Graphical abstract
Keywords: Thyroid carcinogenesis, Liver microtissues, Uridine glucuronosyltransferase, Genomics, Cross species risk assessment
Highlights
-
•
Species differences in UGT induction could mediate thyroid cancer susceptibility.
-
•
The effect of CAR activators on rat thyroid carcinogenesis could be partly explained by differential induction of Ugt 2b17.
-
•
Human UGT changes would likely contribute less to species differences in T4 metabolism than rat UGT changes.
Abstract
Species differences in hepatic metabolism of thyroxine (T4) by uridine diphosphate glucuronosyl transferase (UGT) and susceptibility to thyroid hormone imbalance could underlie differences in thyroid carcinogenesis caused by hepatic enzyme inducers in rats and humans. To investigate this hypothesis we examined profiles of hepatic UGT induction by the prototypical CAR activator phenobarbital (PB) in rat and human liver 3D microtissues. The rationale for this approach was that 3D microtissues would generate data more relevant to humans. Rat and human liver 3D microtissues were exposed to PB over a range of concentrations (500 u M - 2000 u M) and times (24−96 hr). Microarray and proteomics analyses were performed on parallel samples to generate integrated differentially expressed gene (DEG) datasets. Bioinformatics analysis of DEG data, including CAR response element (CRE) sequence analysis of UGT promoters, was used to assess species differences in UGT induction relative to CAR-mediated transactivation potential. A higher proportion of human UGT promoters were found to contain consensus CREs compared to the rat homologs. UGTs 1a6, 2b17 and 2b37 were upregulated by PB in rat liver 3D microtissues, but unaltered in human liver 3D microtissues. By contrast, human UGTs 1A8, 1A10 and 2B10 showed higher levels of induction (RNA and /or protein) compared to the rat homologs. There was general concordance between the presence of CREs and the induction of UGT RNA. As UGT1A and 2B isoforms metabolise T4, these results suggest that differences in UGT induction could contribute to differential susceptibility to CAR-mediated thyroid carcinogenesis in rats and humans.
1. Introduction
Rat thyroid pathology is a common occurrence in 2 year cancer bioassays resulting in thyroid tumours [1]. A frequent mechanism of thyroid carcinogenesis in rats is the response to CAR activators, which has been shown to involve the induction of phase 2 enzymes uridine diphosphate glucuronosyltransferase (UGTs). This effect increases the metabolism of thyroxine which down-regulates a negative feedback loop via the pituitary resulting in TSH-mediated thyroid hyperplasia [[2], [3], [4], [5]].The process underlying this mechanism involves transcriptional activation of UGT gene promoters via CAR response elements (CREs) [6,7].
The mechanism of rat thyroid carcinogenesis downstream of CAR activation and UGT induction, exemplified by the prototypical CAR activator phenobarbital, is not considered relevant to humans in part because rats lack thyroxine-binding globulin, and consequently the half-life of thyroid hormone is much shorter and TSH levels are higher in rats [8], making rats more sensitive to changes in thyroxine metabolism/clearance. However rat hepatocytes also show greater induction of thyroxine glucuronidation in response to enzyme inducers than human primary hepatocytes [9]. Many of the substrates of human UGTs are also glucuronidated by rat UGT family enzymes [10,11], and this is also reflected in the ability of UGT isoforms to metabolise thyroxine [12]. In addition, the UGT isoforms that possess thyroxine glucuronidation activity differ in their kinetic parameters (Km, Vmax) for metabolism of this substrate [9,13]. Hence there is a solid rationale for investigating differences in the profile of UGT induction as a hypothesis to explain in part species differences in thyroid carcinogenesis in response to CAR activators. To investigate this hypothesis, we have for the first time examined the profile of UGT isoform induction at the RNA and protein level in rat and human 3D liver microtissues by investigating the ability of the prototypical CAR activator phenobarbital (PB), a rat thyroid tumour promoter [2], to alter the expression of UGT isoforms. To date most of the historical data on CAR-mediated UGT induction has been reported in rat hepatocytes or rat in vivo studies [13]. We used rat and human 3D liver microtissues in order to investigate possible species differences in this response by utilising a model which displays nuclear hormone receptor induction responses more like the in vivo situation [14]. The idea was to form a 'bridge to humans' in what is considered to be a more physiologically relevant model, in order to assess potential species differences at the level of UGT induction using a comparable 3D culture system.
2. Materials and methods
2.1. Liver microtissues
Human liver microtissues each containing approximately 1000 cells (manufactured by InSphero using a patented 3D select™ technology) consisted of primary hepatocytes mixed with Kupffer cells and human liver endothelial cells (NPCs). Rat liver microtissues also contained hepatocytes, Kupffer cells and rat liver endothelial cells (InSphero white papers: https://insphero.com/science/publications/white-papers/). We chose to perform the study with rat and human liver 3D microtissues in order to utilise a model that had been shown previously to respond to nuclear hormone activators at the level of phase 1 and phase 2 enzyme induction in a manner more representative of the in vivo situation, when compared to 2D cultures of primary hepatocytes [14]. Human primary hepatocytes in the liver 3D microtissues were derived from pooling primary hepatocytes from 5 male and 5 female donors. Rat liver 3D microtissues were made with primary hepatocytes, Kupffer cells and endothelial cells from Sprague Dawley rats. As the same cell types were included in the rat liver microtissues as in the human liver microtissues the models were comparable at the cellular level.
