Background:
Over the past decade, numerous obesity-specific pharmacokinetic (PK) models and dosage regimens have been developed. However, it is unclear whether vancomycin PKs differ between obese and other patients after accounting for weight, age, and kidney function. In this study, the authors investigated whether using obesity-specific population PK models for vancomycin offers any advantage in accuracy and precision over using a recently developed general-purpose model.
Methods:
Vancomycin plasma concentrations in a cohort of 49 obese patients (body mass index [BMI] >30 kg/m2), not previously used in the development of any of the evaluated models, were used to validate the performance of 4 obesity-specific models and a general model. Bias and imprecision were calculated for the a priori and a posteriori predictive performance.
Results:
The bias of the a priori prediction was lowest for one of the obesity-specific models (−1.40%) and that of the general model was a close second (−7.0%). The imprecision was lowest for the general model (4.34 mg/L). The predictive performance for the a posteriori predictions was best for the general model, both for bias (1.96%) and imprecision (2.75 mg/L).
Conclusions:
The results of the external validation of vancomycin PK in obese patients showed that currently available obesity-specific models do not necessarily outperform a broadly supported general-purpose model. Based on these results, the authors conclude that there is no advantage in using vancomycin PK models specifically tailored to obese patients over the general-purpose model reported by Colin et al.
Key Words: modeling and simulation, therapeutic drug monitoring, model-informed precision dosing
BACKGROUND
Population pharmacokinetic (PopPK) models are increasingly being used to guide drug dosing at the point-of-care, particularly in vulnerable patients, such as the critically ill, those with impaired kidney function, and obese patients. Physiological and pathophysiological changes driven by comorbidities alter drug PK in these patients, which raises concerns that they may be put at risk of overexposure or underexposure if they receive dosage regimens derived from the PK of other populations. Over the past decade, these concerns have led to the development of numerous subgroup- and context-specific PK models and dose regimens.
For vancomycin, 4 PopPK models specifically developed for obese patients (further referred to as obesity-specific PopPK models) have been reported.1–4 However, the authors recently developed a pooled PopPK model that covers a broad range of patient populations, including obese patients.5 This model does not require a specific “obesity” covariate for optimal performance after accounting for body weight, age, and kidney function. This model has not been validated in obese individuals, and it is unclear whether it performs comparably to obesity-specific PK models.
In this study, the authors investigated whether using obesity-specific PopPK models for vancomycin offers any advantages over general-purpose models with regard to bias and imprecision by comparing their performance in an independent cohort of obese patients. In addition, the authors also evaluated the performance of the model reported by Goti et al,6 which was recently found to be the best for predicting vancomycin PK in normal-weight hospitalized patients in a large meta-analysis by Broeker et al.7
PATIENTS AND METHODS
Fully anonymized data were available from a previous student project wherein the PKs of vancomycin in obese patients were assessed. Patients aged ≥16 years with a body mass index (BMI) ≥30 kg/m2 and at least one vancomycin concentration measurement were eligible for inclusion. Data were collected retrospectively between October 2015 and June 2016 from therapeutic drug monitoring (TDM) files stored electronically in the computer program OPT.8 Research approval for the original study was obtained from the East of Scotland Research Ethics Service (reference number 15/ES/0148), and local health board approval for handling and storing the data was obtained through the Caldicott Guardian. Patient consent was not required because the study involved fully anonymized, retrospective data generated in the course of routine patient care.
Vancomycin dosage regimens were based on total body weight and renal function according to the national guidelines9 and adjusted to achieve trough concentrations in the range 10–20 mg/L. The following data were available for each patient: age, sex, weight, height, serum creatinine concentration, vancomycin dosage history (including date and time of administration), infusion rate, vancomycin concentrations, and sampling time.
