Abstract
Although many scientists are studying the association between genetic polymorphism of ABCB1 and CR in patients, the molecular mechanism has not been further studied in patients with CHD. This study investigated the relationship between SNP of the ABCB1 gene in patients with CHD and CR, and whether the polymorphism of the ABCB1 gene affects the AS of the gene. 741 patients were enrolled in the study, 316 CR cases and 425 NCR cases. The correlation between CR risk and clinical-pathological characteristics were studied. Additionally, the five SNPs were analysed by PCR and Mass Array genotyping methods. Furthermore, silicon analysis was used to predict whether the polymorphism affects the process of AS. Results showed that there was a significant correlation between rs1045642 polymorphism and CR in genotyping and allele analysis. The rs1045642 polymorphism of the ABCB1 gene of CHD patients carrying the A allele are more likely to develop CR. Silicon analysis showed that rs1045642 generated a new ESE sequence which might affect AS of ABCB1 gene. We hypothesize that the mechanism of CR might be caused by a change in the AS caused by the polymorphism of the gene. Thus, this work provides guidance for the clinical use of clopidogrel.
Keywords: ABCB1, single nucleotide polymorphism, clopidogrel resistance, alternative splicing, silico analysis
INTRODUCTION
Coronary heart disease (CHD) is usually caused by the lipid accumulation in the walls of the heart arteries [1, 2]. CHD is a high-risk cardiovascular disease in the population and the main cause of death in patients with cardiovascular disease [3, 4]. Percutaneous coronary intervention (PCI) is a treatment for CHD, which might lead to stent thrombosis. Therefore, antithrombotic therapy plays a crucial role in the treatment of CHD [5]. Clopidogrel is mainly prescribed for antiplatelet therapy [6] and ultimately converted into active clopidogrel metabolites in the liver [7]. Although clopidogrel is the cornerstone of antiplatelet therapy, some patients will develop into clopidogrel resistance (CR). Genetic polymorphism is one of the main factors affecting the CR of individuals. Therefore, we can use genetic testing to guide the clinical individualization of CR patients [8–10].
At present, there are many genes in clopidogrel pharmacogenomics research, among which CYP2C19 and ATP binding cassette subfamily B member 1 (ABCB1) genes are particularly concerned. We have selected the ABCB1 gene for this experimental study of CR and polymorphisms. The p-glycoprotein (p-gp) encoded by the ABCB 1/MDR 1 (multidrug resistance-1) gene, physiologically, which can transport hydrophobic and hydrophilic compounds through the placenta, intestine and other parts, and protect the blood-brain barrier [11]. In addition, steroid hormones, immunosuppressants and antimetabolites or antibiotics can be discharged, as well as metabolites to protect cells from cytotoxic substances [12]. It causes a variety of drugs to be excreted outside the cancer cells, leading to chemotherapy resistance of tumour [13]. ABCB1 gene is a highly polymorphic gene with multiple single nucleotide polymorphisms (SNPs). Many studies in recent years had shown that genetic polymorphism affects the process of alternative splicing (AS) and leads to the development and progression of many diseases in humans, such as thalassemia, Alzheimer's disease, male infertility, retinitis pigmentosa and cancer [14–24]. More than 95% of protein coding genes occurs AS that increases the diversity of the human genome [25]. The complex process of AS is affected by many elements. Not only the branch point, the sequence of 3′ and 5′ splice sites but also cis-elements and trans-acting elements are required. Mutations or polymorphisms in cis-regulatory factors might affect mechanical damage of the exon splicing complexes and cause many diseases [16].
Although a large number of scientists around the world are studying the association between genetic polymorphism of ABCB1 and CR, the results between the genetic polymorphisms and the risk of clopidogrel are still controversial. Diverse results of the above studies might be caused by differences in sample selection requirements, geographical distribution and ethnic differences, sample size, and different genotyping methods [26, 27]. Recently, mutations at specific sites of the gene have caused certain disease and affected the process of the gene's AS [23, 28–31]. As far as we know, the relationship between gene polymorphism and AS in Chinese population with CR risk has not been studied. Therefore, our present study aims to investigate the molecular mechanism of CR caused by the polymorphism of the gene. Our work might provide theoretical guidance for the clinical individualization of clopidogrel in Chinese population with CHD.
RESULTS
Distributions of selected variables in cases and controls
The demographic information and clinical characteristics of the CR group and NCR groups were showed in Table 1. The relationship between age and sex in the CR group and NCR group has not been found, indicating that frequency matching of age and gender was appropriate. The mean platelet volume (MPV) was different between the CR group and the NCR group (P=0.036). At the same time, the rest of the variables were not found that significant correlation between CR group and NCR group.
Table 1. Distributions of selected variables in CHD cases and controls.
| Characteristics | NCR | CR | P | |
| Total | 425 | 316 | ||
| Sex | M (%) | 233 (55.1) | 190 (44.9) | 0.149 |
| F (%) | 192 (60.4) | 126 (39.6) | ||
| History of smoking | Y (%) | 90 (59.6) | 61 (40.4) | 0.531 |
| N (%) | 335 (56.8) | 255 (43.2) | ||
| Drinking history | Y (%) | 34 (66.7) | 17(33.3) | 0.163 |
| N (%) | 391 (56.7) | 299 (43.3) | ||
| History of diabetes | Y (%) | 35 (57.4) | 26 (42.6) | 0.997 |
| N (%) | 390 (57.4) | 290 (42.6) | ||
| History of hypertension | Y (%) | 114 (56.2) | 89 (43.8) | 0.686 |
| N (%) | 311 (57.8) | 227 (42.2) | ||
| Combined with APC | Y (%) | 354 (56.1) | 277 (43.9) | 0.098 |
| N (%) | 71 (64.5) | 39 (35.5) | ||
| Age | 63.50±10.68 | 64.03±9.28 | 0.479 | |
| BMI/kg/m2 | 24.27±3.43 | 24.16±3.54 | 0.661 | |
| Systolic pressure/mmHg | 122.52±16.18 | 124.38±14.84 | 0.108 | |
| Diastolic pressure/mmHg | 74.78±10.20 | 75.26±9.21 | 0.509 | |
| Glycated hemoglobin/% | 5.48±0.71 | 5.41±0.52 | 0.151 | |
| Triglyceride/mmol/L | 1.26±0.62 | 1.23±0.61 | 0.561 | |
| Total cholesterol/mmol/L | 4.37±0.93 | 4.45±0.96 | 0.241 | |
| HDL-C/mmol/L | 1.34±0.26 | 1.35±0.25 | 0.850 | |
| LDL-C/mmol/L | 2.93±0.64 | 2.98±0.64 | 0.330 | |
| C-reactive protein/mg/L | 2.20±3.97 | 2.54±5.31 | 0.315 | |
| PLT/109 | 215.42±67.11 | 219.00±63.35 | 0.461 | |
| MPV/fL | 12.55±1.73 | 12.28±1.68 | 0.036* | |
| PCT/% | 0.23±0.07 | 0.23±0.07 | 0.831 | |
| PDW/% | 17.99±3.78 | 18.03±3.88 | 0.900 |
Abbreviation: CR, Clopidogrel resistance; M, Male; F, Female; Y, Yes; N, No; PLT, Platelet count; MPV, Mean platelet volume; PCT, Plateletocrit; PWD, Platelet volume distribution width.
