Abstract
Computed Tomography (CT) imaging is widely used for studying body composition, i.e., the proportion of muscle and fat tissues with applications in areas such as nutrition or chemotherapy dose design. In particular, axial CT slices from the 3rd lumbar (L3) vertebral location are commonly used for body composition analysis. However, selection of the third lumbar vertebral slice and the segmentation of muscle/fat in the slice is a tedious operation if performed manually. The objective of this study is to automatically find the middle axial slice at L3 level from a full or partial body CT scan volume and segment the skeletal muscle (SM), subcutaneous adipose tissue (SAT), visceral adipose tissue (VAT) and intermuscular adipose tissue (IMAT) on that slice. The proposed algorithm includes an L3 axial slice localization network followed by a muscle-fat segmentation network. The localization network is a fully convolutional classifier trained on more than 12,000 images. The segmentation network is a convolutional neural network with an encoder-decoder architecture. Three datasets with CT images taken for patients with different types of cancers are used for training and validation of the networks. The mean slice error of 0.87± 2.54 was achieved for L3 slice localization on 1748 CT scan volumes. The performance of five class tissue segmentation network evaluated on two datasets with 1327 and 1202 test samples. The mean Jaccard score of 97% was achieved for SM and VAT tissue segmentation on 1327 images. The mean Jaccard scores of 98% and 83% are corresponding to SAT and IMAT tissue segmentation on the same dataset. The localization and segmentation network performance indicates the potential for fully automated body composition analysis with high accuracy.
Keywords: Third lumbar vertebra, Muscle segmentation, Fat segmentation, CT scan, Convolutional neural network
1. Introduction
Body composition is an important measurement for patients suffering from cancer and/or having conditions such as sarcopenia and cachexia. Studies show that the total body skeletal muscle and adipose tissue volumes are correlated with muscle and fat tissue areas derived from axial (cross-sectional) CT images taken at the third lumbar (L3) (Shen et al., 2004; Mourtzakis et al., 2008) vertebral location. Body composition assessment on L3 slice requires the tedious and time-consuming task of manual expert segmentation. In recent years, several methods have been proposed for automatic L3 slice localization and tissue segmentation. Some recent papers applied convolutional neural networks on maximum intensity projection (MIP) of the CT scan volume to find the location of L3 slice in the spinal column while reducing the input CT scan stack to a 2D dimensional image (Kanavati et al., 2018; Belharbi et al., 2017). Networks trained on the MIP of the CT scan volumes, do not include the significant information of the vertebra, skeletal muscle and other organs’ shape and location for feature learning on the cross-sectional slices. Therefore, while this method might achieve acceptable results for normal cases, it fails when applied on the scans with spine curvature disorders (Kanavati et al., 2018). Bridge et al. presented another approach based on the regression of the axial L3 slice (Bridge et al., 2018). DenseNet (Huang et al., 2017) is the network that is trained to find the off-set of each image with the L3 slice. This approach can result in multiple L3 candidates for one CT volume. Although this issue is resolved by choosing the slice closest to the head as L3, it cannot be employed as a general rule. Here, we address this problem and propose another approach to localize the L3 slice.
Initial papers on automatic L3 CT segmentation proposed shape prior models for body composition analysis at L3 and at T4 vertebra levels (Chung et al., 2009; Popuri et al., 2013, 2016). Recently, deep learning methods have been used for L3 tissue segmentation. Lee et. al applied a fully convolutional network (FCN) for skeletal muscle segmentation on L3 slice (Lee et al., 2017). Later the performance of Unet investigated (Bridge et al., 2018) and an improvement reported. In this work, a convolutional neural network (CNN) is proposed to further improve the generalization and accuracy of muscle and fat segmentation on CT L3 slice.
2. Method
The objective of the proposed framework is to accurately find the middle axial slice at the third lumbar vertebrae from the full or partial body scan of each patient and segment the skeletal muscle (SM), subcutaneous adipose tissue (SAT), visceral adipose tissue (VAT) and intermuscular adipose tissue (IMAT). The algorithm of L3 localization and muscle and fat segmentation is a two-step process shown in Figure 1.