PopPK Models
Four obesity-specific models1–4 and one model for predicting vancomycin PK in normal-weight hospitalized patients6 were evaluated. The models were nonparametric2,3 or parametric1,4,6 and were 1-compartmental1,3, 2-compartmental2,6 or 3-compartmental4 linear PopPK models. The models by Carreno et al2 (approximately 5 samples per individual) and Smit et al4 (a median of 12 samples per individual) were based on rich sampling, the models by Adane et al1 and Crass et al3 were based on peak and trough sampling and the model by Goti et al6 was predominantly based on trough sampling (with two-thirds of patients contributing only 1 sample). The obesity-specific models were based on groups of patients with a median BMI ≥45 kg/m2 (49.5 kg/m2 for the study by Adane et al1).
All evaluated models except that reported by Smit et al4 included a marker for renal function on vancomycin clearance (CL). Creatinine CL estimated according to the Cockcroft–Gault equation was used in the models reported by Adane et al,1 Carreno et al,2 and Goti et al6 (scaled to a power of 0.8). The model reported by Crass et al3 used the individual covariates in the Cockcroft–Gault equation (age, serum creatinine, sex, and total body weight) with the weight scaled allometrically. The model reported by Smit et al4 had total body weight scaled to a power of 0.53 as the sole covariate on vancomycin CL. Total body weight was a linear predictor of the volume of distribution in the models reported by Adane et al,1 Smit et al,4 and Goti et al.6
In addition, the model reported by Smit et al4 showed that the volumes of distribution of the central (V1) and peripheral (V2) compartments decreased with increasing age. The general PopPK model of Colin et al5 was based on a mixture of richly and sparsely sampled studies (in total 8300 vancomycin concentrations in 2554 individuals, including 274 adults with BMI >30 kg/m2) and is a parametric, two-compartmental linear model that uses (postmenstrual) age, weight, and serum creatinine as covariates.
Evaluation of Predictive Performance
Simulations were conducted using NONMEM (version 7.4; GloboMax, Hanover, MD). The “tidyverse” package (version 1.1.1.; Wickham H. 2017) in R (R Foundation for Statistical Computing, Vienna, Austria) was used for all calculations and graphical analyses. Patients' age, weight, height, and/or serum creatinine concentration were used to calculate PK parameters according to the different models. Using these PK parameters, vancomycin plasma concentrations were predicted for all samples in the dataset based on individual dosage history. For the a priori predictions, between-subject variability and residual variability were not considered. For the a posteriori predictions, NONMEM was used to determine individual a posterior PK parameters based on the first TDM sample of each patient.
Plasma concentrations of samples other than those of the first TDM were predicted and used to calculate performance metrics. For the a posteriori predictions, between-subject variability and residual variability were accounted for. For the nonparametric models, a log-normal parametric distribution was used to account for between-subject variability. This was necessary because the distribution of the support points required for the estimation of the maximum a posteriori PK parameters was not available from previous literature. The SD of the surrogate parametric distributions was estimated from the reported standard deviations (or variances) of the support point distribution (ie, PK parameters).
The mean relative prediction error and root mean square error (RMSE), calculated according to Equations 1 and 2, were used to quantify the bias and imprecision of the predictions. To account for the variability in the number of observations per individual, the bias and RMSE were normalized by calculating the values separately for each individual in the dataset and then summarizing the mean values across individuals and obesity classes. Confidence intervals (CIs, 95%) were calculated assuming that the sampling distributions of bias and RMSE followed a normal distribution (ie, the central limit theorem).
| (1) |
| (2) |
RESULTS
A total of 49 patients (18 men, 37%) were included in this study, consisting of 25, 9, and 15 patients who were class I (30–34.9 kg/m2), class II (35–39.9 kg/m2), and class III morbid (≥40 kg/m2) obese, respectively. The median age and serum creatinine concentration was 59 (range, 28–94) years and 70.7 (range, 47.7–76.9) µM, respectively. The median number of samples per patient was 3 (range, 1–7), and 62% of all samples were trough samples collected just before administration of the next dose. Patients received a median of 3 vancomycin doses (range, 1–8) before the first sampling.