*: P value <0.05 was considered as significant.
HWE equilibrium test
Sequenom MassArray typing was performed on the above five polymorphisms. We calculated the specific genotype frequencies of the five SNPs and ensured that each site was in HWE equilibrium (Table 2).
Table 2. Hardy-Weinberg equilibrium test of five SNPs.
| SNP | Frequency n (%) | P(NCR) | P(CR) | |
| NCR | CR | |||
| rs1045642 | GG (37.9%)/GA(47.5%)/AA(14.6%) | GG (29.7%)/GA(47.5%)/AA(22.8%) | 0.916 | 0.413 |
| rs4148727 | TT (81.6%)/TC(17.9%)/CC(0.5%) | TT (82.6%)/TC(17.1%)/CC(0.3%) | 0.315 | 0.302 |
| rs2032582 | CC (28.7%)/CA(51.3%)/AA(20.0%) | CC (30.0%)/CA(47.5%)/AA(22.5%) | 0.487 | 0.422 |
| rs3789243 | GG (43.5%)/GA(46.4%)/AA(10.1%) | GG (44.6%)/GA(42.1%)/AA(13.3%) | 0.369 | 0.236 |
| rs1858923 | AA (27.8%)/AG(50.8%)/GG(21.4%) | AA (26.3%)/AG(49.4%)/GG(24.3%) | 0.671 | 0.827 |
Sequence analysis
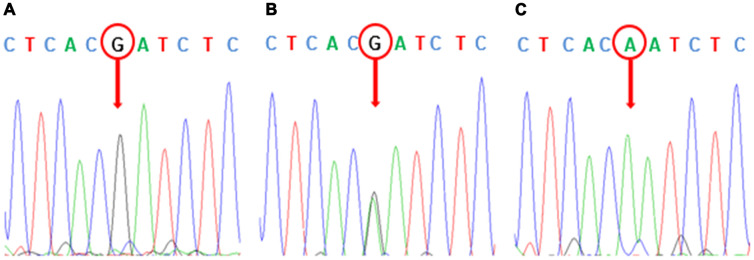
In order to verify the accuracy of the Sequenom MassArray genotyping method, we randomly selected some samples to verify the sequence of 5 SNPs. The image of agarose gel electrophoresis of the polymerase chain reaction (PCR) amplification products of some samples of DNA samples were shown in Figure 1. The wild homozygous sequence, heterozygous sequence and mutant homozygous sequence of the rs1045642 polymorphic site were listed in Figure 2.
Figure 1.
Electropherograms of PCR product fragments verified by sequence verification of 5 SNPs of partial DNA samples. (A) Electropherogram of PCR product of rs1045642 polymorphism. (B) Electropherogram of PCR product of rs4148727 polymorphism. (C) Electropherogram of PCR product of rs2032582 polymorphism. (D) Electropherogram of PCR product of rs3789243 polymorphism. (E) Electropherogram of PCR product of rs1858923 polymorphism.
Figure 2.
Sequence verification of rs1045642 polymorphism. (A) GG-wild type homozygous. (B) AG-heterozygote. (C) AA-SNP homozygous. The arrow in the picture points to the specific bases of the rs1045642 polymorphic sequence.
Statistical analysis of genotyping results of five SNPs
Sequenom MassArray typing technique showed genotypes of 741 participants and we performed a statistical analysis of the genotype results. All clinical data and classification results were entered into SPSS22.0 software. The results were expressed as mean ± standard deviation, and the count data was analysed by chi-square test. Expressed by the number and frequency of distribution, the P value reflects the difference between each variable in the CR group and NCR group. The chi-square test of five polymorphisms of ABCB1 gene was performed to compare the distribution of different genotypes between NCR group and CR group and the results were illustrated in Table 3. Chi-square test showed that the rs1045642 locus of ABCB1 gene was significantly different between the CR group and the NCR group, while the four polymorphisms (rs4148727, rs2032582, rs3789243, rs1858923) were not significantly different between the groups. The rs1045642 of ABCB1 gene genotype was distributed in the NCR group at 37.9% (GG, 161), 47.5% (GA, 202), 14.6% (AA, 62), and the genotype distribution frequency in the CR group was 29.7% (GG, 94), 47.5% (GA, 150), 22.8% (AA, 72). In the genotype analysis, we found that rs1045642 AA genotype and GG genotype were significantly different (OR=1.989, 95% CI=1.301-3.040, P=0.001); AA genotype and GA + AA genotype were also significantly different (OR=1.440, 95% CI=1.965-1.056, P=0.021). In the allele analysis, there was a statistical difference between the allele A and the G allele (OR=1.398, 95% CI=723-1.135, P=0.002). The rs1045642 polymorphism of the ABCB1 gene of CHD patients carrying the A allele is more prone to CR.
Table 3. Statistical analysis of the association between five polymorphisms of the ABCB1 gene and the risk of CR.