Figure 1:
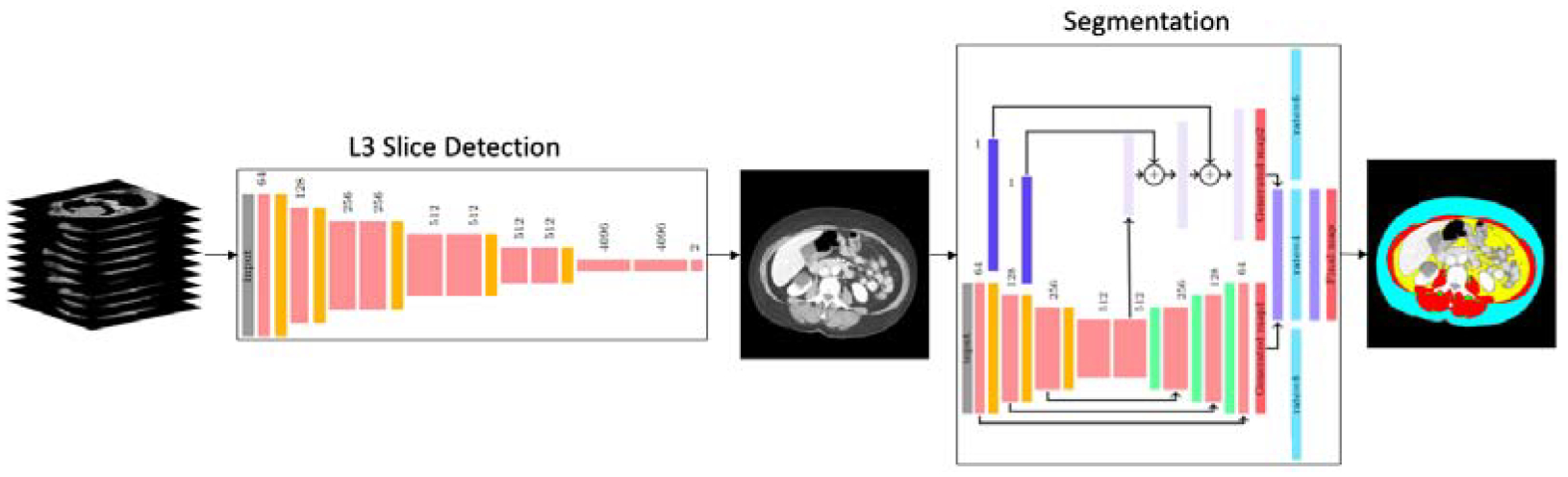
The two-step network pipeline. First step is extracting the middle L3 slice from CT scan volumes and the next step is muscle and fat tissue segmentation on the extracted L3 slice.
In the first step, the group of CT images is fed into the localization network to extract the axial L3 slice from the CT scan volume, details are described in section 2.1. In the next step the extracted L3 image is given to the segmentation network as an input to estimate the muscle, SAT, VAT and IMAT regions, section 2.2 explains the network’s architecture. The images and labels used for the L3 localization and muscle and fat segmentation are described in 3.1.
2.1. L3 localization
Since all the CT scan volumes might not include the full spinal column, designing the L3 localization algorithm based on its location with respect to other vertebrae reduces the model’s robustness. Our approach to the L3 localization problem is to classify the L3 slices from other slices at other vertebrae levels using the features from the 2D axial slices. Therefore, the task is defined as a binary classification of L3 slices and all other slices at other locations. With this procedure, all CT scan volumes with any number of images or field of view can be studied. The L3 localization network learns the L3 slice features and finds the slices regarding the learned features in the input CT volume.
The pre-trained VGG-11 network (Simonyan and Zisserman, 2014) is used for the L3 slice localization. The network consists of 8 convolution layers with rectified linear unit (ReLU) activation function and 3 fully connected layers. Each convolution block is followed by a max-pooling layer with kernel and stride size of 2. The classifier contains two fully connected layers with 4096 nodes and one two nodes layer for binary classification. The weights are initialized with the weights of the pre-trained model on ImageNet (Deng et al., 2009).
2.2. SM, SAT, VAT and IMAT segmentation
Once the middle L3 slice is extracted from the CT scan volume, it is fed to the five class (SM, SAT, VAT, IMAT and back ground) segmentation network. Figure 2 illustrates the segmentation network architecture. This network is the modified version of the binary segmentation network presented in our muscle segmentation study (Dabiri et al., 2019). This multi-label segmentation network is designed to address the Unet and FCN limitations and refine them by proposing a network that leverages from both networks advantages and diminishes their drawbacks. The inputs to this network are the axial L3 images and targets are their corresponding masks showing muscle, SAT, VAT and IMAT regions. The core model consists of 8 convolutional layers and is inspired by the encoder-decoder architecture proposed by Ronneberger et al. in (Ronneberger et al., 2015). This architecture has multiple connections between encoder layers and decoder layers at the same level. These connections translate the fine spatial information on the earlier encoder layers to the decoder which results in fine boundaries in the generated mask. The encoder part consists of four convolution layers each followed by a max-pooling layer. Mirroring that in decoder, four transpose convolution layers follow four convolution layers. The prediction from this model is called mask 1. In the full network, three up-sampling layers and three dilated convolution layers are added to the core model to generate mask 2 and mask 3. Mask 2 is generated following the procedure presented for FCN with 16-pixel stride by Long et al. (Long et al., 2015). In our application of the method, the 2-pixel stride layer is generated and up-sampled once more to get the original image size. The prediction taken from the 2-pixel stride layer is generated by summing the output of the pooling layers with the encoder’s output. This requires up-sampling for upper layers so that they reach the same size as the previous layer. Mask 1 and mask 2 is then concatenated and fed to the three dilated convolutions with 64 channels, kernel size of 3×3 and dilation rate of 4, 6 and 8. The outputs from these layers are concatenated to feed to the prediction layer that gives the final mask.