Table 1 shows the bias and RMSE based on the a priori predictions. Overall, the bias was lowest for the model reported by Carreno et al2 (−1.40%) and that for the model reported by Colin et al5 was a close second (−7.0%). The RMSE was lowest for the model reported by Colin et al5 (4.34 mg/L). When the results were ranked according to the obesity class (Table 1), the bias was lowest for the model reported by Carreno et al2 for classes I (1.81%) and III (−3.75) but was lowest for the model reported by Colin et al5 for class II (−4.12). RMSE was lowest for the model reported by Colin et al,5 except for class I, for which it was lowest for the model reported by Carreno et al2 (4.29 mg·L−1 versus 4.93 mg·L−1). The model reported by Smit et al4 showed the highest absolute bias and RMSE (overall and across BMI classes). The a priori predictions of the model reported by Goti et al6 showed the second highest absolute bias, except for the class III obese group where the model reported by Crass et al3 exhibited the second highest absolute bias (19.1% versus 12.5%).
TABLE 1.
Performance Metrics of Different models
| Bias (%) [95% CI] | RMSE (mg/L) [95% CI] | Bias (%) [95% CI] | RMSE (mg/L) [95% CI] | |
| All patients—a priori predictions (159 observations, 49 patients) | All patients—a posteriori predictions (110 observations, 43 patients) | |||
| Adane et al1 | −20.0 [−30.1 to −9.97] | 5.49 [4.58 to 6.41] | −12.4 [−19.2 to −5.51] | 3.52 [2.89 to 4.15] |
| Carreno et al2 | −1.4 [−11.1 to 8.26] | 4.43 [3.48 to 5.38] | −7.54 [−13.9 to −1.22] | 3.27 [2.60 to 3.94] |
| Colin et al5 | −7.0 [−16.1 to 2.13] | 4.34 [3.40 to 5.27] | 1.96 [−4.03 to 7.94] | 2.75 [2.21 to 3.29] |
| Crass et al3 | 21.1 [7.29 to 34.8] | 5.93 [4.75 to 7.11] | −6.54 [−13.9 to 0.84] | 3.65 [2.87 to 4.43] |
| Goti et al6 | 26.2 [12.2 to 40.2] | 6.12 [4.99 to 7.24] | 17.2 [6.71 to 27.6] | 4.45 [3.55 to 5.36] |
| Smit et al4 | −41.9 [−49.9 to −33.8] | 7.16 [5.86 to 8.45] | −10.8 [−18.4 to −3.12] | 3.71 [2.82 to 4.60] |
| Class I obese—a priori predictions (81 observations, 25 patients) | Class I obese—a posteriori predictions (56 observations, 21 patients) | |||
| Adane et al1 | −28.4 [−41.8 to −14.9] | 5.97 [4.69 to 7.26] | −13.8 [−24.2 to −3.37] | 3.82 [2.73 to 4.92] |
| Carreno et al2 | 1.81 [−12.5 to 16.1] | 4.29 [2.91 to 5.68] | −8.18 [−18.2 to 1.89] | 3.23 [2.26 to 4.21] |
| Colin et al5 | −4.14 [−20.0 to 11.7] | 4.93 [3.42 to 6.44] | 4.56 [−5.27 to 14.4] | 2.96 [2.10 to 3.83] |
| Crass et al3 | 19.2 [−4.76 to 43.1] | 6.73 [4.73 to 8.73] | −6.07 [−16.3 to 4.10] | 3.11 [2.13 to 4.09] |