| SNP | Model | Genotype | NCR | CR | OR (95% CI) | P |
| rs1045642 | Heterozygous model | GG | 161(44.4%) | 94(38.5%) | ||
| GA | 202(55.6%) | 150(61.5%) | 1.272(0.914-1.770) | 0.154 | ||
| Homozygous model | GG | 161(72.2%) | 94(56.6%) | |||
| AA | 62(27.8%) | 72(43.4%) | 1.989(1.301-3.040) | 0.001* | ||
| Dominant model | GG | 161(37.9%) | 94(29.7%) | |||
| GA+AA | 264(62.1%) | 222(70.3%) | 1.440(1.056-1.965) | 0.021* | ||
| Allele genetic model | G | 524(61.6%) | 338(53.5%) | |||
| A | 326(38.4%) | 294(46.5%) | 1.398(1.135-1.723) | 0.002* | ||
| rs4148727 | Heterozygous model | TT | 347(82.0%) | 261(82.9%) | ||
| TC | 76(18%) | 54(17.1%) | 0.945(0.643-1.387) | 0.771 | ||
| Homozygous model | TT | 347(99.4%) | 261(99.6%) | |||
| CC | 2(0.6%) | 1(0.4%) | 0.665(0.060-7.370) | 0.738 | ||
| Dominant model | TT | 347(81.6%) | 261(82.6%) | |||
| TC+CC | 78(18.4%) | 55(17.4%) | 0.937(0.641-1.372) | 0.739 | ||
| Allele genetic model | T | 770(90.6%) | 576(91.1%) | |||
| C | 80(9.4%) | 56(8.9%) | 0.936(0.645-1.339) | 0.716 | ||
| rs2032582 | Heterozygous model | CC | 122(35.9%) | 95(38.8%) | ||
| CA | 218(64.1%) | 150(61.2%) | 0.884(0.629-1.241) | 0.475 | ||
| Homozygous model | CC | 122(58.9%) | 95(57.2%) | |||
| AA | 85(41.1%) | 71(42.8%) | 1.073(0.709-1.622) | 0.740 | ||
| Dominant model | CC | 122(28.7%) | 95(30.1%) | |||
| CA+AA | 303(71.3%) | 221(69.9%) | 0.937(0.681-1.289) | 0.688 | ||
| Allele genetic model | C | 462(54.4%) | 340(53.8%) | |||
| A | 388(45.6%) | 292(46.2%) | 1.023(0.832-1.257) | 0.832 | ||
| rs3789243 | Heterozygous model | GG | 185(48.4%) | 141(51.5%) | ||
| GA | 197(51.6%) | 133(48.5%) | 0.886(0.649-1.208) | 0.444 | ||
| Homozygous model | GG | 185(81.1%) | 141(77.0%) | |||
| AA | 43(18.9%) | 42(23.0%) | 1.282(0.794-2.068) | 0.309 | ||
| Dominant model | GG | 185(43.5%) | 141(44.6%) | |||
| GA+AA | 240(56.5%) | 175(55.4%) | 0.957 (0.714-1.283) | 0.767 | ||
| Allele genetic model | G | 567(66.7%) | 415(65.7%) | |||
| A | 283(33.3%) | 217(34.3 %) | 1.048(0.843-1.302) | 0.678 | ||
| rs1858923 | Heterozygous model | AA | 118(35.3%) | 83(34.7%) | ||
| AG | 216(64.7%) | 156(65.3%) | 0.974(0.688-1.380) | 0.882 | ||
| Homozygous model | AA | 118(56.5%) | 83(51.9%) | |||
| GG | 91(43.5%) | 77(48.1%) | 0.831(0.550-1.257) | 0.381 | ||
| Dominant model | AA | 118(27.8%) | 83(26.3%) | |||
| AG+GG | 307(72.2%) | 233(73.7%) | 0.927(0.667-1.287) | 0.650 | ||
| Allele genetic model | A | 452(53.2%) | 322(50.9%) | |||
| G | 398(46.8%) | 310(49.1%) | 0.915(0.744-1.124) | 0.396 |
*: P value <0.05 was considered as significant.
LD and haplotype analysis
SHEsis software was used to analyze the LD of five polymorphisms of ABCB1. In the Figure 3 and Table 4, LD was detected in the rs3789243 and rs1858923 polymorphisms (r2=0.310, D′=0.815), rs1858923 and rs4148727 polymorphisms (r2=0.070, D′=0.872). Meanwhile, haplotype analysis was performed in CR and NCR groups. In this study, the CR group and NCR group with haplotype estimated frequency below 3% were excluded from further analysis. We found that TGAGG haplotypes (rs4148727, rs1045642, rs2032582, rs3789243, rs1858923.) can appreciably lower the risk of CR (OR=0.663, 95% CI=0.447-0.983, P=0.040), and other haplotypes were not significantly different between CR group and NCR groups (Table 5).
Figure 3.
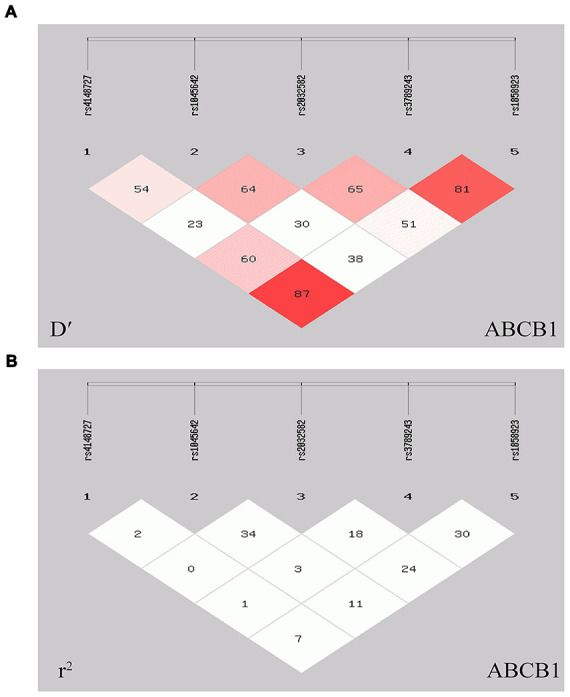
Linkage disequilibrium plot and r2 values for five polymorphisms of the ABCB1 gene. (A) D′ plot of five polymorphic linkage disequilibrium. (B) r2 plot of five polymorphic linkage disequilibrium. Linkage disequilibrium was detected in the rs3789243 and rs1858923 polymorphisms ((r2 = 0.310, D′ = 0.815), rs1858923 and rs4148727 polymorphisms (r2 = 0.070, D′ = 0.872).
Table 4. SHEsis software analyzes the LD relationship of five polymorphisms of ABCB1, and uses D′ and r2 to measure the LD relationship between the five SNPs.
| D′ | SNP | rs1045642 | rs2032582 | rs3789243 | rs1858923 |
| rs4148727 | 0.546 | 0.237 | 0.608 | 0.872 | |
| rs1045642 | - | 0.642 | 0.303 | 0.385 | |
| rs2032582 | - | - | 0.656 | 0.516 | |
| rs3789243 | - | - | - | 0.815 | |
| r2 | SNP | rs1045642 | rs2032582 | rs3789243 | rs1858923 |
| rs4148727 | 0.022 | 0.022 | 0.019 | 0.070 | |
| rs1045642 | - | 0.349 | 0.034 | 0.116 | |
| rs2032582 | - | - | 0.186 | 0.247 | |
| rs3789243 | - | - | - | 0.310 |
Abbreviation: LD, linkage disequilibrium.
Table 5. Results of haplotype analysis of five ABCB1 SNPs in CR and control groups.
| Haplotype* | Frequency n (%) | P | OR (95%CI) | |
| CR | NCR | |||
| C G C G A | 26.78(0.042) | 42.98(0.051) | 0.456 | 0.828 (0.505~1.360) |
| T A A A G | 20.89(0.033) | 14.85(0.017) | 0.053 | 1.924 (0.980~3.776) |
| T A A G A | 29.78(0.047) | 36.67(0.043) | 0.718 | 1.096 (0.667~1.800) |
| T A A G G | 165.44(0.262) | 185.45(0.218) | 0.05 | 1.282 (1.002~1.640) |
| T A C A A | 33.67(0.053) | 27.79(0.033) | 0.05 | 1.667 (0.996~2.789) |
| T G A G G | 40.77(0.065) | 79.64(0.094) | 0.04* | 0.663 (0.447~0.983) |
| T G C A A | 143.43(0.227) | 208.64(0.245) | 0.388 | 0.896 (0.699~1.149) |
| T G C G A | 32.44(0.051 | 59.72(0.070) | 0.132 | 0.713 (0.458~1.109) |
| T G C G G | 63.03(0.100) | 91.22(0.107) | 0.626 | 0.919 (0.653~1.293) |
Abbreviation: CR, Clopidogrel resistance; OR, Odds ratio; CI, Confidence interval.
The order of SNPs in haplotypes: rs4148727, rs1045642, rs2032582, rs3789243, rs1858923.