2.2. Liver microtissue treatments
InSphero rat liver 3D microtissues (primary hepatocytes, co-cultured with NPCs (InSphero #MT-02−00104) and human liver 3D microtissues (multi-donor primary hepatocytes, co-cultured with NPCs (InSphero #MT-02−302-04) were treated with a stock solution of 200 mM PB in fresh 3D Insight rat liver maintenance medium (InSphero #CS-07−002-01) and 3D Insight human liver maintenance medium-AF (InSphero #CS-07001a-01), respectively, to give final concentrations of 500 u M, 750 u M, 1 mM and 2 mM according to previously published methods [15]. The concentrations of PB were chosen based on previous studies examining phase 1 enzyme induction and proliferation in rat primary hepatocytes [16,17]. The choice of this liver 3D microtissue model was based on our previous investigations [15] which showed that the effects of PB on hepatocyte proliferation were consistent with published results from rat and human hepatocytes [16]; specifically phase 1 and phase 2 enzyme induction was observed in both rat and human liver 3D microtissue hepatocytes, consistent with CAR activation, whilst in the rat liver 3D microtissue hepatocytes, there was also a significant dose-dependent increase in hepatocyte proliferation. This indicated that the liver 3D microtissue model responds to PB in a way that is consistent with the known species differences between rat and human [8,18], supporting the contention that this model lends itself to further exploration of the reported species differences in CAR-mediated responses.
Rat and human liver 3D microtissues were exposed, one spheroid per well, in a 96 well gravity trap™ plate to PB dissolved in rat or human liver maintenance medium, respectively, for four different time periods: 24 h, 48 h, 72 h and 96 h. We exposed 4 separate liver 3D microtissues in individual wells and 30 separate liver 3D microtissues, also in separate individual wells, for each treatment and time point for transcriptomics and proteomics analysis, respectively (total 4 × 96 well plates per species). After incubation of the spheroids for the appropriate periods of time, liver 3D microtissues were fixed in 4% paraformaldehyde [Pierce 16 % paraformaldehyde (Thermo Fisher Scientific #28,908) for 30 min at room temperature after which they were rinsed in PBS and then either wax embedded in a microTMA mold prior to sectioning/H&E/histopathology analysis, or harvested for transcriptomics/proteomics analysis using previously published methods [15]. Experiments were performed at least twice on different batches of spheroids.
2.3. Spheroid Tissue Microarray (microTMA) construction, H&E and histopathological analysis
Histopathological analysis of H&E sections was performed from a microTMA constructed using a previously published method [19].Briefly, fixed spheroids were loaded into the wells of a 2% agarose mold containing 96 wells, maintaining the same orientation as used in the 96 well plate, and sealed using molten 0.7 % agarose. We embedded 12 spheroids per treatment in the microTMA mold. The agarose mold containing spheroids was dehydrated for a minimum of 12 h in 70 % ethanol and the microTMA mold was processed to paraffin wax in a tissue processor (Thermo Citadel 1000). Following wax embedding the microTMA block was sectioned (we cut approximately 10 × 6 u M sections per microTMA) using a microtome (Reichert Jung) onto glass microscope slides. Parallel sections of the liver 3D microtissue microTMAs were stained with H&E to facilitate histopathological examination.
2.4. Assessment of liver 3D microtissuehistopathological changes and cytotoxicity
Histopathological changes in the liver 3D microtissues were assessed by examining x20 images of H&E stained sections of the spheroids generated by tile scanning the entire H&E stained microTMA slide. These images were reviewed by a board certified pathologist to assess histopathological changes caused by the treatment.
ATP levels were measured in whole liver 3D microtissue spheroids according to a previously published method [15].
2.5. RNA extraction and transplex labelling
RNA was extracted from 2 to 4 fixed rat or human liver 3D microtissues from each treatment (2 replicates per treatment) using the Qiagen miRNeasy FFPE Kit and labelled using the Agilent SureTag DNA labelling Kit according to previously published methods [15].
2.6. Microarray hybridisation and scanning
Cy3 labelled cDNA purified from the SureTag labelling was hybridised on Agilent Rat or Human Whole genome microarrays in an Agilent G2545A Hybridisation Oven according to previously published methods [15]. The gene specific 60 mer oligonucleotide probes on the Agilent rat and human whole genome arrays span conserved exons across the transcripts of the targeted full-length genes. We chose microarray analysis over q-PCR primarily so that we could efficiently perform whole genome transcript profiling analysis which is impractical to perform using q-PCR. This approach facilitated integration with global proteomics data thus facilitating corroboration of the differential RNA expression data at the protein level. The microarrays were scanned on an Agilent C scanner and Agilent Feature Extraction Software was used to extract raw microarray data from TIFF image files. Raw microarray data was normalised using a quantile algorithm and processed to Log2 fold change data using R according to previously published methods [15].