| Goti et al6 | 31.4 [8.09 to 54.8] | 6.77 [4.75 to 8.80] | 21.8 [3.88 to 39.8] | 5.14 [3.44 to 6.85] |
| Smit et al4 | −39.7 [−53.1 to −26.2] | 7.30 [5.24 to 9.36] | −5.66 [−17.2 to 5.90] | 3.22 [1.93 to 4.51] |
| Class II obese—a priori predictions (32 observations, 9 patients) | Class II obese—a posteriori predictions (23 observations, 9 patients) | |||
| Adane et al1 | −15.1 [−34.8 to 4.73] | 4.16 [2.79 to 5.54] | −11.0 [−20.6 to −1.32] | 3.06 [1.90 to 4.22] |
| Carreno et al2 | −6.40 [−23.8 to 11.0] | 3.36 [1.82 to 4.89] | −12.7 [−25.3 to −0.04] | 3.32 [2.11 to 4.52] |
| Colin et al5 | −4.12 [−23.2 to 15.0] | 3.17 [1.64 to 4.71] | −0.36 [−16.9 to 16.1] | 2.44 [1.35 to 3.53] |
| Crass et al3 | 29.7 [9.10 to 50.2] | 4.56 [2.50 to 6.63] | −12.1 [−27.1 to 2.86] | 3.48 [2.24 to 4.73] |
| Goti et al6 | 34.4 [−1.07 to 69.9] | 6.44 [4.40 to 8.48] | 12.3 [−13.3 to 37.9] | 4.18 [2.82 to 5.54] |
| Smit et al4 | −42.8 [−64.3 to −21.2] | 6.88 [2.92 to 10.8] | −12.0 [−28.4 to 4.49] | 3.64 [2.27 to 5.02] |
| Class III obese—a priori predictions (46 observations, 15 patients) | Class III obese—a posteriori predictions (31 observations, 13 patients) | |||
| Adane et al1 | −9.06 [−32.1 to 14.0] | 5.49 [3.35 to 7.64] | −11.0 [−27.6 to 5.52] | 3.34 [2.27 to 4.42] |
| Carreno et al2 | −3.75 [−25.1 to 17.6] | 5.31 [3.21 to 7.41] | −2.97 [−15.6 to 9.65] | 3.30 [1.65 to 4.95] |
| Colin et al5 | −13.5 [−26.1 to −0.83] | 4.04 [2.36 to 5.72] | −0.64 [−9.53 to 8.24] | 2.63 [1.53 to 3.73] |
| Crass et al3 | 19.1 [−3.5 to 41.7] | 5.41 [3.60 to 7.21] | −3.43 [−21.5 to 14.6] | 4.63 [2.58 to 6.68] |
| Goti et al6 | 12.5 [−6.5 to 31.4] | 5.49 [3.35 to 7.64] | 13.0 [−2.19 to 28.3] | 3.53 [2.51 to 4.56] |
| Smit et al4 | −45.0 [−56.6 to −33.3] | 7.09 [5.20 to 8.98] | −18.2 [−33.8 to −2.53] | 4.54 [2.39 to 6.69] |
Models with the smallest absolute mean bias and lowest root mean square error of each group are indicated in bold.
CI, confidence interval.
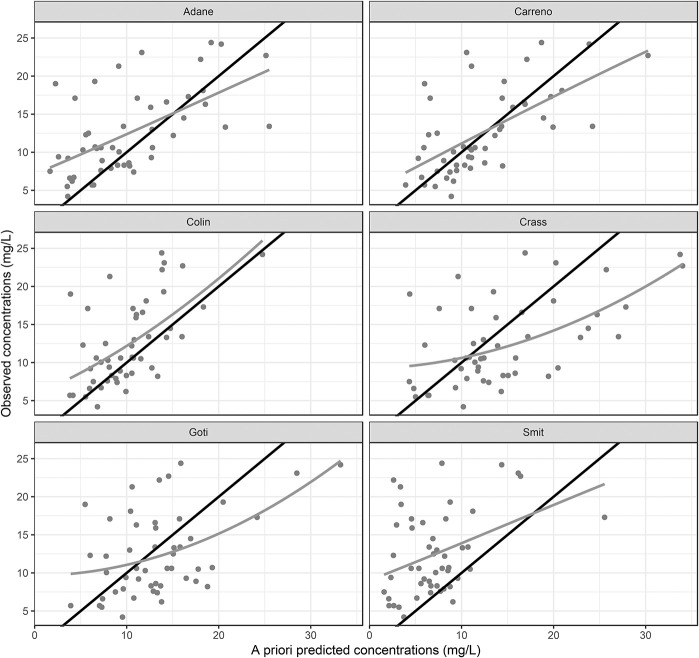
The goodness-of-fit plots (Fig. 1) of the models reported by Carreno et al2 and Colin et al5 were similar and showed good agreement between the a priori model predictions and observed plasma concentrations. The models reported by Crass et al,3 Adane et al,1 and Goti et al6 deviated from the line of unity, whereas those reported by Crass et al3 and Goti et al6 exhibited overprediction at concentrations >15 mg·L−1, and the model reported by Adane et al1 seemed to underpredict concentrations <10 mg·L−1. The model reported by Smit et al4 consistently underpredicted the observed vancomycin concentrations.