*: P value <0.05 was considered as significant.
Silico analysis of rs1045642 polymorphism
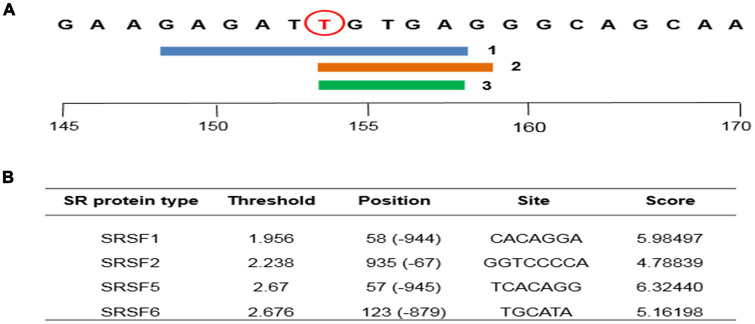
Mutations in cis-elements and splice sites can affect gene splicing. These effects were detected by HSF 3.1 and ESE finder 3.0. The HSF 3.1 software was used to predict whether the mutation of the rs1045642 SNP of the ABCB1 gene has an effect on AS, which predicted that a new ESE sequence was generated by mutation of the rs1045642 SNP of the ABCB1 gene (Figure 4A). Since the ESE sequence is a cis-acting element of AS, we further used ESE-finder software to predict whether the mutation of the gene would alter the ability to bind the trans-acting factor SR proteins in the AS. The ESE-finder software predicted that the new ESE sequence generated by the mutation will bind to the four SR proteins beyond the binding threshold (Figure 4B). Thus, silicon analysis showed that rs1045642 generated a new ESE sequence which might affect AS of ABCB1 gene.
Figure 4.
Human Splicing Finder version 3.1 (HSF) and ESE finder 3.0 bioinformatics analysis softwares are used to predict the effect the mutation of the rs1045642 SNP of the ABCB1 gene on AS. (A) The three algorithms of HSF software predict that polymorphism is an ESE site. (B) ESE finder software predicts the binding ability of ESE site to SRps.
DISCUSSION
ABCB1, P-glycoprotein, is an ABC transporter whose main function of ABCB1 is to expel toxic metabolites and xenobiotics from cells [32, 33]. Therefore, the ABCB1 gene has a great development in the mechanism of anticancer drug resistance [34]. Previous studies had reported gene polymorphisms and CR, and the ABCB1 is one of the hot genes. In our research, we have selected five polymorphisms of rs1045642, rs4148727, rs2032582, rs3789243 and rs1858923 on the ABCB1 gene to study the relationship between ABCB1 gene polymorphisms and the risk of CR. Meanwhile, the polymorphism rs1045642 of ABCB1 gene is more frequently studied in CR. In recent years, there have been different studies between the ABCB1 gene polymorphism and the occurrence of CR in patients with CHD. Our research showed that four polymorphisms of rs4148727, rs2032582, rs3789243 and rs1858923 were not associated with CR. However, the rs1045642 polymorphism was found to be intensively associated with CR.
Previous studies have shown that the concentration of active clopidogrel metabolites in homozygous 3435TT was significantly lower than that of CT/CC, suggesting that the increased expression of p-glycoprotein associated with 3435TT genotype might mediate the increase of clopidogrel efflux in the intestine [35, 36]. Therefore, it showed that ABCB1 gene polymorphism was related to the efficacy of clopidogrel in patients. We further studied the ABCB1 gene polymorphism and CR and found that the TT homozygous rs1045642 mutation has an increased risk of CR compared with CT/CC individuals. People with T alleles have an increased risk of ischemic events. Our results were consistent with those of Jessica L Mega et al. [37]. However, the study on CR and rs1045642 polymorphisms by Jia Su et al. found that there was no significant correlation between rs1045642 mutation and CR [38]. The inconsistency of the research results might be related to different sample selection standards, geographical distribution and ethnic differences, sample size and different genotyping methods. Currently, there are no prescribed standard for CR. Flow cytometry vasodilator-stimulated phosphoprotein phosphorylation (Vasp) analysis and verification-Now P2Y12 detection, pfa-100, whole TEG and impedance aggregation metrology (multi-plate analyzer) can all be used detect platelet function and clopidogrel reaction [39]. The method of evaluating CR is different, which might have an impact on our further research on CR. In this study, we had used TEG method to measure platelet inhibition rate. The VerifyNow analysis P2Y12 method was used by Jia Su et al. Previous studies had confirmed that allelic variants of the CYP2C19 gene have ethnic and geographic diversity, but they were not obvious in Asians or mixed races, which indicated that different genetic backgrounds have different frequencies of mutations [40]. In addition, studies had shown that polymorphisms of enzyme genes that metabolize drugs might change the response to drugs and occur at different frequencies in different ethnic groups [41]. This indicates that genetic background might change the effect of mutations on treatment in a specific group. Our samples were collected in Northwest China, and the samples in the study of Jia Su et al. were recruited in East China. Richard C Crist's study concluded that due to the small sample size, many previous studies were insufficient for retrospective drug genetic studies of African Americans [41]. The sample size is too small to detect experimental effect. The difference in sample size might also affect the results of experimental studies. 741 samples were included in our study, and 180 samples were included in the study of Jia Su et al., which was far less than the sample size of our experiment. At present, there are many methods for genotyping, including PCR-RFLP (polymerase chain reaction-restricted fragment length polymorphism) method, Taq-Man probe, ARMS (amplification difficult mutation system), genechip, Sanger sequencing, Mass spectrometry, etc. Different genotyping methods have their advantages and shortcomings. The typing method used in our experiment was Sequenom MassArray, and the PCR-RFLP genotyping method was used by Jia Su et al. The advantages of the Sequenom MassArray method include low cost, no need for special fluorescent primers, only a pair of PCR primers and amplification primers, convenient detection, high sensitivity, and high data accuracy. However, the PCR-RFLP method requires a specific restriction site for genotyping SNPs, and it is impossible to accurately identify false positives. Therefore, our typing method is better than the typing method used by Jia Su et al.
Compared with previous CR-related studies, our study not only analysed five polymorphisms of the gene, but also used bioinformatic software to predict the relationship between rs1045642 polymorphism and AS. More than 50% of all mutations leading to genetic diseases result in abnormal splicing [42]. Different forms of cis-regulatory elements include exon splicing enhancer (ESE), exon splicing silencer, intron splicing enhancer and intron splicing silencer sequences. The trans-acting factors are mainly SR proteins protein and heterogeneous nuclear ribonucleoproteins [16]. The combination of cis-regulatory element mutation/polymorphism and trans-acting factor leads to mechanical cleavage of exon splicing complex, which is an important mechanism for AS to participate in the development and progression of various diseases. Preliminary studies indicated that exon polymorphism of CD44 gene was analysed by ESE finder 3.0 demonstrated that mutation generated a new SR proteins binding site [43]. Therefore, we used HSF 3.1 to predict the rs1045642 polymorphism to generate a new ESE sequence. Furthermore, it was analysed by software (ESE finder 3.0) analysis that the binding ability of ESE sequence to SR proteins exceeded their binding thresholds. The bioinformatics software predicted that the rs1045642 polymorphism is a potential splice site, and the altered binding ability of the ESE sequence and the SR protein formed by the mutation affects the abnormal process of the AS of the ABCB1 gene. Therefore, we suspect that there is a relationship between the risk of CR in patients with CHD and the rs1045642 polymorphism affecting the AS process of ABCB1 gene.