2.7. Proteomic analysis
Proteins from rat and human liver 3D microtissues (23–30 spheroids/sample, 4 replicates per treatment) were extracted using the Qproteome FFPE Tissue kit (QIAGEN), TMT labelled, and analysed on a Fusion Orbitrap LC MS/MS to raw data according to previously published methods [15]. We used 4 × 96 well plates of rat and 4 × 96 well plates of human liver 3D microtissues for the proteomics analysis, of which 2 plates per species were controls and 2 were PB treated. The analysis was performed for control and PB 750 uM treated samples as we had previously observed pronounced induction of CAR regulated phase 1 and phase 2 genes at this dose in the microarray analysis (see results section below).
Protein abundances were measured from the normalised TMT reporter ion intensities. Peptide mapping was performed using Thermo Proteome Discoverer software against global databases of rat and human proteins.
2.8. Statistical analysis of transcriptomics and proteomics data
Log 2 Fold change values in the transcriptomics analysis were calculated in R software using a linear model for microarray data analysis (LIMMA), and Benjamini Hochberg multiple test correction was applied to p values derived from the T test analysis to derive q values [false discovery rate (FDR) adjusted p values] for these alterations.This process was used to identify a list of differentially expressed genes (DEG list), relative to control, for each of the treatments using a cut off point for the q value derived from the LIMMA T test of q<0.05. Proteomics data was analysed in Proteome Discoverer 2.1 and Microsoft Excel. Raw intensity values for each peptide were normalised by taking the channel (isobaric tag) in the 8 plex analysis with the highest total peptide abundance value and then correcting all the other channels by a constant factor so that all channels had the same total abundance value. These normalised values were then scaled for every peptide so that the average of all channels had an intensity of 100 intensity units. A fold change and p value were calculated from 4 replicate data points for each treatment using t-test in Microsoft Excel.
2.9. UGT promoter analysis
A search of NCBI was performed for the various isoforms of rat and human UGT genes to see if any of the promoter sequences had been fully characterized in the literature. This literature was cross checked with the experimentally validated promoters listed on the Eukaryotic Promoter Database (https://epd.epfl.ch). For those isoforms for which neither of these resources offered UGT promoter sequences, a 1000 base pair sequence directly upstream of the UGT transcription start site was taken from the Ensembl genome database (http://www.ensembl.org/index.html). We chose to examine this region of the UGT promoters because historical data based on genome wide human ChIP-chip data has shown that in general transcription factor binding is focused in a region within 1000 bp of the transcription start site (TSS), [20].
Using experimentally-validated sequences for the CAR response elements (CREs) [21], the promoter sequences obtained (as above) were examined for the presence of CREs using Snapgene gene viewer software.
2.10. Integration of rat and human proteomic and transcriptomic data
Processed transcriptomic and proteomic fold change data were integrated into a single Excel spreadsheet using an R script and uploaded to IPA for bioinformatics analysis according to previously published methods [15].
3. Results
3.1. Liver 3D microtissue histopathology and cell viability
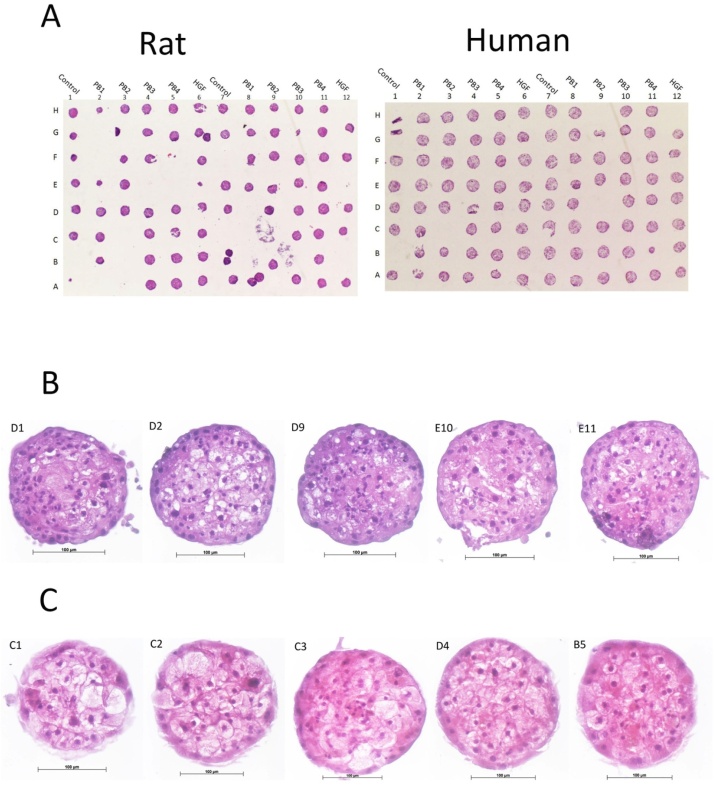
Histopathological examination of H&E stained microTMA slides from control and PB treated (500 u M, 750 u M, 1000 u M, 2000 u M) rat and human liver 3D microtissues revealed some vacuolation of hepatocytes in rat and human control and PB treated liver spheroids, Fig. 1. The reasons for this vaculoation is unknown; however, the finding is not considered of significance because their functional response to PB was unaffected, as evidenced by the induction of CAR regulated genes at the RNA and protein level (rat and human liver 3D microtissues), Table 1, and the expected proliferative response of rat liver 3D microtissues to PB treatment, indicating that the 3D liver microtissue model is suitable for assessing human/rat species differences at this level (BrdU LI% data previously published [15]). The levels of ATP in rat and human liver 3D microtissues were also similar in both controls and in those treated with PB (data previously published [15]).
