form triple complexes with a membrane fusion protein and an outer membrane protein, across the outer membrane of Gram-negative bacteria. Rv2994, Rv3728, Rv2459c, Rv1877, Rv1258c, and Rv1410c are drug resistance-related efflux pumps in the MFS' family expressed in MTB [10, 23, 25, 26].
In this study, HLA-A∗0201-restricted CD8+ CTL epitopes were predicted on 10 efflux pump antigens with the online tools SYFPEITHI (http://www.syfpeithi.de/), BIMAS (https://www-bimas.cit.nih.gov/molbio/hla_bind/hla_motif_search_info.html), and NetCTL-1.2 (http://www.cbs.dtu.dk/services/NetCTL/), and the epitopes with high scores were selected and synthesized. Peptide binding affinity and peptide/HLA-A∗0201complex stability assays were performed using T2A2 cells in vitro, and the abilities of peptide induced CTLs (in vitro and in vivo) to secrete interferon γ (IFN-γ) and lyse the target cells that were detected.
2. Materials and Methods
2.1. Candidate Peptides
First, we used the common online T cell epitope prediction tools, SYFPEITHI, BIMAS, and NetCTL-1.2, to predict candidate 9-mer peptides from 10 antigens of ABC/MFS Efflux Pumps Family [27, 28]. Then, these peptides were synthesized by standard solid-phase Fmoc strategy and purified with reverse-phase high performance liquid chromatography (RP-HPLC) (the purity of each peptide is more than 95%). Subsequently, the molecular weight of each peptide was confirmed with electrospray ionization mass spectrometry (ESI-MS). Peptide cyclooxygenase-2 (COX-2)321-329 (ILIGETIKI) which identified before was used as positive control in T2A2 binding assay and peptide/HLA-A∗0201 complex stability assay [29]. The IAb-restricted HBV core antigen-derived T helper epitope (sequence 128-140: TPPAYRPPNAPIL) was used in the in vivo assay [30]. All the purified peptide with the correct molecular weight were dissolved at the concentration of 10 mg/mL in DMSO and stored at -80°C.
2.2. Cell Line, Human Blood Samples, and Transgenic Mice
The lymphoblastic cell line T2A2 cells (TAP deficient) were a kindly gift by Professor Yuzhang Wu (Third Military Medical University, China) and were cultured in IMDM medium supplemented with 20% fetal bovine serum (FBS, Biological Industries, BI, Israel), 2 mM L-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin, at 37°C in a humidified atmosphere of 5% CO2.
40 mL peripheral blood samples were obtained from HLA-A∗02-positive PPD-positive (HLA-A∗02+PPD+) and HLA-A∗02-positive PPD-negative (HLA-A∗02+PPD−) donors, and verbal informed consent was obtained from all individuals, and the collection was approved by the ethics committee of Zhengzhou University; then, PBLs (peripheral blood lymphocytes) were isolated by using Ficoll-Paque density gradient centrifugation method (lymphocyte separation liquid, TBD, China).
HLA-A2.1/Kb transgenic mice used in this article were purchased from Model Animal Research Center of Nanjing University (Nanjing, China) and bred in our lab in specific pathogen-free facilities (SPF) with SPF food and sterilized water [31]. And mice used in in vivo experiment were 8 weeks; all experiments were performed according to the guidelines of the Institutional Animal Care and Use Committee of Zhengzhou University.
2.3. Binding Affinity and Peptide/HLA-A2 Complex Stability Assay
Candidate peptides have high binding affinity to MHC-I molecule and form a stable peptide/MHC-I complex which is the first step to activate the specific T cell response. So, these assays were performed according to our former study [32]. Briefly, T2A2 cells (1 × 106 cells/mL) resuspended in serum-free IMDM medium were seeded in 24-well plates, added 50 μg/mL peptide and 3 μg/mL human β2-microglobulin (β2-M, Merck, Germany) simultaneously, and cultured at 37°C for 18 h. Then, cells were collected, washed twice with precooled PBS buffer (containing 5% FBS), and incubated with HLA-A2-PE-Cyanine 7 monoclonal antibody (Clone: BB7.2, 25-9876-42, eBioscience, USA) for 30 min at 4°C. Finally, cells were harvested, washed, and analyzed the fluorescence intensity (MFI) of each group by flow cytometer (FACS Calibur, BD Bioscience, USA). The fluorescence index (FI) was calculated as follows: FI = [mean fluorescence intensity (MFI) of the peptide − MFI of the background]/[MFI of the background]. And the background group was added with the corresponding solution buffer.
The peptide/MHC complex stabilization assays were performed similarly [32]. T2A2 cells were incubated with 50 μg/mL peptides and 3 μg/mL β2-M for 18 h. Then, 10 μg/mL brefeldin A (203729, Sigma, USA) were added in medium for another 1 h, subsequently washed by serum-free IMDM medium and incubated at 37°C for 0, 2, 4, and 6 h. Finally, cells were washed twice, stained with HLA-A2-PE-Cyanine 7 monoclonal antibody, and analyzed by flow cytometer (FACS Calibur, BD Bioscience, USA). DC50 was defined as an estimate of the time required for the loss of 50% of the peptide/HLA-A∗0201 complexes stabilized at time 0 h.
2.4. Generation of Autologous DCs from Human PBMCs
Briefly, PBMCs isolated from HLA-A2+ PPD+ and HLA-A2+ PPD− donors were resuspended in RPMI-1640 medium supplemented with 10% FBS and seeded in 6-well plates for 4 h at 37°C, 5% CO2. Adherent cells were washed twice with RPMI-1640 medium, and then cultured in DC medium (RPMI-1640 medium supplement with 10% FBS, 2 mM L-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin, 100 ng/mL IL-4 (200-04, PeproTech), 100 ng/mL GM-CSF (300-03, PeproTech)). On day 3, Fresh DC media (100 ng/mL IL-4 and 100 ng/mL GM-CSF) were replenished. On day 5 of the culture, 10 ng/mL lipopolysaccharide (LPS, Sigma, USA) was added for another two days to induce the maturation of DCs [33].
2.5. Induction of Peptide Specific CTLs in vitro
The CTL induction assay was performed in accordance with the protocols as previously described [32, 33]. Briefly, DCs were pulsed with different peptides at a final concentration of 10 μg/mL in the presence of 3 μg/mL β2-M for 24 h. Then, peptide-pulsed DCs (1 × 105) were cocultured with thawed autologous T cells (1 × 106) in 24-well plates in RPMI-1640 medium supplement with 10% FBS, human recombinant IL-2 (rhIL-2, 50 U/mL, 200-02, PeproTech, USA), and human recombinant IL-7 (rhIL-7, 10 ng/mL, 200-07, PeproTech, USA). Autologous T cells were restimulated three times at weekly intervals to generate peptide-specific CTLs. 50 U/mL rhIL-2 and 10 ng/mL rhIL-7 were added at intervals of 2 days. On day 21, the induced T cells were collected and their IFN-γ secretion was determined with ELISPOT assay, intracellular cytokine staining (ICS) assay, and LDH cytotoxic assay.