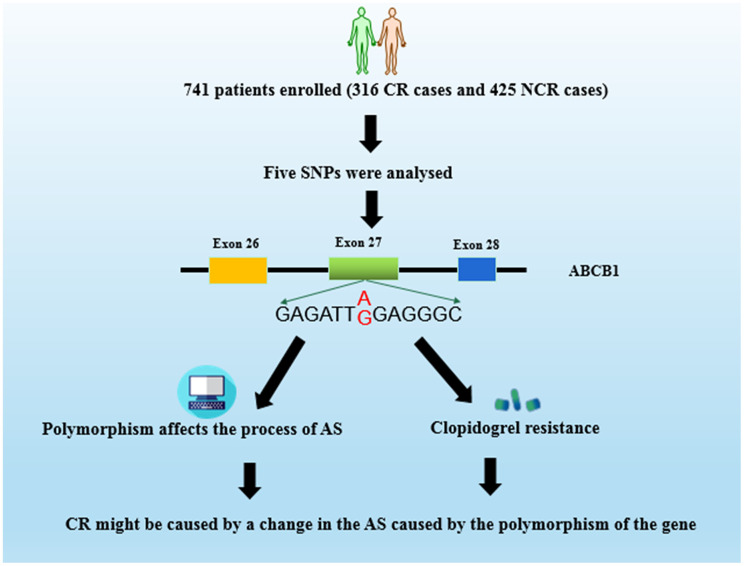
In our research, there was a significant relationship between the risk of CR in CHD patients and the different genotypes carried by individuals in China. Meanwhile, bioinformatics software has predicted that the ABCB1 gene rs1045642 polymorphism affects the process of AS and might have a certain relationship with the risk of CR in CHD patients (Figure 5). This study not only provides theoretical support for the individualized administration of clopidogrel after PCI in clinical CHD patients, but also offers a new basis for CR risk in CHD patients and the change of AS process caused by ABCB1 gene polymorphism.
Figure 5.
Schematic representation of mechanism of genetic polymorphism of ABCB1 and clopidogrel resistance (CR) in patients.
There is still controversial about the study between CR and polymorphism in patients with CHD. Previous studies on CR only performed polymorphic genotyping by collecting basic clinical information, test results, and patient blood samples, and then statistical analysis to obtain genotypic analysis and CR. However, the mechanism of clopidogrel-related drug resistance has not been further studied. In the current study, we not only did the predecessor research, but also predicted the SNP of clopidogrel-related genes through bioinformatic software. It was predicted that rs1045642 SNP site will potentially affect the AS of the ABCB1 gene. Therefore, we hypothesize that the mechanism of CR might be caused by a change in the AS caused by the polymorphism of the gene. Our research not only provides theoretical guidance for the clinical individualization of clopidogrel in patients with CHD, but also provides new idea for the study of the mechanism of CR. Meanwhile, specific experiments are needed to support our findings.
MATERIALS AND METHODS
Study population
The subjects of this case-control study were 741 patients with CHD or taking clopidogrel hydrogen sulfate after PCI in the Second Hospital of Lanzhou University and Gansu Provincial People's Hospital in Lanzhou City, Gansu Province, China. Briefly, there were 316 patients in the CR group and 425 patients in the NCR group. The exclusion criteria of this study were as follows: (1) the subjects were under 18 years old; (2) patients with a history of clopidogrel allergy; (3) severe liver and kidney insufficiency; (4) peptic ulcer; (6) major surgery; (7) cerebrovascular accident or recent bleeding history. Clinical data were collected from the electronic records, including demographic characteristics, history of smoking, history of drinking, history of hypertension, history of diabetes and previous history of major medical conditions. Laboratory tests, including triglycerides, total cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, glycosylated hemoglobin, high-sensitivity C-reactive protein and platelet count.and other biochemical variables, were performed after admission. This study was approved by the Ethics Committee of Lanzhou University School of Basic Medicine.
Thromboelastography test
The platelet inhibition rate was detected by thromboelastography (TEG) method according to the following steps: (1) 1 ml of citrated blood was mixed with kaolin by inversion and 340 μl of the activated blood was added to the 20 μl of calcium chloride; (2) 360 μl heparin anticoagulated blood was added to 10 μl batroxobin mixture; (3) 10 μl batroxobin and 10 μl ADP mixture were added to 360 μl heparin blood; (4) the CFMSLEPU-8800 instrument was used to detect the blood clot elasticity map. The test was performed using the CFMSLEPU-8800 Analyzer System. At present, there is no standardized baseline for CR. In this study, ADP inhibition rate <20% was classified as CR, and ADP inhibition rate ≥50% was defined as NCR [44].
SNPs selection
The ABCB1 gene is located on human chromosome 7 at q21 [45]. This study referred to NCBI (http://www.ncbi.nlm.nih.gov/ncbisearch), Ensembl (http://asia.ensembl.org/index.html/), the HapMap database of Han Chinese in Beijing, China (http://hapmap.ncbi.nlm.nih.gov) Databases and published articles related to CR when selecting the SNPs of the ABCB1 gene [46]. Ultimately, five polymorphisms rs1045642, rs4148727, rs2032582, rs3789243 and rs1858923 were selected in the current study.
Extraction of genomic DNA
Genomic DNA was extracted from EDTA-treated peripheral blood using the phenol/chloroform method. The extracted DNA was dissolved in 50 μl of 1 × TE buffer and frozen (−80° C) until required. The NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Lenexa, KS, USA) was used to detect the concentration and purity of DNA. Samples with DNA concentration exceeding 20 ng/ul were sent to biological company (Bomiao Biotechnology Co., Ltd., Beijing, China) for genotyping.
PCR amplification
The extracted genomic DNA was amplified by PCR. The PCR primers were designed using Assay design 3.1 software and submitted to Shanghai Invitrogen Biotechnology Co., Ltd. for synthesis. The amplification primers were shown in Table 6. For amplification of the DNA, the thermal cycling involved initial denaturation at 95° C for 5 min, 45 cycles of 95° C for 20 s, 56° C for 30 s, and 72° C for 1 min, and a final extension at 72° C for 7 min.