FIGURE 1.
Goodness-of-fit plots showing observed versus a priori predicted vancomycin concentrations. Grey solid line indicates locally weighted smoothing (LOESS) of the data. Line of identity is shown as solid black line.
Table 1 shows the bias and RMSE based on a posteriori predictions. The model reported by Colin et al5 outperformed the other models in both bias and RMSE. The absolute bias of the a posteriori prediction was highest for the model reported by Goti et al6 and those for the models reported by Adane et al1 and Smit et al4 were a close second and third (17.2% versus 12.4% and 10.8%). The RMSE was highest for the model reported by Goti et al,6 except for the class III obese group for which the model reported by Crass3 showed the highest RMSE and that of Smit et al4 exhibited the second highest RMSE (4.63 mg·L−1 and 4.54 mg·L−1, respectively).
DISCUSSION
The results of the external validation using obese patients in this study showed that the obesity-specific models did not perform better than the model reported by Colin et al5 did. In contrast, the data collectively indicated that the a priori predictions of the model reported by Colin et al5 exhibited the lowest RMSE (4.34 mg·mg·L−1) and the second lowest bias (−7.0%; 95% CI, −16.1% to 2.13%) among the tested models. The bias was consistently lower for the model reported by Carreno et al,2 but the differences were not statistically significant (based on the overlapping 95% CI) and likely had limited clinical importance because the absolute bias was low for both models (<20%). Further, the a priori underprediction of the model reported by Colin et al5 for class III obesity was completely attenuated when a single TDM sample was used to forecast PK parameters (bias: −13.5% [95% CI, −26.1% to −0.83%] and −0.64% [95% CI, −9.53% to 8.24%] for the a priori and a posteriori predictions, respectively). Moreover, the a posteriori predictive performance was best for the model reported by Colin et al,5 irrespective of obesity class, both in bias and RMSE.
The motivation for developing obesity-specific models is that after accounting for differences related to weight, age, and kidney function, PK parameters of the obese group may be sufficiently different from those of other groups such that specialization will improve model predictive performance. General-purpose models involve a different approach of treating individuals of all groups on a single continuum, smoothly interpolating data across diverse age, weight, and kidney function ranges. In the authors' previously described vancomycin model,5 PK parameters of obese patients are based on the same set of covariates (age, weight, and kidney function) that are used for other patients. This study showed that the general-purpose approach results in accuracy and bias comparable with, and in some cases better than that of, those of obesity-specific models.
Model-informed precision dosing (MIPD) tools use PopPK models to suggest individualized dosage regimens based on patient covariates and/or measurements from TDM programs or both. Keizer et al10 recently described the challenges that hinder the widespread implementation of MIPD tools, which includes model selection. Keizer et al10 state that “one needs to select a model that matches the intended population. In practice, this usually means matching age groups, body composition, indications and comorbidities, and potentially genetic makeup and dose levels studied and analytical assay(s) used.”10 This poses a challenge to the user of the MIPD tool, who must consider the limitations of the models used and might have to switch models when treating different patients.
The results of this evaluation of the model reported by Goti et al6 illustrate the consequences of not matching a model and its supporting population and the intended target population. As expected, the results of this study showed that extrapolation of a PK model developed for nonobese patients to an obese population leads to poor predictive performance. In addition, the results of the experiments using the model reported by Smit et al4 demonstrated that a marker for renal function is a pivotal component of a model when attempting to predict vancomycin PK in obese patients other than those undergoing elective bariatric surgery.