2.6. Generation of CTLs in HLA-A2.1/Kb Transgenic Mice
Activity of CTLs induced with IFA-emulsified peptide in HLA-A2.1/Kb transgenic mice was performed as previously described [31]. Firstly, HLA-A2.1/Kb transgenic mice (n = 5) were subcutaneously immunized at the base of tail with each peptide (100 μg/mice) and T helper epitope (140 μg/mice) emulsified with incomplete Freund's adjuvant (IFA, F5506, Sigma, USA) at 1 : 1 three times (on days 0, 5, and 10).
To establish a similar immune effect of PPD+ donor, we considered whether the complete Freund's adjuvant (CFA, F5881, Sigma, USA) which contains inactivated mycobacterium tuberculosis could use for enhancing the immune response of the peptide. Activity of CTLs induced with peptide after preimmunization with CFA in HLA-A2.1/Kb transgenic mice was performed. Emulsified complete Freund's adjuvant (CFA, 200 μL) was subcutaneously injected into HLA-A2.1/Kb transgenic mice (n = 5) on day-14. The mice were then subcutaneously immunized at the base of the tail with each peptide (100 μg/mice) and T helper epitope (140 μg/mice) emulsified with incomplete Freund's adjuvant (IFA) at 1 : 1 (on days 0, 5, and 10).
On day 11, all mice were sacrificed and the serum was collected from each mouse to measure the IFN-γ concentration with an ELISA assay. The splenic lymphocytes were isolated from each mouse and restimulated once with each peptide (10 μg/mL) and murine recombinant IL-2 (rmIL-2, 50 U/mL, 212-12-20, PeproTech, USA) in vitro for another 6 days. On day 7, the IFN-γ release in each group was measured with an intracellular cytokine staining (ICS) assay and the lysis activity was measured with an LDH assay, and the E : T ratios were 20 : 1, 40 : 1, and 80 : 1. The body weight change of each mouse was recorded [31].
2.7. Enzyme-Linked Immunospot (ELISPOT) Assay
ELISPOT assay was performed using a commercial kit according to previous studies (Dakewe, China) [32]. In brief, on day 21, peptide-induced T cells were used as effector cells, and effector cells (1 × 105) and stimulator target cells (T2A2 cells loaded with/without corresponding peptide, 1 × 105) were seeded into the 96-well plates which were precoated with anti-human IFN-γ antibody and coincubated at 37°C. After incubation for 18 h, cells in plate were lysed by ice deionized water and washed thrice by provided buffer of the kit, followed by incubating with detection antibodies and enzyme-linked avidin every other hour at 37°C. After the termination of drying, the number of spots was determined automatically by using a computer-assisted video image analyzer (Dakewe, China).
2.8. Intracellular Cytokine IFN-γ Staining (ICS) Assay
IFN-γ release of induced CD8+ T cells was measured by the intracellular cytokine staining assay according to our former study [31, 34]. Briefly, the induced CTLs (1 × 106) and T2A2 cells loaded with/without corresponding peptide (1 × 106) were incubated by adding protein transport inhibitor (containing brefeldin A) for 5 hours at 37°C. Afterward, cells were harvested, transferred to 1.5 mL EP tubes, and washed by PBS (contain 2% FBS). Cells were then stained for cell surface markers (anti-human CD3 PerCP-eFlour (Clone: OKT3, 46-0037-42, eBioscience, USA)/anti-mouse CD3 PerCP-eFlour710 (Clone: 17A2, 46-0032-80, eBioscience, USA) and anti-human CD8α APC (Clone: SK1, 17-0087-42, eBioscience, USA)/anti-mouse CD8α PE (Clone: 53-6.7, 12-0081-82, eBioscience, USA)) for 30 min at 4°C. Afterward, the cells were fixed by fixation buffer for another 30 min at room temperature and washed twice with permeabilization wash buffer. Anti-human IFN-γ PE (Clone: 4S.B3, 12-7319-42, eBioscience, USA) or anti-mouse IFN-γ APC (Clone: XMG1.2, 17-7311-81, eBioscience, USA) was added to those tubes for intracellular staining for 30 min at 4°C. After being washed twice with permeabilization wash buffer, the IFN-γ release of CD8+ T cells was analyzed by flow cytometry (FACS Calibur, BD Bioscience, USA).
2.9. ELISA Assay
ELISA assay was performed using a commerciall kit following the manufacturer's instructions (121002, Dakewe, China) [34, 35]. In brief, the serum of each mouse was diluted and added into the 96-well plates which were precoated with anti-human IFN-γ antibody and coincubated at room temperature, then incubated with detection antibodies at room temperature for 90 min, washed quartic, and added enzyme-linked avidin every other hour. After adding TMB and stop solution, the OD450 was detected and the concentration of IFN-γ in each group was calculated.
2.10. LDH Cytotoxicity Assay
Cytotoxic activity was tested based on the measurement of LDH release using the nonradioactive cytotoxicity assay kit (G1780, Promega, USA) at gradient E : T (effector cells/target cells) ratio [32]. T2A2 cells (2 × 106/mL) were pulsed by each peptide (50 μg/mL) at 37°C for 4 h and used as the target cells. CTLs induced by each group were used as the effector cells. Then, target cells (1 × 105/well) were cocultured with different amount effector cells (E : T ratio was 12.5 : 1, 25 : 1, and 50 : 1 in vitro and 20 : 1, 40 : 1, and 80 : 1 in vivo, respectively) at 37°C for 4 h. Subsequently, the plate was centrifuged, and the supernatant of each well was transferred to a new plate, added the substrate mixture, and incubated out of light at room temperature. Finally, the stop solution was added and OD490 was measured. The percentage of specific lysis of the target cells was determined as follows: percentage of specific lysis = [(experimental release − effector spontaneous release − target spontaneous release)/(target maximum release − target spontaneous release)] × 100.
2.11. Statistical Analysis
All data were presented as means ± SD. Comparisons between experimental groups and relevant controls were analyzed by 1-tailed Student's t-test. P < 0.05, P < 0.01, and P < 0.001 represent significance of differences relative to the control group.
3. Results
3.1. CTL Epitope Prediction and Synthesis
First, 29 HLA-A∗0201-resticted candidate 9-mer peptides with high prediction scores (SYFPEITHI > 24, BIMAS > 20, and NetCTL − 1.2 > 0.9) were synthesized, as shown in Table 1, and purified with RP-HPLC, and their molecular weights were confirmed with ESI-MS (Table 2).