Figure 2:
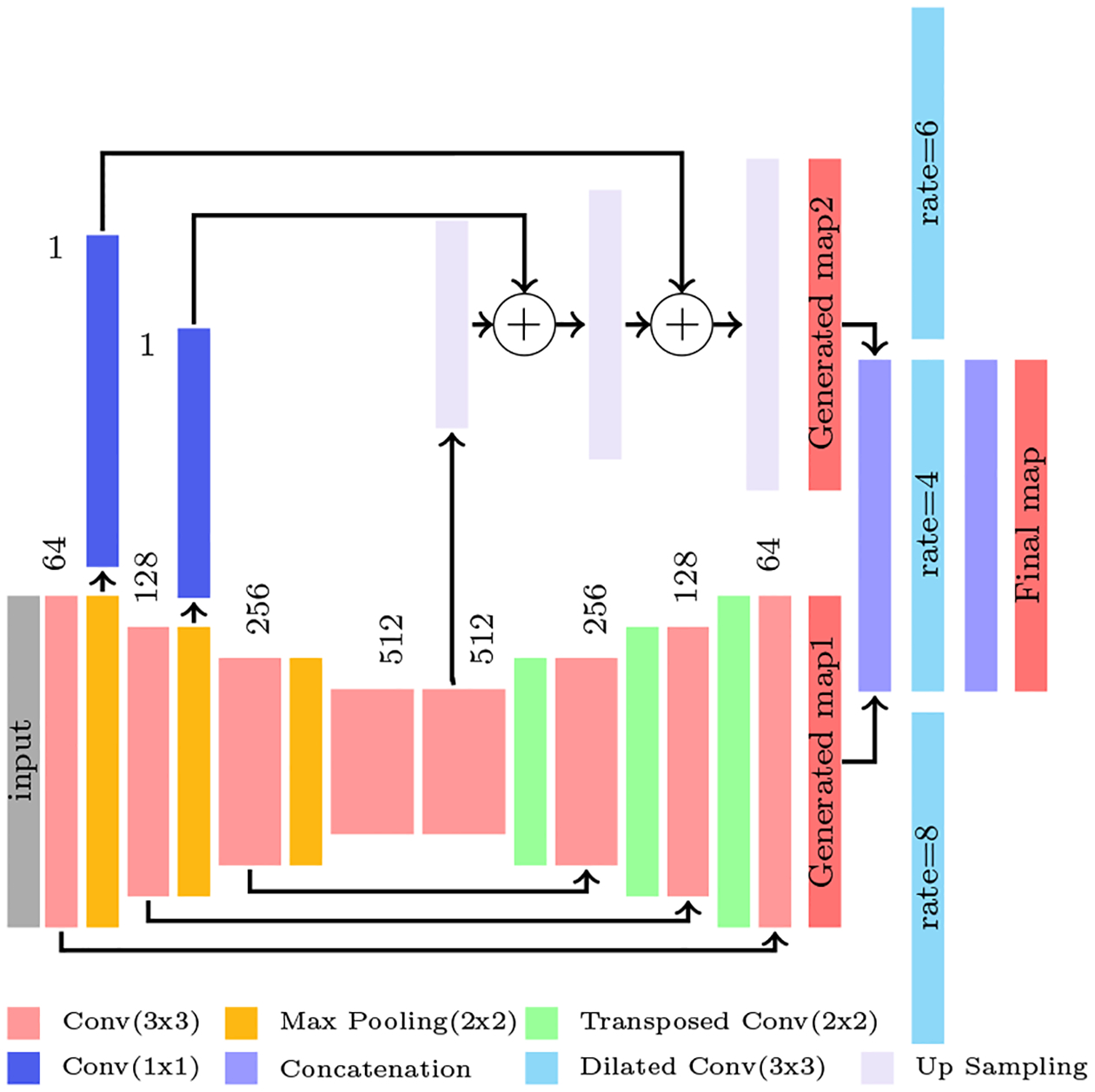
The illustration of the full network architecture. The prediction from Pooling layers 1 and 2 and also the prediction from the Convolution layer 5 from the core model is extracted and fed to up sampling layers shown in purple to achieve the second map. Then, map1 and map 2 is concatenated and fed to three dilated convolution layer shown in cyan. Finally the outputs of these three layers are concatenated and generates the final map. The circles with + symbol means the feature maps summation.