General-purpose PK models might be more useful in this context with expectations of being more generalizable than other models and, as shown in this study, could replace subgroup-specific models without compromising performance. Notably, Cunio et al11 recently showed that the authors' general-purpose vancomycin model outperformed several ICU-specific PK models for vancomycin. Most notably, in line with the results of this study, Cunio et al11 found that the a posteriori predictions of the authors' model showed a clinically acceptable performance (ie, relative bias between −20% and 20% and 95% CI including zero) in ICU patients.
A limitation of the present study is the retrospective nature of the PK samples used to validate the different vancomycin models. A prospective study using a more diverse sampling scheme that does not predominantly involve collecting trough samples would allow a more granular comparison between the different models. In addition, a longer follow-up spanning multiple TDM and dose adjustment cycles would allow the comparison of the performance of different models in handling within-subject variability, such as that due to alterations in renal function.
In conclusion, the results of the external validation of vancomycin PKs in obese patients in this study demonstrated that currently available obesity-specific models do not necessarily outperform a broadly supported general-purpose model. Based on these results, the authors conclude that there is no advantage in using vancomycin PK models specifically tailored for obese patients over using the general-purpose model reported by Colin et al.5
Footnotes
Supported by departmental funding.
The authors declare no conflict of interest.
REFERENCES
- 1.Adane ED, Herald M, Koura F. Pharmacokinetics of vancomycin in extremely obese patients with suspected or confirmed Staphylococcus aureus infections. Pharmacotherapy. 2015;35:127–139. [DOI] [PubMed] [Google Scholar]
- 2.Carreno JJ, Lomaestro B, Tietjan J, et al. Pilot study of a Bayesian approach to estimate vancomycin exposure in obese patients with limited pharmacokinetic sampling. Antimicrob Agents Chemother. 2017;61:e02478–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crass RL, Dunn R, Hong J, et al. Dosing vancomycin in the super obese: less is more. J Antimicrob Chemother. 2018;73:3081–3086. [DOI] [PubMed] [Google Scholar]
- 4.Smit C, Wasmann RE, Goulooze SC, et al. Population pharmacokinetics of vancomycin in obesity: finding the optimal dose for (morbidly) obese individuals. Br J Clin Pharmacol. 2020;86:303–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colin PJ, Allegaert K, Thomson AH, et al. Vancomycin pharmacokinetics throughout life: results from a pooled population analysis and evaluation of current dosing recommendations. Clin Pharmacokinet. 2019;58:767–780. [DOI] [PubMed] [Google Scholar]
- 6.Goti V, Chaturvedula A, Fossler MJ, et al. Hospitalized patients with and without hemodialysis have markedly different vancomycin pharmacokinetics: a population pharmacokinetic model-based analysis. Ther Drug Monit. 2018;40:212–221. [DOI] [PubMed] [Google Scholar]
- 7.Broeker A, Nardecchia M, Klinker KP, et al. Towards precision dosing of vancomycin: a systematic evaluation of pharmacometric models for Bayesian forecasting. Clin Microbiol Infect. 2019;25:1286.e1–1286.e7. [DOI] [PubMed] [Google Scholar]
- 8.Kelman AW, Whiting B, Bryson SM. Opt: a package of computer programs for parameter optimisation in clinical pharmacokinetics. Br J Clin Pharmacol. 1982;14:247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scottish Antimicrobial Prescribing Group. Scottish Medicines Consortium: Vancomycin. Available at: https://www.sapg.scot/quality-improvement/hospital-prescribing/gentamicin-and-vancomycin/vancomycin/. [Google Scholar]
- 10.Keizer RJ, Ter Heine R, Frymoyer A, et al. Model-informed precision dosing at the bedside: scientific challenges and opportunities. CPT Pharmacometrics Syst Pharmacol. 2018;7:785–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunio CB, Uster DW, Carland JE, et al. Towards precision dosing of vancomycin in critically ill patients: an evaluation of the predictive performance of pharmacometric models in ICU patients. Clin Microbiol Infect. 2020. doi: 10.1016/j.cmi.2020.07.005. [DOI] [PubMed] [Google Scholar]