Table 1.
Prediction scores of HLA-A2-restricted peptides derived from efflux pump antigens.
| Antigen | Position | Sequence | Scores | ||
|---|---|---|---|---|---|
| SYFPEITHI | BIMAS | NetCTL-1.2 | |||
| Rv0194 | p160 | LLAVLLVPV | 31 | 271.948 | 1.1865 |
| Rv0933 | p186 | LLDEPTSAL | 28 | 59.558 | 1.1356 |
| p208 | SLADRLTVI | 27 | 40.336 | 1.28 | |
| Rv1218c | p24 | ALDGLDLTV | 28 | 27.821 | 1.2377 |
| p200 | ALCEKVTII | 27 | 40.336 | 0.9255 | |
| p158 | LLDEPSSGL | 26 | 59.558 | 0.988 | |
| p280 | SLVSQPPTL | 25 | 21.362 | 1.0547 | |
| p165 | GLDPLMENV | 24 | 31.354 | 1.1238 | |
| Rv1819c | p74 | LLLSVVLAV | 30 | 1006.209 | 1.3652 |
| p70 | MLGVLLLSV | 28 | 271.948 | 1.2016 | |
| p122 | WMSIGVFSV | 26 | 3082.436 | 1.2808 | |
| Rv2209 | p158 | VLATGVTLV | 29 | 271.948 | 1.2373 |
| Rv2477c | p182 | LLDEPTNHL | 28 | 59.558 | 0.9542 |
| p316 | RLGNVVVEV | 28 | 159.97 | 1.1527 | |
| p362 | TLFKTIVGL | 28 | 181.794 | 1.1419 | |
| p265 | RLTEELAWV | 27 | 3607.314 | 1.2421 | |
| p173 | LLLSKPDLL | 25 | 65.841 | 0.989 | |
| Rv1877 | p71 | YLLGGTVVV | 32 | 485.348 | 1.4044 |
| p95 | LLGSVVVFV | 28 | 1495.716 | 1.166 | |
| p215 | YLGILVIAV | 28 | 735.86 | 1.217 | |
| p295 | AMLGALTFV | 28 | 2351.109 | 1.3441 | |
| Rv2994 | p223 | LMMPQTVTV | 29 | 315.959 | 1.407 |
| p100 | SLLWIGVFL | 26 | 434.725 | 1.2626 | |
| p307 | VLLMIAISV | 25 | 437.482 | 1.2895 | |
| Rv3728 | p264 | LLAAAVAMV | 30 | 271.948 | 1.2172 |
| p313 | YLLTHVLFL | 29 | 4599.389 | 1.5011 | |
| p110 | ILGVALFTV | 28 | 1577.3 | 1.2034 | |
| p54 | FLDSTIVNV | 27 | 294.344 | 1.5013 | |
| Rv2459c | p469 | VLAADAVFV | 25 | 650.311 | 1.1335 |
Table 2.
The data of ESI-MS and the binding affinity and stability of peptide/HLA-A∗0201 complex.
| Antigen | Peptide | ESI − MS[M ± H]± | MFI | FIb | DC50c | |
|---|---|---|---|---|---|---|
| Calculated | Observed | |||||
| Cell | - | - | - | 5.64 | - | |
| Background | - | - | - | 131 | - | - |
| Rv0194 | p160 | 936.25 | —a | ND | ND | ND |
| Rv0933 | p186 | 958.08 | 959.90 | 465 | 2.52 | <2 h |
| p208 | 987.16 | 988.57 | 479 | 2.59 | <2 h | |
| Rv1218c | p24 | 916.04 | 916.89 | 569 | 3.32 | >4 h |
| P200 | 989.24 | 989.78 | 366 | 1.75 | >4 h | |
| p158 | 930.02 | 930.85 | 439 | 2.35 | >4 h | |
| p280 | 941.09 | 939.98 | 353 | 1.62 | <2 h | |
| p165 | 987.14 | 987.69 | 479 | 2.6 | > 2 h | |
| Rv1819c | p74 | 926.21 | —a | ND | ND | ND |
| p70 | 944.25 | —a | ND | ND | ND | |
| p122 | 1025.24 | 1026.6 | 177 | 0.35 | ND | |
| Rv2209 | p158 | 872.07 | 873.4 | 462 | 2.54 | >4 h |
| Rv2477c | p182 | 1051.16 | 1051.77 | 379 | 1.87 | >6 h |
| p316 | 984.16 | 984.8 | 348 | 1.58 | >2 h | |
| p362 | 991.24 | 991.85 | 367 | 1.72 | > 2 h | |
| p265 | 1116.28 | 1116.87 | 315 | 1.36 | ND | |
| p173 | 1011.27 | 1011.89 | 272 | 1.15 | ND | |
| Rv1877 | p71 | 920.12 | 919.59 | 419 | 2.17 | >2 h |
| p95 | 932.17 | —a | ND | ND | ND | |
| p215 | 960.22 | —a | ND | ND | ND | |
| p295 | 922.16 | —a | ND | ND | ND | |
| Rv2994 | p223 | 1019.29 | 1020.62 | 430 | 2.28 | <2 h |
| p100 | 1047.3 | —a | ND | ND | ND | |
| p307 | 958.27 | —a | ND | ND | ND | |
| Rv3728 | p264 | 858.11 | —a | ND | ND | ND |
| p313 | 1118.38 | —a | ND | ND | ND | |
| p110 | 932.17 | —a | ND | ND | ND | |
| p54 | 1007.15 | —a | ND | ND | ND | |
| Rv2459c | p469 | 904.07 | 905.42 | 440 | 2.36 | >4 h |
| COX-2d | p321 | 999.6 | 1000.3 | 408 | 2.11 | >4 h |
aPurification failed. bFI = [mean fluorescence intensity (MFI) of the peptide − MFI of the background]/[MFI of the background]. cDC50: estimate of the time required for the loss of 50% of the peptide/HLA-A∗0201 complexes stabilized at time 0 h. dPositive control peptide. ND: not determined. Background group was added with the corresponding solution buffer.
3.2. Binding Affinity and Peptide/HLA-A∗0201 Complex Stability
Because the solubility of some peptides was low, the binding affinity of only 17 peptides was determined, as shown in Table 2 and Figure 1(a), and 14 peptides with high binding affinity (FI > 1.5) were selected to measure the stability of their peptide/HLA-A∗0201 complexes. Six peptides, Rv1218c-p24, Rv1218c-p200, Rv1218c-p158, Rv2209-p158, Rv2477c-p182, and Rv2459c-p469, formed stable peptide/HLA-A∗0201 complexes (DC50 > 4 h) (Figure 1(b)) and were selected for further study.
Figure 1.