Fig. 1.
H&E histopathological analysis of rat and human liver 3D microtissue microTMAs.
The liver microtissues were treated with increasing concentration of phenobarbital (see methods section). (A) H&E ‘maps’ of one example of the rat and human liver microtissue experiments; the microTMAs were ∼90 % efficient in that greater than 90 % of all the spheroids in the 96 well plate are contained on each section of the microTMA; (B) Enlarged (x25) H&E images of control and PB treated rat liver 3D microtissues and (C) Enlarged (x25) H&E images of control and PB treated human liver 3D microtissues. Histopathological analysis revealed some vacuolation of hepatocytes in both species in both control and PB treated liver spheroids, however since there was the expected phase1 and phase 2 enzyme induction and hepatocyte proliferation response to PB this effect was not considered significant.
Table 1.
Summary of bioinformatics, transcriptomics and proteomics data showing species differences (yellow highlights) between rat and human liver 3D microtissues at the level of UGT promoters, RNA and protein induction.
| 1. Gene Name* | 2. Species | 3. Upregulated by CAR/PXR Activator in Literature? | 4. Study Reference | 5. Promoter Characterisation Method | 6. Sequence contains CRE? | 7. CRE is AGGTCA or AGTTCA? | 8. RNA Fold Change at 48 h/PB3 | 9. RNF P Val at 48 h/PB3 | 10. RNF Q Val at 48 h/PB3 | 11. Protein Fold Change** | 12. Protein Change P Val** |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UGT1A1 | Human | Yes - Upregulated | [26,27] | Ensembl | Yes | Both | NF | NF | NF | ND | ND |
| Ugt1a1 | Rat | Yes - Upregulated | [22,28] | NCBI | No | NF | NF | NF | 1.2 | 0 | |
| Ugt1a2 | Rat | NA | Ensembl | Yes | AGGTCA | NF | NF | NF | 1.14 | 0.29 | |
| UGT1A4 | Human | Yes - Upregulated | [28] | EPD | Yes | AGTTCA | NF | NF | NF | 0.99 | 0.99 |
| UGT1A6 | Human | Yes - Upregulated | [28] | EPD | Yes | AGTTCA | 1.08 | 0.17 | 0.43 | 1.24 | 0.04 |
| Ugt1a6 | Rat | Yes - Upregulated | [7] | Ensembl | No | 7.33 | 0.00001 | 0.001 | ND | ND | |
| UGT1A8 | Human | NA | EPD | Yes | AGGTCA | 1.53 | 0.00001 | 0.0002 | ND | ND | |
| Ugt1a8 | Rat | Yes - Upregulated | [13] | Ensembl | No | 0.88 | 0.28 | 0.57 | ND | ND | |
| UGT1A9 | Human | Yes - Upregulated | [29] | EPD | Yes | AGGTCA | NF | NF | NF | ND | ND |
| Ugt1a9 | Rat | NA | NCBI | No | 1.25 | 0.03 | 0.15 | 1.37 | 0.2 | ||
| UGT1A10 | Human | NA | EPD | Yes | AGGTCA | NF | NF | NF | 1.43 | 0.01 | |
| UGT2A1 | Human | NA | EPD | Yes | AGTTCA | 0.98 | 0.65 | 0.85 | ND | ND | |
| Ugt2a1 | Rat | Yes - Upregulated | [22] | Ensembl | Yes | AGTTCA | 0.97 | 0.74 | 0.89 | ND | ND |
| UGT2A3 | Human | NA | EPD | Yes | AGTTCA | 1.05 | 0.37 | 0.65 | 1.22 | 0.07 | |
| Ugt2a3 | Rat | NA | EPD | Yes | Both | NF | NF | NF | ND | ND | |
| UGT2B4 | Human | NA | EPD | Yes | AGGTCA | 1.45 | 0.0001 | 0.002 | 1.61 | 0.01 | |
| Ugt2b4 | Rat | NA | EPD | Yes | AGGTCA | NF | NF | NF | 1.15 | 0.14 | |
| UGT2B7 | Human | NA | EPD | No | NF | NF | NF | 1.46 | 0.07 | ||
| Ugt2b7 | Rat | NA | Ensembl | Yes | AGTTCA | 1.09 | 0.30 | 0.59 | ND | ND | |
| UGT2B10 | Human | NA | EPD | Yes | AGGTCA | 0.55 | 0.0001 | 0.002 | ND | ND | |
| Ugt2b10 | Rat | NA | Ensembl | Yes | AGTTCA | 1.32 | 0.03 | 0.15 | 1.2 | 0.48 | |
| UGT2B11 | Human | NA | EPD | Yes | AGGTCA | 0.62 | 0.001 | 0.009 | ND | ND | |
| UGT2B17 | Human | NA | EPD | Yes | Both | NF | NF | NF | 1.13 | 0.47 | |
| Ugt2b17 | Rat | Yes - Upregulated | [22] | EPD | Yes | AGTTCA | 10.48 | 0.000001 | 0.001 | 1.97 | 0.002 |
| UGT2B15 | Human | NA | EPD | Yes | Both | 1.00 | 0.93 | 0.97 | 2.25 | 0.01 | |
| Ugt2b37 | Rat | NA | Ensembl | Yes | Both | 2.62 | 0.00003 | 0.002 | 1.23 | 0.16 |
*A bold gene name indicates this gene has a direct rat equivalent (homolog). **Proteomics data obtained from 96 h at PB2 (750 u M) dose.