Table 6. Primers for polymerase chain reaction.
| Gene | SNP | Group | Base sequence (5′ → 3′) | Products size |
| ABCB1 | rs1045642 | Forward primer | ACGTTGGATGTTGCCTATGGAGACAACAGC | |
| Reverse primer | ACGTTGGATGAAGGCATGTATGTTGGCCTC | 99bp | ||
| rs4148727 | Forward primer | ACGTTGGATGCTTCAATGCTTTGGAGCCAT | ||
| Reverse primer | ACGTTGGATGTAAGTTTGGGTGGAGGAAGG | 98bp | ||
| rs2032582 | Forward primer | ACGTTGGATGCATATTTAGTTTGACTCACC | ||
| Reverse primer | ACGTTGGATGTGTTGTCTGGACAAGCACTG | 92bp | ||
| rs3789243 | Forward primer | ACGTTGGATGATAAGCCCAAGATCCTGTCC | ||
| Reverse primer | ACGTTGGATGCTGACTGCTTCAGTTCCAAC | 107bp | ||
| rs1858923 | Forward primer | ACGTTGGATGTCTGACTCCTGTGATAAGGG | ||
| Reverse primer | ACGTTGGATGTGCTGATTTTCCTCCAGCTC | 109bp |
Identification of ABCB1 SNPs by Sequenom MassArray
Extracted DNA samples were sent to a biological company (Bomiao Biotechnology Co., Ltd., Beijing, China) for genotyping. Sequenom MassArray typing technique was used to determine the all individual genotypes. The specific steps of the Sequenom MassArray genotyping method are as follows: (1) The PCR amplification primer sequences and extension primer sequences of genotyping SNPs were designed, and the primer sequences were sent Shanghai Invitrogen Biotechnology Co., Ltd for synthesis; (2) 4 μl PCR master mix and 1 μl template DNA were added to each well of the 384-well plate. The PCR reaction program was 94° C, 5 min; 45 cycles: 94° C, 20 s, 56° C, 30 s, 72° C, 1 min; 72° C, 3 min; 4° C, ∞; (3) The PCR product was treated with shrimp alkaline phosphatase (SAP) to remove free dNTPs in the system, and 2 μl SAP mixture and 5 μl PCR product were added to each well of 384-well plate for reaction. The SAP reaction program was37° C, 20 min; 85° C, 5 min; 4° C, ∞; (4) 2 μl single base mixture and 7 μl SAP-PCR reaction product were added to each well of 384-well plate. The extension reaction program was 90° C, 30 s; 40 cycles: 94° C, 5 s, (52° C, 5 s, 80° C, 5 s) × 5 cycles; 72° C, 3 min; 4° C, ∞; (5) Purification reaction; (6) Chip spotting; (7) Mass spectrometry detection and data output. After the MALDI-TOF mass spectrometer analyzer detects the SpectroCHIP chip, TYPER4.0 software was used to further analyze and process the raw data.
Sequencing verification
Conclusively, partial PCR products were randomly selected for direct sequencing verification. PRIMER 3.0 software was used for primer design and sent to General Biosystems (Anhui) Co., Ltd. for synthesis. The synthesized primer sequence and PCR product size were shown in Table 7. The PCR reaction program was pre-denaturation 94° C for 3 min, 31 cycles of 94° C for 30 s, 58.6° C for 30 s, and 72° C for 25 s, and a final extension at 72° C for 5 min. In the 1.1% agarose gel pool in 1×TAE electrophoresis solution, the PCR products (5 μl/well) of each sample were electrophoresed at 110 V for 25 min, and the agarose was treated with GoldView I. The target bands were detected on the Tanon 1600 gel imaging system (Tanon Science and Technology Co., Ltd.), and the amplified products were sent to General Biosystems (Anhui) Co., Ltd. for sequencing verification. The sequencing results were analysed by SeqManII software (DNASTAR, Madison, WI, USA).
Table 7. Primers for polymerase chain reaction.
| Gene | SNP | Group | Base sequence (5′ → 3′) | Products size |
| ABCB1 | rs1045642 | Forward primer | GCTGAGAACATTGCCTATGG | |
| Reverse primer | GGAAGTGTGGCCAGATGCTTGT | 259bp | ||
| rs4148727 | Forward primer | CATTCTGCCTATTCTGGCTA | ||
| Reverse primer | GGATAAGTTTGGGTGGAGGA | 232bp | ||
| rs2032582 | Forward primer | GCTATAGGTTCCAGGCTTGC | ||
| Reverse primer | GTCCAAGAACTGGCTTTGCT | 390bp | ||
| rs3789243 | Forward primer | CCATTCCTTGAGGACTTTGG | ||
| Reverse primer | CCCAGACAATAAGCCCAAGA | 202bp | ||
| rs1858923 | Forward primer | TGTCTTGATTTCGCCATTGA | ||
| Reverse primer | CCTGTGATAAGGGGTTGAGG | 246bp |
Statistical analysis
SPSS 22.0 statistical software (IBM Corp. Armonk, NY, USA) was used for data analysis. For the continuous variables, analysis of variance techniques or Kruskal-Wallis tests were used and expressed as the means ± SD. Categorical variables are summarised by frequency and percentage. Associations between SNPs and CR in CHD were assessed using the Chi-square test. Hardy-Weinberg equilibrium (HWE), linkage disequilibrium (LD) and haplotype analysis tests were evaluated using SHEsis software (http://analysis.bio-x.cn/myAnalysis.php) in case and control groups. Odds ratio (OR) with 95% Confidence intervals (CI) were calculated to estimate the strength of relationship between individual genotypes of five polymorphisms and CR. P value < 0.05 for the difference was significant.
Silico analysis
In order to determine whether rs1045642 affects the splicing efficiency, computer analysis was performed using HSF 3.1 and ESE finder 3.0 bioinformatics analysis software [42, 43, 45]. HSF 3.1 detects receptor (3'ss) and donor (5'ss) positions by a position weight matrix. ESE finder 3.0 software was used to estimate ESE sequences that change due to mutations. The default threshold was used to determine the locus responsible for the four Serine/arginine-rich (SR) proteins, including AS factor/splicing factor 2 (ASF / SF2), SR splicing factor 5 (SRp40), SR splicing factor 3 (SC35), SR splicing factor 6 (SRp55). The default thresholds for HSF were 1.956, 2.67, 2.383, and 2.6676, respectively. Only sequences with scores above or equal to the threshold were selected.
Availability of data and materials
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ABCB1
ATP binding cassette subfamily B member 1
- CR
Clopidogrel resistance
- CHD
Coronary heart disease
- SNP
Single nucleotide polymorphism
- AS
Alternative splicing
- PCR
Polymerase chain reaction
- PCI
Percutaneous coronary intervention
- ESE
exon splicing enhancer
- MPV
mean platelet volume
- SAP
Shrimp alkaline phosphatase
- CI
Confidence intervals
- OR
Odds ratio
- HWE
Hardy-Weinberg equilibrium
- LD
Linkage disequilibrium
- SR
Serine/arginine-rich
- ASF / SF2
AS factor/splicing factor 2
Footnotes
AUTHOR CONTRIBUTIONS: SSZ, JW, CXD and XDX conceived the manuscript. SSZ, JW, AAZ, WKY, YHC and XTZ did experiments and wrote the initial draft of the manuscript. XWZ, TY and DXX participated in the collection of blood samples. All authors reviewed the manuscript.
CONFLICTS OF INTEREST: The authors declare that they have no conflicts of interest.
FUNDING: The national Key R&D project of the Chinese Ministry of Science and Technology (2018YFE0205100), the current study was supported by the National Natural Science Foundation of China (31670379), the Key Program of the National Natural Science Foundation of China (U1632270), and the National Natural Science Foundation of China (11675234).