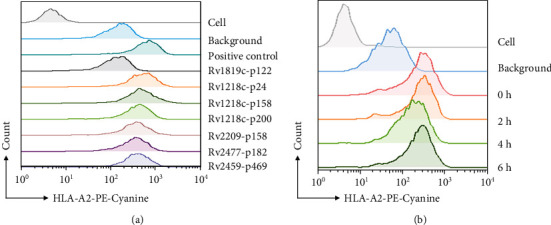
Representative flow chart of binding affinity and peptide/HLA-A2 complex stability assay. Representative flow chart of (a) the binding affinity of the peptides and (b) peptide/HLA-A2 complex stability assay of peptide Rv2477c-p182.
3.3. IFN-γ Secretion of Peptide-Induced CTLs in vitro
First, IFN-γ secretion of peptide-induced CTLs from four HLA-A∗02+PPD+ donors was measured with an ELISPOT assay (Figures 2(a) and 2(b)). The results showed that the Rv1218c-p158-, Rv2459-p469-, Rv1218c-p24-, and Rv2477c-p182-induced CTLs secreted IFN-γ most potently in at least three of the five PPD+ donors. To determine whether these peptides could distinguish between HLA-A∗02+PPD+ donors and HLA-A∗02+PPD− donors, we performed an intracellular cytokine staining (ICS) assay. As shown in Figures 3(a) and 3(b), the Rv2459c-p469-, Rv1218c-p24-, Rv1218c-p158-, and Rv2477c-p182-induced CTLs from HLA-A∗02+PPD+ donors contained significantly more CD8+ IFN-γ+ T cells than those from HLA-A∗02+PPD− donors. Furthermore, the Rv2459c-p469-, Rv1218c-p24-, and Rv2477c-p182-induced CTLs from HLA-A∗02+PPD+ donors contained more CD8+ IFN-γ+ T cells than the PBS-treated group. Therefore, Rv2459c-p469, Rv1218c-p24, and Rv2477c-p182 were selected to assess their cytotoxic activity in vitro in HLA-A∗02+PPD+ donors.
Figure 2.
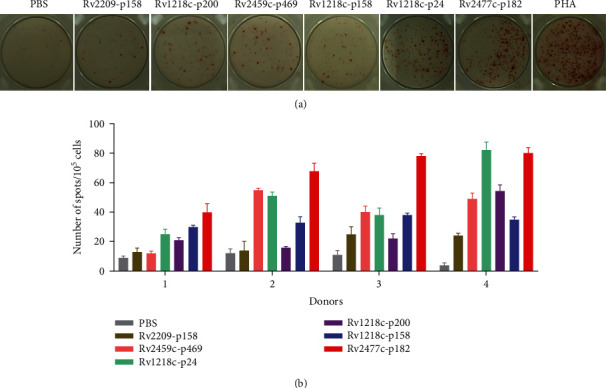
IFN-γ secretion of peptide-induced CTLs was determined with ELISPOT assay. Candidate peptides were used to induce CTLs with peptide-loaded DCs from four healthy PPD+ donors in vitro. T cells were stimulated three times at weekly intervals, and rhIL-2 (50 U/mL) and rhIL-7 (10 ng/mL) were added at intervals of 2 days. On day 21, the induced T cells were collected to measure IFN-γ secretion with an ELISPOT assay. (a) Typical chart for each group. (b) Statistical analysis of each peptide in four donors (n = 4). Peptide-loaded T2A2 cells or T2A2 cells only were used as the stimulated cells. PBS group was the negative control; PHA was used as the positive control.
Figure 3.
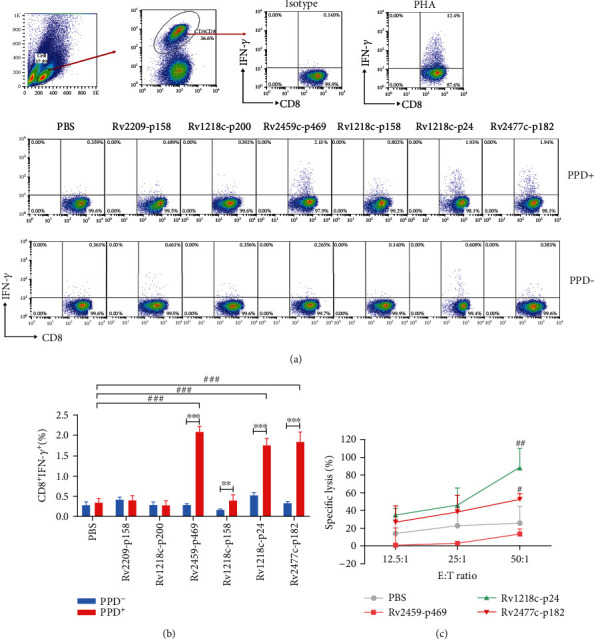
Immune activity of peptide-induced CTLs in vitro was determined with intracellular cytokine staining (ICS) assay and LDH assay. CTLs were induced in vitro with peptide-loaded DCs from three healthy PPD+ donors and three PPD− donors. T cells were stimulated three times at weekly intervals, and rhIL-2 (50 U/mL) and rhIL-7 (10 ng/mL) were added at intervals of 2 days. On day 21, the induced T cells were collected and their IFN-γ secretion and lysis activity were measured with intracellular cytokine staining (ICS) and LDH assays. (a) Typical flow chart for each group of PPD+ and PPD− donors. (b) Statistical results of ICS assay for each peptide (n = 3). (c) LDH assay (n = 4). Peptide-loaded T2A2 cells were used as the stimulated cells and target cells. PBS group was the negative control; PHA was used as the positive control. Statistical significance was determined with Student's t-test. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 represent the significance of the differences between the PPD+ donors and PPD− donors. #P < 0.05, ##P < 0.01, and ###P < 0.001 represent the significance of the differences between the peptide-treated groups and the PBS group.
3.4. In vitro Cytotoxic Activity of Peptide-Induced CTLs
An LDH cytotoxicity assay was used to determine whether the CTLs induced by these three selected peptides lysed the target cells. PBMCs from HLA-A∗02+PPD+ donors were stimulated with each candidate peptide as described above. T2A2 cells pulsed with the corresponding peptide were used as the target cells. As shown in Figure 3(c), the Rv1218c-p24 and Rv2477c-p182 peptide-induced CTLs lysed the target cells at an E : T ratio of 50 : 1, and the lysis rates were 88.6% and 52.5%, respectively, which differed significantly from that of the PBS group. Therefore, these two peptides (Rv1218c-p24 and Rv2477c-p182) were selected for an in vivo assay.