EPD = Eukaryotic promoter database: https://epd.epfl.ch//index.php. NA = no data available; NF = probe not found on the microarray; ND = not detected.
3.2. UGT promoter analysis
Promoter sequences obtained for all rat and human UGT1 and 2 isoforms were characterized to assess the presence or absence of CREs, Table 1, Column 4. Within these sequences, CAR response elements (CREs) were found in all human UGT isoform promoters (with the exception of UGT2B7), whilst the rat promoter sequences showed a smaller proportion of isoforms containing CREs, Table 1 Column 5. A higher proportion of human UGT promoters were also found to contain multiple CREs compared to the rat promoters Table 1, Column 6.
3.3. Species differences in UGT RNA induction
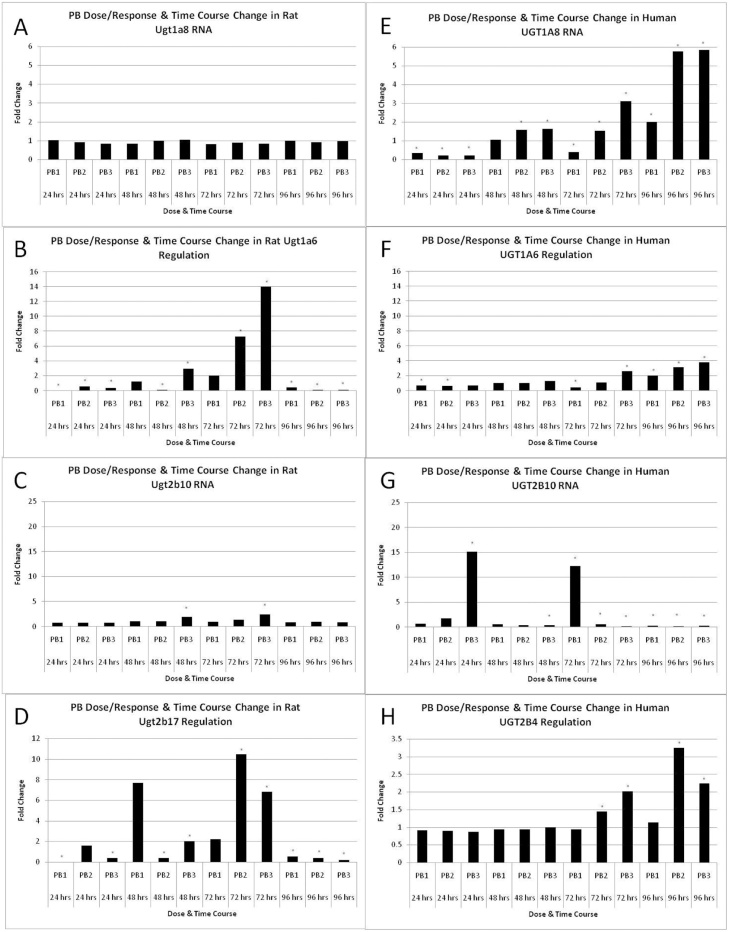
There was a marked species difference in PB treatment-mediated Ugt2b10, Ugt2b17, Ugt1a6 and Ugt2b37 RNA and/or protein induction between rat and human liver 3D microtissues both in terms of fold change and temporality, Table 1, Fig. 2. Ugt1a6, Ugt2b10 and Ugt2b17 RNA induction in rat liver 3D microtissues was dose-dependent at the 48 h/72 h time points, however the induction of these genes returned to approximately control levels by 96 h, Fig. 2. The magnitude of UGT1A6 induction in human liver 3D microtissues was lower than in rat, Fig. 2. UGT1A8, UGT2B4 and UGT2B10 were up-regulated relative to control in human liver 3D microtissues compared to rat, where they were either unchanged or only slightly induced, Fig. 2. Ugt2b7 and UGT2A3 RNA levels were unaltered relative to control by PB treatment, Table 1.
Fig. 2.
Effects of PB treatment over time on the induction (fold change) of UGT genes (RNA).
Histograms show fold change data (relative to control) derived from microarray analysis of control and PB-treated liver 3D microtissues. Results are mean fold change values (n = 2).
(A) rat Ugt1a8; (B) rat Ugt1a6; (C) rat Ugt2b10; (D) rat Ugt2b17; (E) human UGT1A8; (F) human UGT1A6; (G) human UGT2B10; (H) human UGT2B4. * fold change values significantly different from control (q<0.05). PB1 = phenobarbital 500 u M; PB2 = phenobarbital 750 u M; PB3 = phenobarbital 1000 u M.