3. Experiments
3.1. Dataset
Three different datasets were used for training the L3 localization and segmentation networks. Dataset-1: The first dataset consists of abdominal axial CT images taken at the L3 level from patients with head, neck and lung cancers. This dataset was acquired at the Cross Cancer Institute (CCI), University of Alberta, Canada. The manual segmentation was performed by a single expert operator using Slice-O-Matic V4.3 software. Dataset-2: The second dataset is the 3D CT scan volumes from the ”C-scan Study” of patients diagnosed with stage IIII invasive colorectal cancer who had a surgical resection at Kaiser Permanente Northern California (KPNC) between 2006 and 2011 (Caan et al., 2017). Participants (male/female) were taken from a range of race/ethnicity and body mass index (BMI) categories. A trained researcher quantified the muscle tissue discriminating components using Slice-O-Matic software version 5.0 (Tomovision, Montreal, Canada). Dataset-3: The third dataset consists of CT images at third lumbar level and is from female patients in the ”B-scan Study” aged 18 to 80 years diagnosed with stage II or III invasive breast cancer at KPNC between January 2005 and December 2013 (Caan et al., 2018). Two trained researchers quantified the cross-sectional area of muscle using Slice-O-Matic software version 5.0.
3.1.1. Data preparation
CT images are stored in DICOM format. The normal images’ sizes are 512 by 512. In this step, the DICOM images are converted to grey scale PNG images. All images were standardized so that their pixel values lie in the range of [0,1]. To reduce the possibility of over-fitting and improve the generalization of the network some transformation is done on the available data. Random rotation, horizontal and vertical flip are the transformations used on the training dataset.
3.2. L3 Localization Network Training
The L3 localization network is trained to classify the inputs as L3 or non-L3 images. The inputs to the network are the 2D axial slices at L3 and non-L3 vertebrae levels. While Dataset-2 consists of 3D CT image volumes, Dataset-3 and Dataset-1 only include the 2D axial slices at L3 level. Therefore the L3 localization network is trained on 60% of the subjects in Dataset-2 (L3 and non-L3 slices) and all L3 slices from Dataset-3 and Dataset-1. In total, more than 6600 slices for each class of L3 and non-L3 were available for training the network. The loss function is defined as the binary cross-entropy and stochastic gradient descent is the optimizer of localization network. In the test stage, the network evaluated on the rest 40% of the subjects from Dataset-2.
3.3. L3 Segmentation Network Training
The segmentation network is trained in two steps. At first, the core encoder-decoder part is trained to find a good weight initialization for training the full network. Afterwards, the full network is trained to achieve the final prediction. The network was trained on approximately 60% of subjects in each dataset and evaluated on the remaining 40% kept as unseen test data. The model is optimized using Adam optimizer. The loss function for this step is the multi-class cross entropy. For training the full network the weighted loss of mask 1, mask 2 and final mask with the weight vector of [0.2, 0.2, 1] was considered. While ReLU was the activation function of all hidden layers, softmax was applied on the output prediction layer.
The Hounsfield unit (HU) range for each tissue was used in the postprocessing of the predicted final segmentation mask. The SM, VAT, SAT and IMAT on CT images are found to respectively have the range between [−29, 150], [−150 −50], [−190 −30] and [−190 −30] HU in intensity. These ranges were applied as the threshold to the segmentation mask to remove pixels that classified as the tissue of interest but are outside its intensity range.
4. Results
4.1. Results of L3 slice localization
To evaluate the L3 slice localization model, its performance on 1748 unseen 3D CT image volumes investigated. The 3D CT images consist of 2D axial slices at different vertebrae levels. These 2D slices are fed into the network to find the L3-candidate slices of each 3D CT image. L3-candidate images were extracted based on the classifier’s output. Then the slice with the highest probability among the L3-candidates of each CT image volume was chosen as the middle L3 slice.
The mean slice error for 1748 volumes is 0.87 ± 2.54 slice. For 1149 of the test volumes the labeled L3 slice predicted correctly. In 435 of the incorrect cases, one of the L3-candidate images is the true middle L3 slice and the other one is the slice before or after the true middle L3-slice. Therefore we ran an experiment to study the affect of choosing either of these two slices as L3 on the body-composition analysis results. For this experiment the ratio of the tissue difference between the true middle L3 and predicted L3 slice to the tissue in the true L3 slice calculated. Respectively the mean values of 0.049, 0.174, 0.084 and 0.096 were achieved for SM, IMAT, SAT and VAT.
The proposed method is also compared to two other methods of L3 localization applied on the frontal MIP image of the CT scan volumes. The fully convolutional network proposed in (Kanavati et al., 2018) and the regression algorithm using VGG16 in (Belharbi et al., 2017) were trained and tested on our dataset. The mean slice error of FCN with frontal MIP images as input and 1D confidence map as output (Kanavati et al., 2018) and regression algorithm using VGG16 network (Belharbi et al., 2017) on 1748 CT scan test images is reported in table 1.
Table 1:
The comparison of three L3-localization algorithm performances on 1748 CT images.
| Method | Training Data | Test Error (mean±standard deviation) |
|---|---|---|
| FCN (Kanavati et al., 2018) | Frontal MIP images | 0.87 ± 4.66 |
| VGG16 (Belharbi et al., 2017) | Frontal MIP images | 2.04 ± 2.54 |
| VGG11 (Proposed method) | Axial images | 0.87 ± 2.54 |
As shown in table 1, while the mean slice error of the method presented in Kanavati et al. (2018) and our approach is the same, our proposed classification method has lower standard deviation. To evaluate the generalization of our proposed algorithm and Kanavati et al. model we further tested them on the same dataset but with the partial field of view. To generate the partial field of view scans, a number of slices from the coccyx location of the vertebral column to the bottom of the scans were removed. While the performance of our model remained the same, the FCN model error in terms of the number of slices increased to 3.37 ± 11.89. These results demonstrate the importance of the features extracted from the individual axial slices such as the vertebrae structure shape, muscle shape and other organs presented on the axial slices of each vertebrae location.