3.5. Activity of CTLs Induced with IFA-Emulsified Peptide in HLA-A2.1/Kb Transgenic Mice
To investigate whether the selected peptides induced CTLs in HLA-A2.1/Kb mice, HLA-A2.1/Kb transgenic mice (n = 5) were immunized subcutaneously at the base of the tail with each peptide (100 μg/mice) and T helper epitope (140 μg/mice), emulsified with incomplete Freund's adjuvant (IFA) at a ratio of 1 : 1, three times (on days 0, 5, and 10). On day 11, the serum was collected from each mouse to determine the IFN-γ concentration with an ELISA, and the results are shown in Figure 4(a). The Rv1218c-p24- and Rv2477c-p182-induced T cells produced significantly more IFN-γ (363 pg/mL and 303.5 pg/mL, respectively) than the NS group. The splenic lymphocytes were isolated from each mouse and stimulated once with each peptide (10 μg/mL) and mIL-2 (50 U/mL) in vitro for 6 days. On day 7, the IFN-γ release in each group was measured with an intracellular cytokine staining (ICS) assay (Figure 4(b)). We found that the Rv1218c-p24 and Rv2477c-p182 groups induced more CD8+IFN-γ+ T cells than the NS group. In a lysis assay (Figure 4(c)), the lysis rate of the Rv1218c-p24 group at an E : T of 80 : 1 was 26.4%, which differed significantly from that of the NS group, and the lysis rates of the Rv2477c-p182 group at E : T of 40 : 1 and 80 : 1 were 21.8% and 24.6%, respectively, which also differed significantly from that of the NS group.
Figure 4.
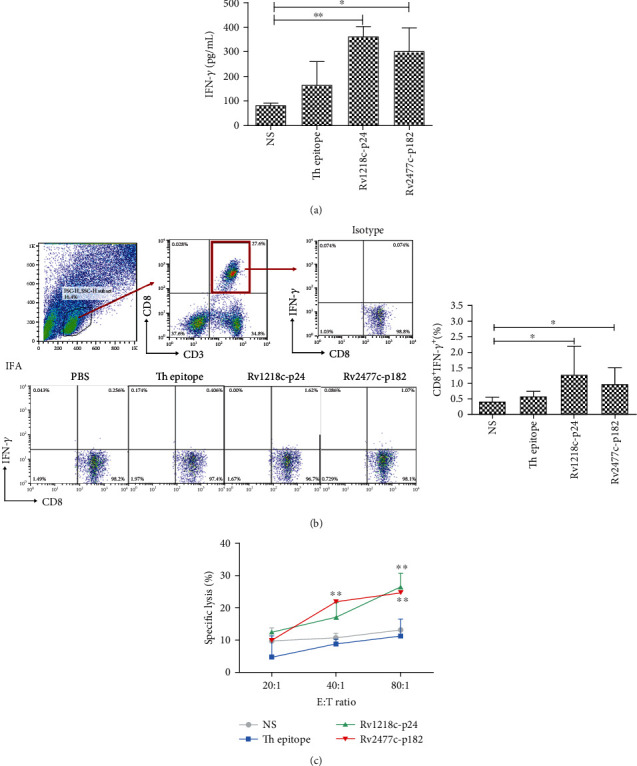
Activity of CTLs induced with IFA-emulsified peptide in HLA-A2.1/Kb transgenic mice. HLA-A2.1/Kb transgenic mice (n = 5) were subcutaneously immunized at the base of the tail with each peptide (100 μg/mice) and T helper epitope (140 μg/mice) emulsified with incomplete Freund's adjuvant (IFA) at 1 : 1, three times (on days 0, 5, and 10). On day 11, all the mice were sacrificed and the serum was collected from each mouse to measure the IFN-γ concentration with an ELISA assay (a). The splenic lymphocytes were isolated from each mouse and restimulated once with each peptide (10 μg/mL) and mIL-2 (10 U/mL) in vitro for another 6 days. On day 7, the IFN-γ release in each group was measured with an intracellular cytokine staining (ICS) assay (b) and the lysis activity was measured with an LDH assay (c). Peptide-loaded T2A2 cells were used as the target cells. Normal saline (NS) group and Th epitope only group were the negative controls. Statistical significance was determined with Student's t-test (n = 5). ∗P < 0.05 and ∗∗P < 0.01 represent the significance of differences relative to the NS group.
3.6. Activity of CTLs Induced with Peptide after Preimmunization with CFA in HLA-A2.1/Kb Transgenic Mice
The results of our in vitro and in vivo experiments indicated that these two peptides, Rv1218c-p24 and Rv2477c-p182, induced the strongest immune response in PPD+ donors. Therefore, we investigated whether MTB in the complete Freund's adjuvant (CFA) could be used to mimic the symptoms of PPD+ donors to enhance the immune response to the peptide. Emulsified complete Freund's adjuvant (CFA, 200 μL) was subcutaneously injected into HLA-A2.1/Kb transgenic mice (n = 5) on day-14. The mice were then immunized subcutaneously at the base of tail with each peptide (100 μg/mice) and T helper epitope (140 μg/mice) emulsified with incomplete Freund's adjuvant (IFA) (1 : 1) three times (on days 0, 5, and 10). On day 11, the serum was collected from each mouse to detect the IFN-γ concentration with an ELISA assay, and the results are shown in Figure 5(a). The peptides Rv1218c-p24 and Rv2477c-p182 triggered the production of significantly more IFN-γ (1817.0 pg/mL and 1587.8 pg/mL, respectively) than was produced in the NS group. The splenic lymphocytes isolated from each mouse were then stimulated once with each peptide (10 μg/mL) and mIL-2 (50 U/mL) in vitro for 6 days. On day 7, the IFN-γ release in each group was measured by intracellular cytokine staining (ICS) assay (Figure 5(b)). We found that the Rv1218c-p24 and Rv2477c-p182 groups produced more CD8+ IFN-γ+ T cells than the NS group. In the lysis assay (Figure 5(c)), the lysis rates of the Rv1218c-p24 group at E : T of 40 : 1 and 80 : 1 were 24.6% and 41.2%, respectively, which were significantly higher than that of the NS group, and the lysis rates of Rv2477c-p182 group at E : T of 40 : 1 and 80 : 1 were 25.8% and 38.9%, respectively, which were also significantly higher than that of the NS group. The combined results shown in Figures 4 and 5 indicate that preimmunization with CFA before peptide immunization enhanced the immune response of the peptide.
Figure 5.