3.4. Species differences in UGT protein induction
Ugt2b17 was upregulated at the protein level in rat but not in human. UGT1A10, UGT2B4, UGT1A6 and UGT2B15 were upregulated at the protein level in human but not in rat liver 3D microtissues, Table 1.
4. Discussion
The finding that Ugt1a6, Ugt2b17, Ugt2b37, UGT1A10, UGT2B4 and UGT2B15 (RNA and/or protein) were differentially induced by PB treatment in rats and humans suggests that UGT induction could contribute to species differences in thyroid carcinogenesis observed with this compound. Previous investigations have shown that rat hepatocytes produce higher basal levels of thyroxine-glucuronide metabolites than human hepatocytes and PCB153, a phenobarbital-like PCB, induces T4G metabolism (∼2 fold) in rat but not in human hepatocytes [9]. Hence historical data would suggest that rat hepatocytes are more inducible than human hepatocytes with regards to T4 glucuronidation.
Rat and human Ugt2b17/UGT2B17 promoters were both found to contain CREs. There were no probes for UGT2B17 on the human expression microarray so we were unable to determine whether or not this isoform was induced at the RNA level in human liver 3D microtissues. However there was no change in the level of this isoform at the protein level in human liver 3D microtissues indicating the species difference for the PB-mediated induction of this isoform was at the protein level. Ugt2b17 was also induced in rat liver by a series of conazole antifungal agents which are known CAR activators [22]. As UGT2B17 glucuronidates thyroxine, this difference could account in part, for differences in thyroxine clearance observed previously between rat and human hepatocytes [9] in response to exposure to CAR activators.
Ugt1a6 was also markedly induced by PB in rat liver 3D microtissues but only slightly induced in human liver 3D microtissues. The absence of rat Ugt1a6 protein induction following PB treatment suggests that RNA induction of this gene in rats is not translated to protein. UGT1A6 has been found to metabolise thyroxine to a glucuronide conjugate [9] so the present data suggests that species differences in the induction of this isoform could also contribute to differences in thyroxine metabolism caused by PB treatment.
Studies with recombinant UGT enzymes have shown that UGT2b17 has a higher activity for metabolising T4 than Ugt1A6 [23]. As the basal level of Ugt2b17 in rat liver is higher than other Ugt isoforms (Vassel and Klassen 2002), it is plausible that the effect of PB treatment on the levels of T4 metabolism caused by induction of this isoform could be more profound in rat compared to human hepatocytes. Hence the greater effects of PB and other CAR activators at the level of thyroid carcinogenesis could be partly explained by differential induction of Ugt2b17.
As the induction of human UGTs 2B4 and 2B15 was of a lower magnitude than observed for rat isoforms Ugt2b17 and 1a6, the human UGT changes would likely contribute less to species differences in T4 metabolism than the rat UGT changes.
Our previous investigations have shown that rat and human liver 3D microtissues respond to PB treatments at the level of phase 1 metabolic enzyme induction and hepatocyte proliferation in a similar way to liver in vivo and primary hepatocytes in vitro [15,24,25]. Hence as it appears that liver 3D microtissues recapitulate in vivo responses to CAR activators, it is feasible to consider using these in vitro models for screening/risk reduction in compound selection for early key events in terms of potential liver/thyroid carcinogenesis. As 3D cultures of primary hepatocytes retain responsiveness to enzyme inducers for longer than 2D hepatocytes, it is possible that liver 3D microtissues may provide a more efficient screening model that better represents the in vivo situation [14,25], and one that is more cost effective (InSphero online webinar: https://insphero.com/science/digital-media/webinars/#dili). Processing liver 3D microtissues within a microTMA offers the potential of running multiple assays from a single experiment. By analysing parallel sections it is possible to look at histological/phenotypic and genomic endpoints [19].
In conclusion, we have found that differential induction of rat and human UGT isoforms by the CAR activator PB could help to explain species differences in susceptibility to thyroid carcinogenesis in response to PB and other CAR activators. This approach offers the potential for cross-species risk assessment of such compounds. The use of microTMA-based assays can also be used to assess this pathway (AOP162 https://aopwiki.org/aops/162) and other adverse outcome pathways (AOPs) in a similar time frame, offering the potential for effective screening at early stages in the discovery process, reducing later stage attrition rates.
5. Author statement
S.P., J.W., E.M., R.C., and D.C., conceived and planned the experiments. S.P., S.W., B.B., and M.E. carried out the experiments. All authors discussed the results and contributed to the final manuscript.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We are grateful to Dr Cristina Vasquez Martin for technical assistance with the proteomics analysis and Ruth Plummer for editorial assistance on the manuscript.