4.2. Results of tissue segmentation on L3 slices
The generalization of the segmentation network to unseen images was investigated by training the proposed network on two datasets separately and using subsets of the two datasets set aside for testing. The Jaccard score, Dice coefficient, sensitivity and specificity of each experiment are reported for bench marking performance in Table 2. Figure 3 displays the two trained models as the distribution of the Jaccard score for each segmented tissue. The red and green histograms are the results of testing the model trained on Dataset-2 and Dataset-3, respectively. Based on the violin plot the Jaccard score of SM, SAT and VAT are above 90 percent for all experiments. The poorest network performance is for the IMAT segmentation. One possible reason is the fact the area of IMAT tissue is small and one mis-classified pixel penalizes the average performance more. Different classification of adipose tissue (Mitsiopoulos et al., 1998; Gorgey and Dudley, 2007) leads to difficulty in accurately segmenting this adipose tissue on CT scan images in the labeling process. The reduction in performance in some cross-database experiments could be attributed to different manual labeling protocols followed for the datasets.
Table 2:
The quantitative results of the two trained model on two test sets of L3 images. The top row of the table shows the results of testing each of the two networks on the test samples set aside from Dataset-2. The images used for the two trained networks are shown in the second column. The bottom row displays the results of applying each of the two networks on the test samples set aside for Dataset-3.
| Test set (Number of samples) |
Training set (Number of samples) |
Tissue | Jaccard Score | Dice Coefficient | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| Dataset-2 (1327) | Dataset-2(3774) | Muscle | 97.82 ± 2.51 | 98.88 ± 1.37 | 99.02 ± 1.34 | 99.89 ± 0.16 |
| VAT | 97.47 ± 5.39 | 98.6 ± 4.31 | 98.63 ± 4.90 | 99.92 ± 0.09 | ||