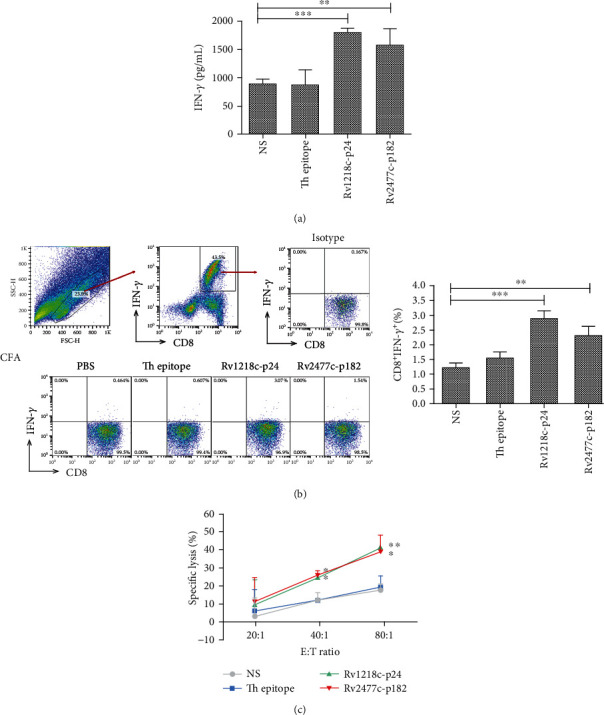
Activity of CTLs induced with peptide after preimmunization with CFA in HLA-A2.1/Kb transgenic mice. Complete Freund's adjuvant (CFA, 200 μL) was injected subcutaneously into HLA-A2.1/Kb transgenic mice (n = 5) on day-14. The mice were then subcutaneously immunized at the base of the tail with each peptide (100 μg/mice) and T helper epitope (140 μg/mice) emulsified with incomplete Freund's adjuvant (IFA) at 1 : 1 three times (on days 0, 5, and 10). On day 11, all mice were sacrificed and the serum was collected from each mouse to measure the IFN/-γ concentration (a). The splenic lymphocytes were isolated from each mouse and restimulated once with each peptide (10 μg/mL) and mIL-2 (10 U/mL) in vitro for another 6 days. On day 7, the IFN-γ release in each group was measured with an intracellular cytokine staining (ICS) assay (b) and the lysis activity were measured with an LDH assay (c). Peptide-loaded T2A2 cells were used as the target cells. Normal saline (NS) group and Th epitope only group were the negative controls. Statistical significance was determined with Student's t-test (n = 5). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 represent the significance of differences relative to the NS group.
4. Discussion
Tuberculosis (TB) is an infectious disease caused by MTB that remains a major cause of death worldwide. With the development of genomic techniques, increasing numbers of proteins associated with the drug resistance of MTB have been identified. However, the prevalence of MDR-TB, XDR-TB, and TDR-TB infections is increasing alarmingly in many countries. Only 56% of patients with MDR-TB can be treated successfully, and there are no appropriate medicines for treating XDR-TB [36]. To meet the evolving challenge of TB, new anti-TB drugs and vaccines against drug-resistant TB are urgently required.
Studies have shown that the efflux systems play an important role in MTB drug resistance, and five efflux pump families and many efflux pump proteins have been shown to be associated with it. In recent years, immunotherapy has played an important role in fighting tumors and infectious diseases. CD8+ T lymphocytes, which are activated by the epitopes of target antigens, are among the most important immune cells and play a critical role in cell-mediated immunity to control latent MTB infections. CD8+ T cells specifically recognize and lyse MTB-infected cells in an HLA-I-restricted manner, so a key step in this research is to identify the epitopes of these antigens.
The majority of anti-TB drugs are pumped out by the efflux pumps of MTB, and some efflux pump proteins are highly expressed or mutated in drug-resistant MTB, including MDR-TB, XDR-TB, and TDR-TB. To combat drug-resistant MTB, several epitopes of the proteins involved in TB drug resistance have been identified. In our previous study, we identified several epitopes of MTB proteins, such as the secreted protein CFP21, an RD-11 region-encoded protein Rv3425, overexpression efflux pump proteins Rv1410c, Rv1258c, Rv2686c-Rv2687c-Rv2688c, and Rv2937 (DrrB), and the mutant proteins KatG and InhA [32, 37, 38]. Previous studies have also reported that the overexpression of different efflux pump genes is associated with the resistance of clinical isolates of MTB to many drugs [25]. The overexpression of efflux pump Rv1218c is responsible for the efflux of a wide variety of drugs, including isoniazid (INH) and rifampicin (RIF) [21, 39]. Studies have shown that Rv2477c has strong sensitive ATPase activity to orthovanadate, so Rv2477c may be involved in mycobacterial protein translation and in its resistance to tetracyclines and macrolides [40]. In the present study, we identified the HLA-A2-restricted epitopes on 10 selected antigens (Rv0194, Rv0933 (PstB), Rv1218c, Rv1819c, Rv2209, Rv2477c, Rv1877, Rv2459c, Rv2994, and Rv3728, members of the ABC and the MFS families). 29 epitopes were selected and synthesized. After the exclusion of several epitopes based on hydrophobicity, steric hindrance, and temperature, 17 epitopes were selected for further analysis (Tables 1 and 2). The T2A2 binding affinity of the epitopes was measured, and FI > 1.5 was determined for 14 of the 17 peptides. The stability of the 14 peptide/MHC complexes was then tested, and the DC50 values for 6 of 14 peptides exceeded 4 h (Table 2 and Figure 1). Based on the results for T2A2 binding affinity and stability, the immunogenic activity of these six peptides (Rv1218c-p24, Rv1218c-p200, Rv1218c-p158, Rv2209-p158, Rv2477c-p182, and Rv2459c-p469) was tested in vitro and in vivo.
The PPD test (tuberculin skin test) is broadly used to diagnose whether the human body is infected with MTB and the degree of the immunological response to infection, but the sensitivity and specificity of the PPD test can be reduced by previous BCG vaccination, severe malnutrition, malignant tumors, immunodeficiency diseases, and so on. Lalvani et al. first applied the enzyme-linked immunospot experimental technique to the diagnosis of TB [41]. In the present study, we examined whether these peptides can distinguish PPD+ from PPD− donors. The results of an in vitro ELISPOT assay (Figures 3(a) and 3(b)) showed that Rv2459-p469-, Rv1218c-p24-, and Rv2477c-p182-induced CTLs from PPD+ donors contained more CD8+ IFN-γ+ T cells than those from PPD− donors, indicating that the IFN-γ production triggered by these three peptides (Rv2459c-p469, Rv1218c-p24, and Rv2477c-p182) distinguished the PPD+ donors from the PPD− donors. This phenomenon may have utility as a more accurate and convenient index of MTB infection than the PPD test. Analysis of the in vitro LDH lysis activity of these three peptides (Figure 3(c)) showed that Rv1218c-p24 and the Rv2477c-p182-induced T cells lysed peptide-loaded T2A2 cells, so these two peptides were selected for an in vivo assay. Several studies have shown that complete Freund's adjuvant (CFA) significantly enhances the phagocytosis by macrophage in mice, and that this is closely associated with the improved immune response conferred by adjuvants [42, 43]. In an in vivo experiment in mice, the CFA adjuvant was subcutaneously injected first into the mouse, then the antigenic peptide was subcutaneously emulsified with IFA. The immunogenic effect of the antigenic peptide was greater in the mice injected with CFA than in the mice injected with only IFA. This suggests that CFA effectively enhanced the immune activity of the antigenic peptides in the study of the epitope peptide immunization of MTB. In addition, the IFA and CFA were used to immunize the HLA-A2.1/Kb transgenic mouse, and for further human beings use, other adjuvants should be used for TB vaccine.