Edited by Dr. A.M Tsatsaka
References
- 1.Martin M.T., Judson R.S., Reif D.M., Kavlock R.J., Dix D.J. Profiling chemicals based on chronic toxicity results from the U.S. EPA ToxRef Database. Environ. Health Perspect. 2009;117(3):392–399. doi: 10.1289/ehp.0800074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClain R.M. Thyroid gland neoplasia: non-genotoxic mechanisms. Toxicol. Lett. 1992:397–408. doi: 10.1016/0378-4274(92)90213-4. 64-65 Spec No. [DOI] [PubMed] [Google Scholar]
- 3.McClain R.M., Levin A.A., Posch R., Downing J.C. The effect of phenobarbital on the metabolism and excretion of thyroxine in rats. Toxicol. Appl. Pharmacol. 1989;99(2):216–228. doi: 10.1016/0041-008x(89)90004-5. [DOI] [PubMed] [Google Scholar]
- 4.Qatanani M., Zhang J Fau - Moore D.D., Moore D.D. Role of the constitutive androstane receptor in xenobiotic-induced thyroid hormone metabolism. Endocrinology. 2005;146(3):995–1002. doi: 10.1210/en.2004-1350. [DOI] [PubMed] [Google Scholar]
- 5.Vansell N.R., Klaassen C.D. Increase in rat liver UDP-glucuronosyltransferase mRNA by microsomal enzyme inducers that enhance thyroid hormone glucuronidation. Drug Metab. Dispos. 2002;30(3):240–246. doi: 10.1124/dmd.30.3.240. [DOI] [PubMed] [Google Scholar]
- 6.Frank C., Gonzalez M.M., Oinonen C., Dunlop T.W., Carlberg C. Characterization of DNA complexes formed by the nuclear receptor constitutive androstane receptor. J. Biol. Chem. 2003;278(44):43299–43310. doi: 10.1074/jbc.M305186200. [DOI] [PubMed] [Google Scholar]
- 7.Xie W., Yeuh Mei-Fei, Radominska-Pandya Anna, Saini Simrat P.S., Negishi Yoichi, Bottroff Bobbie Sue, Cabrera Geraldine Y., Tukey Robert H., Evans Ronald M. Control of steroid, heme, and carcinogen metabolism by nuclear pregnane X receptor and constitutive androstane receptor. Proc Natl Acad Sci U S A. 2003;100(7):4150–4155. doi: 10.1073/pnas.0438010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meek M.E., Bucher J.R., Cohen S.M., Dellarco V., Hill R.N., Lehman-McKeeman L.D., Longfellow D.G., Pastoor T., Seed J., Patton D.E. A framework for human relevance analysis of information on carcinogenic modes of action. Crit. Rev. Toxicol. 2003;33(6):591–653. doi: 10.1080/713608373. [DOI] [PubMed] [Google Scholar]
- 9.Richardson V.M., Ferguson Ss Fau - Sey Y.M., Sey Ym Fau - Devito M.J., Devito M.J. In vitro metabolism of thyroxine by rat and human hepatocytes. Xenobiotica. 2014;44(5):391–403. doi: 10.3109/00498254.2013.847990. [DOI] [PubMed] [Google Scholar]
- 10.King C., Tang W., Ngui J., Tephly T., Braun M. Characterization of rat and human UDP-glucuronosyltransferases responsible for the in vitro glucuronidation of diclofenac. Toxicol. Sci. 2001;61(1):49–53. doi: 10.1093/toxsci/61.1.49. [DOI] [PubMed] [Google Scholar]
- 11.Orzechowski A., Schrenk D., Bock-Hennig B.S., Bock K.W. Glucuronidation of carcinogenic arylamines and their N-hydroxy derivatives by rat and human phenol UDP-glucuronosyltransferase of the UGT1 gene complex. Carcinogenesis. 1994;15(8):1549–1553. doi: 10.1093/carcin/15.8.1549. [DOI] [PubMed] [Google Scholar]
- 12.Tong Z., Li H Fau - Goljer I., Goljer I Fau - McConnell O., McConnell O Fau - Chandrasekaran A., Chandrasekaran A. In vitro glucuronidation of thyroxine and triiodothyronine by liver microsomes and recombinant human UDP-glucuronosyltransferases. Drug Metab. Dispos. 2007;35(12):2203–2210. doi: 10.1124/dmd.107.016972. [DOI] [PubMed] [Google Scholar]
- 13.Shelby M.K., Klaassen C.D. Induction of rat UDP-glucuronosyltransferases in liver and duodenum by microsomal enzyme inducers that activate various transcriptional pathways. Drug Metab. Dispos. 2006;34(10):1772–1778. doi: 10.1124/dmd.106.010397. [DOI] [PubMed] [Google Scholar]
- 14.Messner S., Fredriksson L., Lauschke V., Roessger K., Escher c., Bober M., Kelm J., Ingelman-Sundberg M., Moritz W. Transcriptomic, proteomic, and functional long-term characterization of multicellular three-dimensional human liver microtissues. Appl. In Vitro Toxicol. 2018;4(1):1–12. doi: 10.1089/aivt.2017.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plummer S., Cassidy B., Wallace S., Ball G., Wright J., McInnes L., Currie R., Peffer R., Cowie D. Cross-species comparison of CAR-mediated procarcinogenic key events in a 3D liver microtissue model. Toxicol. Rep. 2019;6 doi: 10.1016/j.toxrep.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soldatow V., Peffer R.C., Trask O.J., Cowie D.E., Andersen M.E., LeCluyse E., Deisenroth C. Development of an in vitro high content imaging assay for quantitative assessment of CAR-dependent mouse, rat, and human primary hepatocyte proliferation. Toxicol. In Vitro. 2016;36:224–237. doi: 10.1016/j.tiv.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Yamada T., Kikumoto H., Lake B.G., Kawamura S. Lack of effect of metofluthrin and sodium phenobarbital on replicative DNA synthesis and Ki-67 mRNA expression in cultured human hepatocytes. Toxicology Research doi. 2015;4:1–15. [Google Scholar]