REFERENCES
- 1.Wardoku R, Blair C, Demmer R, Prizment A. Association between physical inactivity and health-related quality of life in adults with coronary heart disease. Maturitas. 2019; 128:36–42. 10.1016/j.maturitas.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Känel R, Carney RM, Zhao S, Whooley MA. Heart rate variability and biomarkers of systemic inflammation in patients with stable coronary heart disease: findings from the heart and soul study. Clin Res Cardiol. 2011; 100:241–47. 10.1007/s00392-010-0236-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin X, Pan B, Wu H, Wu B, Li Y, Wang X, Liu G, Dang X, Xu D. The efficacy and safety of shenzhu guanxin recipe granules for the treatment of patients with coronary artery disease: protocol for a double-blind, randomized controlled trial. Trials. 2019; 20:520. 10.1186/s13063-019-3629-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikrani R, Li C, Naveed M, Li C, Baig MM, Zhang Q, Wang Y, Peng J, Zhao L, Zhou X. Pharmacokinetic advantage of ASD device promote drug absorption through the epicardium. Pharm Res. 2020; 37:173. 10.1007/s11095-020-02898-6 [DOI] [PubMed] [Google Scholar]
- 5.Moulias A, Alexopoulos D. Long-term outcome of percutaneous coronary intervention: the significance of native coronary artery disease progression. Clin Cardiol. 2011; 34:588–92. 10.1002/clc.20929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patti G, Micieli G, Cimminiello C, Bolognese L. The role of clopidogrel in 2020: a reappraisal. Cardiovasc Ther. 2020; 2020:8703627. 10.1155/2020/8703627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox KA, Chelliah R. Clopidogrel: an updated and comprehensive review. Expert Opin Drug Metab Toxicol. 2007; 3:621–31. 10.1517/17425225.3.4.621 [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Wang Y. [Status quo and countermeasure of clopidogrel resistance predicted by gene testing]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2019; 36:649–53. 10.3760/cma.j.issn.1003-9406.2019.06.030 [DOI] [PubMed] [Google Scholar]
- 9.Pereira NL, So D, Bae JH, Chavez I, Jeong MH, Kim SW, Madan M, Graham J, O’Cochlain F, Pauley N, Lennon RJ, Bailey K, Hasan A, et al. International survey of patients undergoing percutaneous coronary intervention and their attitudes toward pharmacogenetic testing. Pharmacogenet Genomics. 2019; 29:76–83. 10.1097/FPC.0000000000000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su J, Zheng N, Li Z, Huangfu N, Mei L, Xu X, Zhang L, Chen X. Association of GCK gene DNA methylation with the risk of clopidogrel resistance in acute coronary syndrome patients. J Clin Lab Anal. 2020; 34:e23040. 10.1002/jcla.23040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin-Orozco E, Sanchez-Fernandez A, Ortiz-Parra I, Ayala-San Nicolas M. Wnt signaling in tumors: the way to evade drugs and immunity. Front Immunol. 2019; 10:2854. 10.3389/fimmu.2019.02854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cario E. P-glycoprotein multidrug transporter in inflammatory bowel diseases: more questions than answers. World J Gastroenterol. 2017; 23:1513–20. 10.3748/wjg.v23.i9.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, Chen XY, Dong XD, Wang JQ, Feng W, Teng QX, Cui Q, Li J, Li XQ, Chen ZS. NVP-CGM097, an HDM2 inhibitor, antagonizes ATP-binding cassette subfamily B member 1-mediated drug resistance. Front Oncol. 2020; 10:1219. 10.3389/fonc.2020.01219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mijajlovic MD, Shulga O, Bloch S, Covickovic-Sternic N, Aleksic V, Bornstein NM. Clinical consequences of aspirin and clopidogrel resistance: an overview. Acta Neurol Scand. 2013; 128:213–19. 10.1111/ane.12111 [DOI] [PubMed] [Google Scholar]
- 15.Huang BY, Zeng Y, Li YJ, Huang XJ, Hu N, Yao N, Chen MF, Yang ZG, Chen ZS, Zhang DM, Zeng CQ. Uncaria alkaloids reverse ABCB1-mediated cancer multidrug resistance. Int J Oncol. 2017; 51:257–68. 10.3892/ijo.2017.4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Yuan Y. Alternative RNA splicing and gastric cancer. Mutat Res. 2017; 773:263–73. 10.1016/j.mrrev.2016.07.011 [DOI] [PubMed] [Google Scholar]
- 17.Thein SL. The molecular basis of β-thalassemia. Cold Spring Harb Perspect Med. 2013; 3:a011700. 10.1101/cshperspect.a011700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasquez JB, Simpson JF, Harpole R, Estus S. Alzheimer’s disease genetics and ABCA7 splicing. J Alzheimers Dis. 2017; 59:633–41. 10.3233/JAD-170872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rafaee A, Mohseni Meybodi A, Yaghmaei P, Hosseini SH, Sabbaghian M. Single-nucleotide polymorphism c.474G>A in the SEPT12 gene is a predisposing factor in male infertility. Mol Reprod Dev. 2020; 87:251–59. 10.1002/mrd.23310 [DOI] [PubMed] [Google Scholar]
- 20.Shankar SP, Birch DG, Ruiz RS, Hughbanks-Wheaton DK, Sullivan LS, Bowne SJ, Stone EM, Daiger SP. Founder effect of a c.828+3A>T splice site mutation in peripherin 2 (PRPH2) causing autosomal dominant retinal dystrophies. JAMA Ophthalmol. 2015; 133:511–17. 10.1001/jamaophthalmol.2014.6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Littink KW, Pott JW, Collin RW, Kroes HY, Verheij JB, Blokland EA, de Castro Miró M, Hoyng CB, Klaver CC, Koenekoop RK, Rohrschneider K, Cremers FP, van den Born LI, den Hollander AI. A novel nonsense mutation in CEP290 induces exon skipping and leads to a relatively mild retinal phenotype. Invest Ophthalmol Vis Sci. 2010; 51:3646–52. 10.1167/iovs.09-5074 [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Freedman JA, Liu H, Moorman PG, Hyslop T, George DJ, Lee NH, Patierno SR, Wei Q. Associations between RNA splicing regulatory variants of stemness-related genes and racial disparities in susceptibility to prostate cancer. Int J Cancer. 2017; 141:731–43. 10.1002/ijc.30787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao F, Zhang P, Wang Y, Tian Y, James M, Huang CC, Wang L, Wang L. Single-nucleotide polymorphism rs13426236 contributes to an increased prostate cancer risk via regulating MLPH splicing variant 4. Mol Carcinog. 2020; 59:45–55. 10.1002/mc.23127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao D, Zhang C, Jiang M, Wang Y, Liang Y, Wang L, Qin K, Rehman FU, Zhang X. Survival-associated alternative splicing signatures in non-small cell lung cancer. Aging (Albany NY). 2020; 12:5878–93. 10.18632/aging.102983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallo CM, Ho A, Beffert U. ApoER2: functional tuning through splicing. Front Mol Neurosci. 2020; 13:144. 10.3389/fnmol.2020.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsantes AE, Ikonomidis I, Papadakis I, Bonovas S, Gialeraki A, Kottaridi C, Kyriakou E, Kokori S, Douramani P, Kopterides P, Karakitsos P, Lekakis J, Kapsimali V. Impact of the proton pump inhibitors and CYP2C19*2 polymorphism on platelet response to clopidogrel as assessed by four platelet function assays. Thromb Res. 2013; 132:e105–11. 10.1016/j.thromres.2013.06.015 [DOI] [PubMed] [Google Scholar]