| SAT | 98.07 ± 4.58 | 98.94 ± 3.96 | 98.98 ± 4.32 | 99.88 ± 0.15 | ||
| IMAT | 83.63 ± 7.70 | 90.87 ± 5.01 | 90.09 ± 7.41 | 99.94 ± 0.05 | ||
| Dataset-3(1801) | Muscle | 94.61 ± 3.67 | 97.19 ± 2.10 | 95.5 ± 2.98 | 99.91 ± 0.12 | |
| VAT | 96.43 ± 6.26 | 98.03 ± 4.85 | 98.52 ± 5.36 | 99.85 ± 0.13 | ||
| SAT | 96.59 ± 5.25 | 98.16 ± 4.25 | 98.9 ± 4.36 | 99.69 ± 0.34 | ||
| IMAT | 61.23 ± 12.13 | 75.21 ± 9.8 | 63.08 ± 12.32 | 99.98 ± 0.02 | ||
| Dataset-3 (1202) | Dataset-2(3774) | Muscle | 94.34 ± 4.23 | 97.02 ± 3.38 | 99.03 ± 3.46 | 99.63 ± 0.23 |
| VAT | 95.28 ± 6.01 | 97.46 ± 4.27 | 96.94 ± 4.92 | 99.92 ± 0.11 | ||
| SAT | 97.74 ± 1.97 | 98.85 ± 1.04 | 98.65 ± 1.45 | 99.83 ± 0.23 | ||
| IMAT | 63.25 ± 12.13 | 76.76 ± 9.87 | 93.86 ± 7.45 | 99.79 ± 0.16 | ||
| Dataset-3(1801) | Muscle | 96.28 ± 2.73 | 98.08 ± 1.50 | 98.34 ± 1.87 | 99.84 ± 0.14 | |
| VAT | 96.12 ± 5.39 | 97.92 ± 3.90 | 98.17 ± 4.00 | 99.91 ± 0.12 | ||
| SAT | 98.36 ± 1.72 | 99.16 ± 0.91 | 99.36 ± 0.85 | 99.83 ± 0.25 | ||
| IMAT | 81.96 ± 10.73 | 89.65 ± 7.56 | 88.95 ± 9.23 | 99.96 ± 0.04 |
Figure 3:
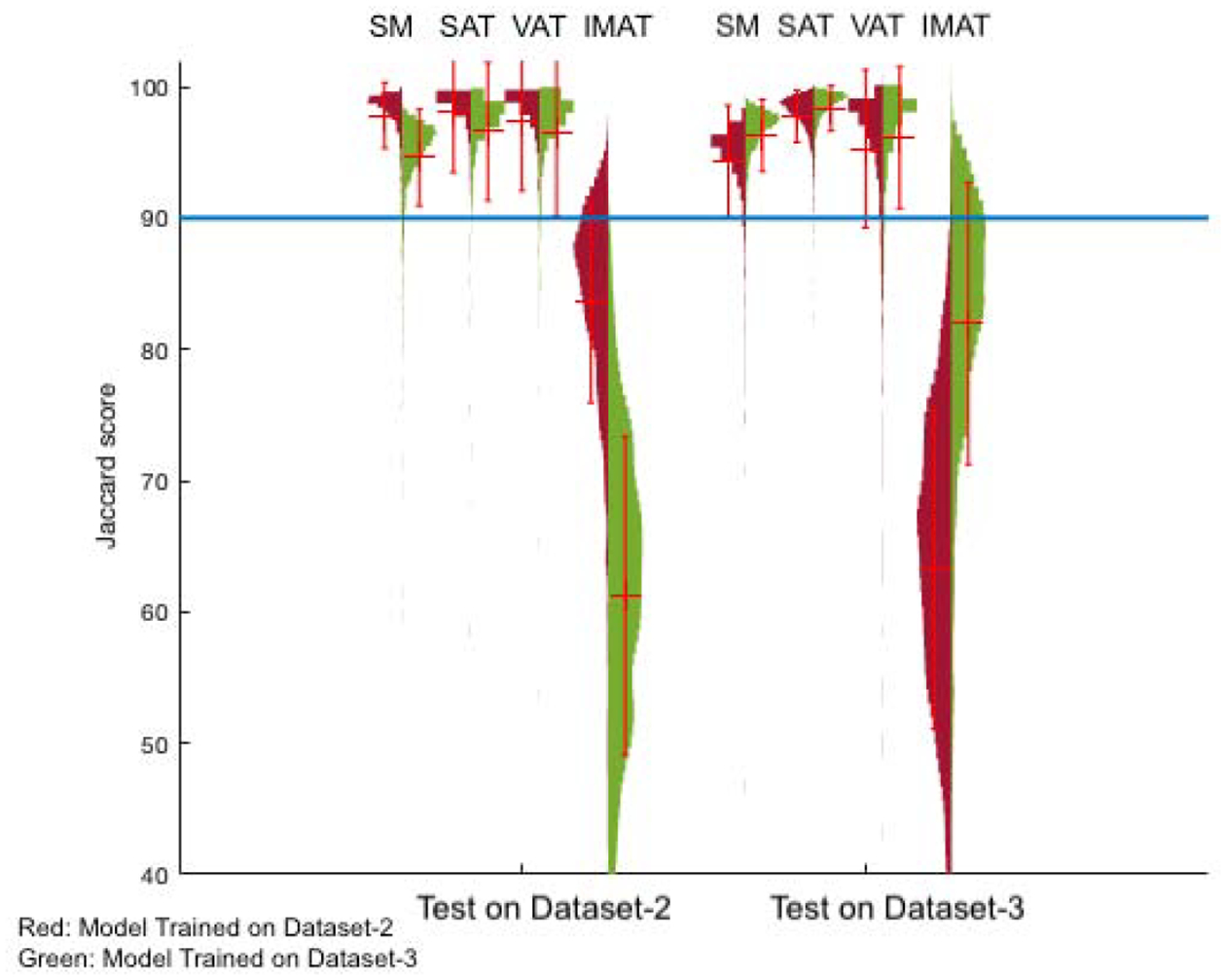
Violin plot demonstrating the segmentation network’s performance. The group of four graphs from the left are the performance of the two trained networks tested on Dataset-2 and Dataset-3, respectively. Each group shows the distribution of Jaccard scores for SM, SAT, VAT and IMAT for testing the model trained on Dataset-2 (Showing in red) and Dataset-3 (showing in green).
Figures 4 represents some of the best and poor predicted segmentation maps for SM, VAT, SAT and IMAT from Dataset-2 and Dataset-3. Note that the test set is identical in bench marking the performance of the two separately trained networks. For comparison purposes, Unet is also trained and tested on Dataset-2. Its performance on 1327 L3 test samples from Dataset-2 resulted in 96.21, 96.82, 94.39 and 60.17 Jaccard score for SM, SAT, VAT and IMAT respectively. Comparing these scores with the proposed network performance from Table 2 an improvement for all the four regions is achieved.