- 18.Elcombe C.R., Peffer R.C., Wolf D.C., Bailey J., Bars R., Bell D., Cattley R.C., Ferguson S.S., Geter D., Goetz A., Goodman J.I., Hester S., Jacobs A., Omiecinski C.J., Schoeny R., Xie W., Lake B.G. Mode of action and human relevance analysis for nuclear receptor-mediated liver toxicity: a case study with phenobarbital as a model constitutive androstane receptor (CAR) activator. Crit. Rev. Toxicol. 2014;44(1):64–82. doi: 10.3109/10408444.2013.835786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plummer S., Wallace S., Ball G., Lloyd R., Schiapparelli P., Quiñones-Hinojosa A., Hartung T., Pamies D. A Human iPSC-derived 3D platform using primary brain cancer cells to study drug development and personalized medicine. Sci. Rep. 2019;9(1):1407. doi: 10.1038/s41598-018-38130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koudritsky M., Domany E. Positional distribution of human transcription factor binding sites. Nucleic Acids Res. 2008;36(21):6795–6805. doi: 10.1093/nar/gkn752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosoda K., Kanno Y., Sato M., Inajima J., Inouye Y., Yanai K. Identification of CAR/RXR α hetero-dimer binding sites in the human genome by a modified yeast one-hybrid assay. Adv. Biol. Chem. 2015;05:83–97. doi: 10.4236/abc.2015.52008. [DOI] [Google Scholar]
- 22.Goetz A.K., Dix D.J. Mode of action for reproductive and hepatic toxicity inferred from a genomic study of triazole antifungals. Toxicol. Sci. 2009;110(2):449–462. doi: 10.1093/toxsci/kfp098. [DOI] [PubMed] [Google Scholar]
- 23.Kato Y., Ikushiro S.-i., Emi Y., Tamaki S., Suzuki H., Sakaki T., Yamada S., Degawa M. Hepatic UDP-Glucuronosyltransferases responsible for glucuronidation of thyroxine in humans. Drug Metab. Dispos. 2008;36(1):51. doi: 10.1124/dmd.107.018184. [DOI] [PubMed] [Google Scholar]
- 24.Peffer R.C., Moggs J.G., Pastoor T., Currie R.A., Wright J., Milburn G., Waechter F., Rusyn I. Mouse liver effects of cyproconazole, a triazole fungicide: role of the constitutive androstane receptor. Toxicol. Sci. 2007;99(1):315–325. doi: 10.1093/toxsci/kfm154. [DOI] [PubMed] [Google Scholar]
- 25.Plant N.J., Horley N.J., Dickins M., Hasmall S., Elcombe C.R., Bell D.R. The coordinate regulation of DNA synthesis and suppression of apoptosis is differentially regulated by the liver growth agents, phenobarbital and methylclofenapate. Carcinogenesis. 1998;19(9):1521–1527. doi: 10.1093/carcin/19.9.1521. [DOI] [PubMed] [Google Scholar]
- 26.Assenat E., Gerbal-Chaloin S., Larrey D., Saric J., Fabre J.M., Maurel P., Vilarem M.J., Pascussi J.M. Interleukin 1beta inhibits CAR-induced expression of hepatic genes involved in drug and bilirubin clearance. Hepatology. 2004;40(4):951–960. doi: 10.1002/hep.20387. [DOI] [PubMed] [Google Scholar]
- 27.Chen C., Staudinger J.L., Klassen C.D. Nuclear receptor, pregname X receptor, is required for induction of UDP-glucuronosyltranferases in mouse liver by pregnenolone-16 alpha-carbonitrile. Drug Metab Dispos. 2003;31(7):908–915. doi: 10.1124/dmd.31.7.908. [DOI] [PubMed] [Google Scholar]
- 28.Gardner-Stephen D., Heydel J.M., Goyal A., Lu Y., Xie W., Lindblom T., Mackenzie P., Radominska-Pandya A. Human PXR variants and their differential effects on the regulation of human UDP-glucuronosyltransferase gene expression. Drug Metab Dispos. 2004;32(3):340–347. doi: 10.1124/dmd.32.3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartley D.P., Dai X., Yabut J., Chu X., Cheng O., Zhang T., He Y.D., Roberts C., Ulrich R., Evers R., Evans D.C. Identification of potential pharmacological and toxicological targets differentiating structural analogs by a combination of transcriptional profiling and promoter analysis in LS-180 and Caco-2 adenocarcinoma cell lines. Pharmacogenet Genomics. 2006;16(8):579–599. doi: 10.1097/01.fpc.0000220561.59972.7a. [DOI] [PubMed] [Google Scholar]