- 27.Akram N, Mustafa G, Hanif AA, Tawwab S, Hussain S, Kaul H, Mohsin S. Cytochrome 2C19 and paraoxonase-1 polymorphisms and clopidogrel resistance in ischemic heart disease patients. Per Med. 2019. [Epub ahead of print]. 10.2217/pme-2018-0030 [DOI] [PubMed] [Google Scholar]
- 28.Xue LL, Wang F, Xiong LL, Du RL, Zhou HL, Zou Y, Wu MX, Yang MA, Dai J, He MX, Wang TH. A single-nucleotide polymorphism induced alternative splicing in Tacr3 involves in hypoxic-ischemic brain damage. Brain Res Bull. 2020; 154:106–15. 10.1016/j.brainresbull.2019.11.001 [DOI] [PubMed] [Google Scholar]
- 29.Mascarenhas JB, Tchourbanov AY, Fan H, Danilov SM, Wang T, Garcia JG. Mechanical stress and single nucleotide variants regulate alternative splicing of the MYLK gene. Am J Respir Cell Mol Biol. 2017; 56:29–37. 10.1165/rcmb.2016-0053OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh RK, Cooper TA. pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012; 18:472–82. 10.1016/j.molmed.2012.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan W, Wang W, Ma Q. Physiological and pathological function of serine/arginine-rich splicing factor 4 and related diseases. Biomed Res Int. 2018; 2018:3819719. 10.1155/2018/3819719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ofverholm A, Einbeigi Z, Manouchehrpour S, Albertsson P, Skrtic S, Enerbäck C. The ABCB1 3435 T allele does not increase the risk of paclitaxel-induced neurotoxicity. Oncol Lett. 2010; 1:151–54. 10.3892/ol_00000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang YW, Lin CY, Tsai HC, Fong YC, Han CK, Huang YL, Wu WT, Cheng SP, Chang HC, Liao KW, Wang SW, Tang CH. Amphiregulin promotes cisplatin chemoresistance by upregulating ABCB1 expression in human chondrosarcoma. Aging (Albany NY). 2020; 12:9475–88. 10.18632/aging.103220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parle-McDermott A, Pangilinan F, O’Brien KK, Mills JL, Magee AM, Troendle J, Sutton M, Scott JM, Kirke PN, Molloy AM, Brody LC. A common variant in MTHFD1L is associated with neural tube defects and mRNA splicing efficiency. Hum Mutat. 2009; 30:1650–56. 10.1002/humu.21109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taubert D, von Beckerath N, Grimberg G, Lazar A, Jung N, Goeser T, Kastrati A, Schömig A, Schömig E. Impact of p-glycoprotein on clopidogrel absorption. Clin Pharmacol Ther. 2006; 80:486–501. 10.1016/j.clpt.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 36.Stokanovic D, Nikolic VN, Konstantinovic SS, Zvezdanovic JB, Lilic J, Apostolovic SR, Pavlovic M, Zivkovic VS, Jevtovic-Stoimenov T, Jankovic SM. P-glycoprotein polymorphism C3435T is associated with dose-adjusted clopidogrel and 2-oxo-clopidogrel concentration. Pharmacology. 2016; 97:101–06. 10.1159/000442712 [DOI] [PubMed] [Google Scholar]
- 37.Mega JL, Close SL, Wiviott SD, Shen L, Walker JR, Simon T, Antman EM, Braunwald E, Sabatine MS. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet. 2010; 376:1312–19. 10.1016/S0140-6736(10)61273-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su J, Yu Q, Zhu H, Li X, Cui H, Du W, Ji L, Tong M, Zheng Y, Xu H, Zhang J, Zhu Y, Xia Y, et al. The risk of clopidogrel resistance is associated with ABCB1 polymorphisms but not promoter methylation in a Chinese Han population. PLoS One. 2017; 12:e0174511. 10.1371/journal.pone.0174511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aradi D, Storey RF, Komócsi A, Trenk D, Gulba D, Kiss RG, Husted S, Bonello L, Sibbing D, Collet JP, Huber K, and Working Group on Thrombosis of the European Society of Cardiology. Expert position paper on the role of platelet function testing in patients undergoing percutaneous coronary intervention. Eur Heart J. 2014; 35:209–15. 10.1093/eurheartj/eht375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhuo ZL, Xian HP, Long Y, Liu C, Sun YY, Ma YT, Gao H, Zhao JZ, Zhao XT. Association between CYP2C19 and ABCB1 polymorphisms and clopidogrel resistance in clopidogrel-treated Chinese patients. Anatol J Cardiol. 2018; 19:123–29. 10.14744/AnatolJCardiol.2017.8097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crist RC, Clarke TK, Ang A, Ambrose-Lanci LM, Lohoff FW, Saxon AJ, Ling W, Hillhouse MP, Bruce RD, Woody G, Berrettini WH. An intronic variant in OPRD1 predicts treatment outcome for opioid dependence in African-Americans. Neuropsychopharmacology. 2013; 38:2003–10. 10.1038/npp.2013.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003; 31:3568–71. 10.1093/nar/gkg616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esmaeili R, Abdoli N, Yadegari F, Neishaboury M, Farahmand L, Kaviani A, Majidzadeh-A K. Unique CD44 intronic SNP is associated with tumor grade in breast cancer: a case control study and in silico analysis. Cancer Cell Int. 2018; 18:28. 10.1186/s12935-018-0522-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bliden KP, DiChiara J, Tantry US, Bassi AK, Chaganti SK, Gurbel PA. Increased risk in patients with high platelet aggregation receiving chronic clopidogrel therapy undergoing percutaneous coronary intervention: is the current antiplatelet therapy adequate? J Am Coll Cardiol. 2007; 49:657–66. 10.1016/j.jacc.2006.10.050 [DOI] [PubMed] [Google Scholar]
- 45.Razi B, Anani Sarab G, Omidkhoda A, Alizadeh S. Multidrug resistance 1 (MDR1/ABCB1) gene polymorphism (rs1045642 C > T) and susceptibility to multiple myeloma: a systematic review and meta-analysis. Hematology. 2018; 23:456–62. 10.1080/10245332.2018.1443897 [DOI] [PubMed] [Google Scholar]
- 46.Hung CC, Chiou MH, Teng YN, Hsieh YW, Huang CL, Lane HY. Functional impact of ABCB1 variants on interactions between p-glycoprotein and methadone. PLoS One. 2013; 8:e59419. 10.1371/journal.pone.0059419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.