Figure 4:
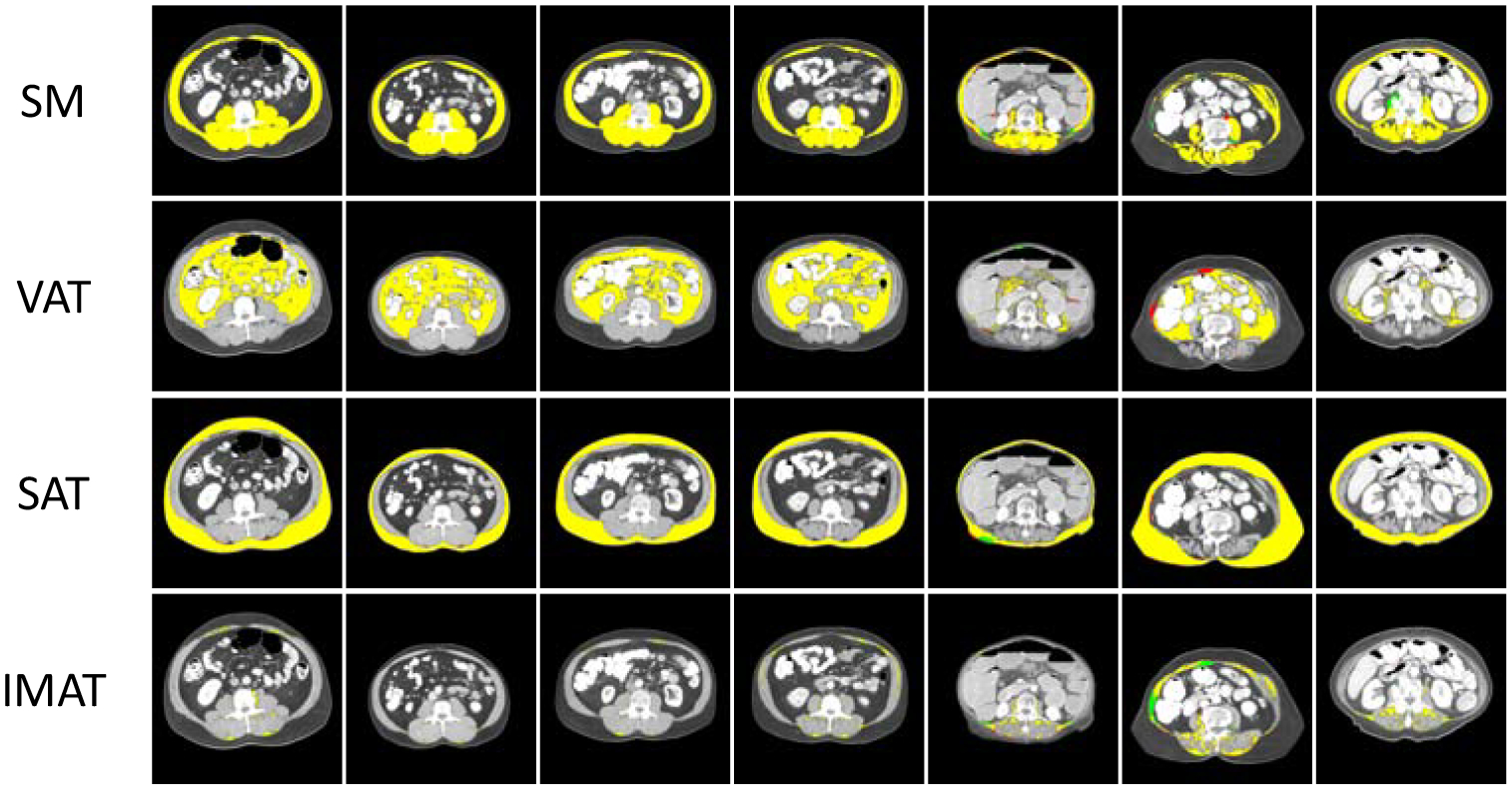
Illustration of some of the best and poor performances. Each row represents the SM, VAT, SAT and IMAT of one sample respectively. Yellow regions are the pixels that correctly classified. Red pixels are the pixels that mis-classified and green pixels are the pixels which are missed from predicted map.
5. Conclusion
In this paper, we developed an algorithm to automatically extract the third lumbar middle slice from the volume of full-body or partial-body CT scans and segment the SM, VAT, SAT and IMAT tissues. The generalization of the model was investigated using experiments conducted on three datasets. The results from these experiments suggest that L3 slice localization is possible employing only the 2D axial slices for feature learning. Moreover, the network is applicable for CT scans of various field of view and device settings. Regarding the segmentation network, the features were carefully studied considering our goals to retain high spatial resolution, the smoothness of labeling within a given region and the accuracy of the boundary for small degenerated areas in the muscle. Results show the high performance of the segmentation model on datasets from different CT devices and data acquisition settings.
Highlights:
An algorithm is proposed to automatically extract the CT slice at the third lumbar (L3) vertebra level and segment the muscle and adipose tissue regions from that.
The L3 localization network is a binary classification model trained on the 2D axial slices of a partial or full CT scan image to classify the input images as L3 or non-L3 slices.
The five class segmentation network is a convolutional neural network trained to segments the skeletal muscle (SM), subcutaneous adipose tissue (SAT), visceral adipose tissue (VAT) and intermuscular adipose tissue (IMAT) on the middle L3 slice.
Models are validated on three CT scan datasets. The mean slice error of 0.87±2.54 achieved for L3 slice localization on 1748 CT scan volumes. The performance of the tissue segmentation network evaluated on two datasets and high Jaccard scores in the range of 0.97–0.98 is achieved for SM, SAT and VAT. The Jaccard score for IMAT segmentation is 0.85.