In conclusion, using a binding affinity experiment, stability analysis, and the assessment of immune activity in vivo and in vitro, we identified two advantages of epitopes Rv1218c-p24 and Rv2477c-p182 that effectively stimulate the immune response to MTB. These two peptides may be useful as accurate and convenient indices of MTB infection and may lay a foundation for developing multivalent peptide vaccines against drug-resistant MTB.
Acknowledgments
This work was supported by grants from the National Science Foundation of China (81571547 and 81601448) and the Foundation of Henan Province (19A180007).
Data Availability
Correspondence and requests for materials should be addressed to Prof. Yahong Wu (yahongwu@zzu.edu.cn).
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Yan Lin and Yu Dong contributed equally to this work.
References
- 1.Sable S. B., Posey J. E., Scriba T. J. Tuberculosis vaccine development: progress in clinical evaluation. Clinical microbiology reviews. 2019;33(1) doi: 10.1128/CMR.00100-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chai Q., Lu Z., Liu C. H. Host defense mechanisms against Mycobacterium tuberculosis. Cellular and Molecular Life Sciences: CMLS. 2020;77(10):1859–1878. doi: 10.1007/s00018-019-03353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Safi H., Gopal P., Lingaraju S., et al. Phase variation in Mycobacterium tuberculosis glpK produces transiently heritable drug tolerance. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(39):19665–19674. doi: 10.1073/pnas.1907631116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lange C., Dheda K., Chesov D., Mandalakas A. M., Udwadia Z., Horsburgh C. R., Jr. Management of drug-resistant tuberculosis. The Lancet. 2019;394(10202):953–966. doi: 10.1016/S0140-6736(19)31882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becerra M. C., Huang C. C., Lecca L., et al. Transmissibility and potential for disease progression of drug resistant Mycobacterium tuberculosis: prospective cohort study. BMJ. 2019;367:p. l5894. doi: 10.1136/bmj.l5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moorlag S., Arts R. J. W., van Crevel R., Netea M. G. Non-specific effects of BCG vaccine on viral infections. Clinical microbiology and infection. 2019;25(12):1473–1478. doi: 10.1016/j.cmi.2019.04.020. [DOI] [PubMed] [Google Scholar]
- 7.Patankar Y. R., Sutiwisesak R., Boyce S., et al. Limited recognition of Mycobacterium tuberculosis-infected macrophages by polyclonal CD4 and CD8 T cells from the lungs of infected mice. Mucosal Immunology. 2020;13(1):140–148. doi: 10.1038/s41385-019-0217-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mupfumi L., Mpande C. A. M., Reid T., et al. Immune phenotype and functionality of Mtb-specific T-cells in HIV/TB co-infected patients on antiretroviral treatment. Pathogens. 2020;9(3):p. 180. doi: 10.3390/pathogens9030180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey R. P., Kumar S., Ahmad S., et al. Use Chou’s 5-steps rule to evaluate protective efficacy induced by antigenic proteins of Mycobacterium tuberculosis encapsulated in chitosan nanoparticles. Life Sciences. 2020;256:p. 117961. doi: 10.1016/j.lfs.2020.117961. [DOI] [PubMed] [Google Scholar]
- 10.AlMatar M., Var I., Kayar B., Köksal F. Differential expression of resistant and efflux pump genes in MDR-TB isolates. Endocrine, metabolic & immune disorders drug targets. 2020;20(2):271–287. doi: 10.2174/1871530319666191009153834. [DOI] [PubMed] [Google Scholar]
- 11.Merker M., Kohl T. A., Barilar I., et al. Phylogenetically informative mutations in genes implicated in antibiotic resistance in Mycobacterium tuberculosis complex. Genome Medicine. 2020;12(1):p. 27. doi: 10.1186/s13073-020-00726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright J., Muench S. P., Goldman A., Baker A. Substrate polyspecificity and conformational relevance in ABC transporters: new insights from structural studies. Biochemical Society Transactions. 2018;46(6):1475–1484. doi: 10.1042/BST20180146. [DOI] [PubMed] [Google Scholar]
- 13.Cao R., Teskey G., Islamoglu H., et al. Flavonoid mixture inhibits Mycobacterium tuberculosis survival and infectivity. Molecules. 2019;24(5):p. 851. doi: 10.3390/molecules24050851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X., Szewczyk P., Karyakin A., et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467(7318):991–994. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellmann-Sickert K., Stone T. A., Poulsen B. E., Deber C. M. Efflux by small multidrug resistance proteins is inhibited by membrane-interactive helix-stapled peptides. The Journal of Biological Chemistry. 2015;290(3):1752–1759. doi: 10.1074/jbc.M114.616185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma S., Huang Y., Xie F., et al. Transport mechanism of Mycobacterium tuberculosis MmpL/S family proteins and implications in pharmaceutical targeting. Biological Chemistry. 2020;401(3):331–348. doi: 10.1515/hsz-2019-0326. [DOI] [PubMed] [Google Scholar]
- 17.Gupta S., Kumar A., Singh K., et al. Rv1273c, an ABC transporter of Mycobacterium tuberculosis promotes mycobacterial intracellular survival within macrophages via modulating the host cell immune response. International Journal of Biological Macromolecules. 2020;142:320–331. doi: 10.1016/j.ijbiomac.2019.09.103. [DOI] [PubMed] [Google Scholar]
- 18.Yimer S. A., Birhanu A. G., Kalayou S., et al. Comparative proteomic analysis of Mycobacterium tuberculosis lineage 7 and lineage 4 strains reveals differentially abundant proteins linked to slow growth and virulence. Frontiers in Microbiology. 2017;8:p. 795. doi: 10.3389/fmicb.2017.00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Målen H., De Souza G. A., Pathak S., Søfteland T., Wiker H. G. Comparison of membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains. BMC Microbiology. 2011;11(1):p. 18. doi: 10.1186/1471-2180-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh T. S., Kim Y. J., Kang H. Y., Kim C. K., Cho S. Y., Lee H. J. RNA expression analysis of efflux pump genes in clinical isolates of multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis in South Korea. Infection, genetics and evolution. 2017;49:111–115. doi: 10.1016/j.meegid.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Wang K., Pei H., Huang B., et al. The expression of ABC efflux pump, Rv1217c-Rv1218c, and its association with multidrug resistance of Mycobacterium tuberculosis in China. Current Microbiology. 2013;66(3):222–226. doi: 10.1007/s00284-012-0215-3. [DOI] [PubMed] [Google Scholar]