Acknowledgements
National Cancer Institute grants R01CA175011 and R01CA184953 supported the C- and B-SCANS studies, respectively. Dr. Cespedes Feliciano was supported by K01CA226155.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Belharbi S, Chatelain C, Hérault R, Adam S, Thureau S, Chastan M, Modzelewski R, 2017. Spotting l3 slice in ct scans using deep convolutional network and transfer learning. Computers in biology and medicine 87. [DOI] [PubMed] [Google Scholar]
- Bridge CP, Rosenthal M, Wright B, Kotecha G, Fintelmann F, Troschel F, Miskin N, Desai K, Wrobel W, Babic A, et al. , 2018. Fully-automated analysis of body composition from ct in cancer patients using convolutional neural networks, in: OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis. Springer, pp. 204–213. [Google Scholar]
- Caan BJ, Feliciano EMC, Prado CM, Alexeeff S, Kroenke CH, Bradshaw P, Quesenberry CP, Weltzien EK, Castillo AL, Olobatuyi TA, et al. , 2018. Association of muscle and adiposity measured by computed tomography with survival in patients with nonmetastatic breast cancer. JAMA oncology 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caan BJ, Meyerhardt JA, Kroenke CH, Alexeeff S, Xiao J, Weltzien E, Feliciano EC, Castillo AL, Quesenberry CP, Kwan ML, et al. , 2017. Explaining the obesity paradox: the association between body composition and colorectal cancer survival (c-scans study). Cancer Epidemiology and Prevention Biomarkers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Cobzas D, Birdsell L, Lieffers J, Baracos V, 2009. Automated segmentation of muscle and adipose tissue on ct images for human body composition analysis, in: Medical Imaging 2009: Visualization, Image-Guided Procedures, and Modeling, International Society for Optics and Photonics. p. 72610K. [Google Scholar]
- Dabiri S, Popuri K, Feliciano EMC, Caan BJ, Baracos VE, Beg MF, 2019. Muscle segmentation in axial computed tomography (ct) images at the lumbar (l3) and thoracic (t4) levels for body composition analysis. Computerized Medical Imaging and Graphics 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L, 2009. Imagenet: A large-scale hierarchical image database, in: Computer Vision and Pattern Recognition, 2009. CVPR 2009. IEEE Conference on, IEEE. pp. 248–255. [Google Scholar]
- Gorgey A, Dudley G, 2007. Skeletal muscle atrophy and increased intramuscular fat after incomplete spinal cord injury. Spinal cord 45. [DOI] [PubMed] [Google Scholar]
- Huang G, Liu Z, Van Der Maaten L, Weinberger KQ, 2017. Densely connected convolutional networks, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 4700–4708. [Google Scholar]
- Kanavati F, Islam S, Aboagye EO, Rockall A, 2018. Automatic l3 slice detection in 3d ct images using fully-convolutional networks. arXiv preprint arXiv:1811.09244. [Google Scholar]
- Lee H, Troschel FM, Tajmir S, Fuchs G, Mario J, Fintelmann FJ, Do S, 2017. Pixel-level deep segmentation: artificial intelligence quantifies muscle on computed tomography for body morphometric analysis. Journal of digital imaging 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Shelhamer E, Darrell T, 2015. Fully convolutional networks for semantic segmentation, in: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- Mitsiopoulos N, Baumgartner R, Heymsfield S, Lyons W, Gallagher D, Ross R, 1998. Cadaver validation of skeletal muscle measurement by magnetic resonance imaging and computerized tomography. Journal of applied physiology 85. [DOI] [PubMed] [Google Scholar]
- Mourtzakis M, Prado CM, Lieffers JR, Reiman T, McCargar LJ, Baracos VE, 2008. A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Applied Physiology, Nutrition, and Metabolism 33. [DOI] [PubMed] [Google Scholar]
- Popuri K, Cobzas D, Esfandiari N, Baracos V, Jägersand M, 2016. Body composition assessment in axial ct images using fem-based automatic segmentation of skeletal muscle. IEEE transactions on medical imaging 35. [DOI] [PubMed] [Google Scholar]
- Popuri K, Cobzas D, Jägersand M, Esfandiari N, Baracos V, 2013. Fem-based automatic segmentation of muscle and fat tissues from thoracic ct images, in: ISBI, Citeseer; pp. 149–152. [Google Scholar]
- Ronneberger O, Fischer P, Brox T, 2015. U-net: Convolutional networks for biomedical image segmentation, in: International Conference on Medical image computing and computer-assisted intervention, Springer; pp. 234–241. [Google Scholar]
- Shen W, Punyanitya M, Wang Z, Gallagher D, St.-Onge MP, Albu J, Heymsfield SB, Heshka S, 2004. Total body skeletal muscle and adipose tissue volumes: estimation from a single abdominal cross-sectional image. Journal of applied physiology 97. [DOI] [PubMed] [Google Scholar]
- Simonyan K, Zisserman A, 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]