- 22.Padiadpu J., Vashisht R., Chandra N. Protein-protein interaction networks suggest different targets have different propensities for triggering drug resistance. Systems and Synthetic Biology. 2010;4(4):311–322. doi: 10.1007/s11693-011-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X. Z., Zhang L., Nikaido H. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrobial Agents and Chemotherapy. 2004;48(7):2415–2423. doi: 10.1128/AAC.48.7.2415-2423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dos Santos S. C., Teixeira M. C., Dias P. J., Sá-Correia I. MFS transporters required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: understanding their physiological function through post-genomic approaches. Frontiers in Physiology. 2014;5:p. 180. doi: 10.3389/fphys.2014.00180#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta A. K., Katoch V. M., Chauhan D. S., et al. Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common anti-tuberculous drugs. Microbial drug resistance. 2010;16(1):21–28. doi: 10.1089/mdr.2009.0054. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X., Zhang W., Zhang Y., et al. Assessment of efflux pump gene expression in a clinical isolate Mycobacterium tuberculosis by real-time reverse transcription PCR. Microbial drug resistance. 2008;14(1):7–11. doi: 10.1089/mdr.2008.0772. [DOI] [PubMed] [Google Scholar]
- 27.Rammensee H., Bachmann J., Emmerich N. P., Bachor O. A., Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3-4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 28.Singh M., Bhatt P., Sharma M., Varma-Basil M., Chaudhry A., Sharma S. Immunogenicity of late stage specific peptide antigens of Mycobacterium tuberculosis. Infection, genetics and evolution. 2019;74, article 103930 doi: 10.1016/j.meegid.2019.103930. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y. H., Gao Y. F., He Y. J., et al. A novel cytotoxic T lymphocyte epitope analogue with enhanced activity derived from cyclooxygenase-2. Scandinavian Journal of Immunology. 2012;76(3):278–285. doi: 10.1111/j.1365-3083.2012.02738.x. [DOI] [PubMed] [Google Scholar]
- 30.Shi T. D., Wu Y. Z., Jia Z. C., Zou L. Y., Zhou W. Therapeutic polypeptides based on HBV core 18-27 epitope can induce CD8+ CTL-mediated cytotoxicity in HLA-A2+ human PBMCs. World Journal of Gastroenterology. 2004;10(13):1902–1906. doi: 10.3748/wjg.v10.i13.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y., Zhai W., Zhou X., et al. HLA-A2-restricted epitopes identified from MTA1 could elicit antigen-specific cytotoxic T lymphocyte response. Journal of Immunology Research. 2018;2018:11. doi: 10.1155/2018/2942679.2942679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv H., Gao Y., Wu Y., et al. Identification of a novel cytotoxic T lymphocyte epitope from CFP21, a secreted protein of Mycobacterium tuberculosis. Immunology Letters. 2010;133(2):94–98. doi: 10.1016/j.imlet.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Thomas R., Shaath H., Naik A., Toor S. M., Elkord E., Decock J. Identification of two HLA-A∗0201 immunogenic epitopes of lactate dehydrogenase C (LDHC): potential novel targets for cancer immunotherapy. Cancer immunology, immunotherapy. 2020;69(3):449–463. doi: 10.1007/s00262-020-02480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan Z., Wu Y., Du J., et al. A novel peptide targeting Clec9a on dendritic cell for cancer immunotherapy. Oncotarget. 2016;7(26):40437–40450. doi: 10.18632/oncotarget.9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X. M., Li W. Q., Wu Y. H., et al. Intrinsic expression of immune checkpoint molecule TIGIT could help tumor growth in vivo by suppressing the function of NK and CD8(+) T cells. Frontiers in Immunology. 2018;9:p. 2821. doi: 10.3389/fimmu.2018.02821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q., Liu Y., Tang S., Sha W., Xiao H. Clinical benefit of delamanid (OPC-67683) in the treatment of multidrug-resistant tuberculosis patients in China. Cell Biochemistry and Biophysics. 2013;67(3):957–963. doi: 10.1007/s12013-013-9589-5. [DOI] [PubMed] [Google Scholar]
- 37.Chen F., Zhai M. X., Zhu Y. H., Qi Y. M., Zhai W. J., Gao Y. F. In vitro and in vivo identification of a novel cytotoxic T lymphocyte epitope from Rv3425 of Mycobacterium tuberculosis. Microbiology and Immunology. 2012;56(8):548–553. doi: 10.1111/j.1348-0421.2012.00470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y. H., Gao Y. F., Chen F., et al. Identification of novel T cell epitopes from efflux pumps of Mycobacterium tuberculosis. Immunology Letters. 2011;140(1-2):68–73. doi: 10.1016/j.imlet.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 39.Balganesh M., Kuruppath S., Marcel N., Sharma S., Nair A., Sharma U. Rv1218c, an ABC transporter of Mycobacterium tuberculosis with implications in drug discovery. Antimicrobial Agents and Chemotherapy. 2010;54(12):5167–5172. doi: 10.1128/AAC.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniel J., Abraham L., Martin A., Pablo X., Reyes S. Rv2477c is an antibiotic-sensitive manganese-dependent ABC-F ATPase in Mycobacterium tuberculosis. Biochemical and Biophysical Research Communications. 2018;495(1):35–40. doi: 10.1016/j.bbrc.2017.10.168. [DOI] [PubMed] [Google Scholar]
- 41.LALVANI A., PATHAN A. . A., McSHANE H., et al. Rapid detection of Mycobacterium tuberculosis infection by enumeration of antigen-specific T cells. American Journal of Respiratory and Critical Care Medicine. 2001;163(4):824–828. doi: 10.1164/ajrccm.163.4.2009100. [DOI] [PubMed] [Google Scholar]
- 42.Jin X. W., Liu B. K., Zhang X., Zhao Z. H., Shao Y. Establishment of a novel autoimmune experimental model of bladder pain syndrome/interstitial cystitis in C57BL/6 mice. Inflammation. 2017;40(3):861–870. doi: 10.1007/s10753-017-0531-7. [DOI] [PubMed] [Google Scholar]
- 43.Dubé J. Y., McIntosh F., Zarruk J. G., David S., Nigou J., Behr M. A. Synthetic mycobacterial molecular patterns partially complete Freund’s adjuvant. Scientific Reports. 2020;10(1):p. 5874. doi: 10.1038/s41598-020-62543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Correspondence and requests for materials should be addressed to Prof. Yahong Wu (yahongwu@zzu.edu.cn).


