Abstract
Natural products have had a major impact upon quality of life, with antibiotics as a classic example of having a transformative impact upon human health. In this contribution, we will highlight both historic and emerging methods of natural product bio-manufacturing. Traditional methods of natural product production relied upon native cellular host systems. In this context, pragmatic and effective methodologies were established to enable widespread access to natural products. In reviewing such strategies, we will also highlight the development of heterologous natural product biosynthesis, which relies instead on a surrogate host system theoretically capable of advanced production potential. In comparing native and heterologous systems, we will comment on the base organisms used for natural product biosynthesis and how the properties of such cellular hosts dictate scaled engineering practices to facilitate compound distribution. In concluding the article, we will examine novel efforts in production practices that entirely eliminate the constraints of cellular production hosts. That is, cell free production efforts will be introduced and reviewed for the purpose of complex natural product biosynthesis. Included in this final analysis will be research efforts made on our part to test the cell free biosynthesis of the complex polyketide antibiotic natural product erythromycin.
Keywords: Natural product, Bio-manufacturing, Native host, Heterologous host, Biosynthesis
1. Introduction
Natural products span a wide range of compounds that demonstrate similarly broad bioactivity [[1], [2], [3], [4], [5]]. Though generally small molecules, these compounds often derive from complex biosynthetic systems that feature fascinating enzymatic pathways [[6], [7], [8]]. It is the genetic and biochemical pathways responsible for natural products that then provide the basis for various production efforts. Resulting natural product classifications include polyketides, nonribosomal peptides, ribosomally synthesized and post-translationally modified peptides (RiPPs), and isoprenoids [9].
As the name implies, natural products derive from environmental cellular hosts that also span a variety of morphological features [[10], [11], [12], [13]]. Microbial sources are common, including bacterial and fungal species, which are traditional hosts for polyketide and peptide natural products [14]. However, plants are also prolific producers of natural products (especially isoprenoids) [15,16]. In conjunction with the underlying natural biosynthetic pathway, the native host system will also greatly influence the directions in scaled production efforts.
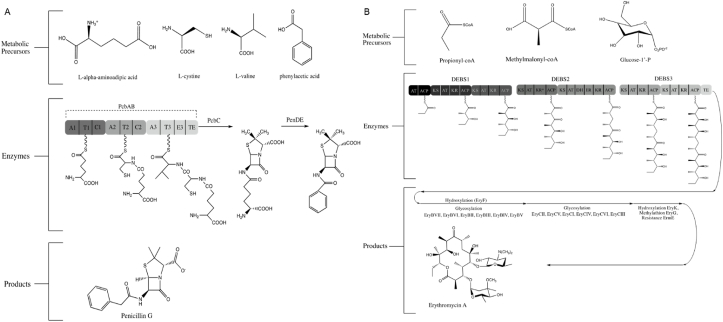
Fig. 1 depicts the biosynthetic production sequence for two historically important natural products. The first is penicillin, produced natively by Penicillium fungal species, which ushered in the widespread use and impact of antibiotics and is representative of a nonribosomal peptide natural product [[17], [18], [19]]. The second example provided is erythromycin, natively generated by the soil bacterium Saccharopolyspora erythaea, another antibiotic but derived from a more complex modular polyketide biosynthetic pathway [[20], [21], [22], [23], [24]]. Both pathways feature metabolite precursors required to support the enzymatic conversion to the final natural product. The enzymes help to distinguish the classification of the natural product and, as indicated in Fig. 1, highlight the complexity of the biosynthetic process. The penicillin case presents a relatively simple process requiring three enzymes in the construction of a beta-lactam peptide product, with one of these enzymes featured as a nonribosomal peptide synthetase. The erythromycin example highlights a more complex biosynthetic process that can be divided between core polyketide synthase enzymes (here, a multi-domain and high molecular weight complex) and tailoring enzymes responsible for adornment of the polyketide intermediate to functionalize and activate the final antibiotic product. Though only representative examples, the penicillin and erythromycin cases demonstrate key features of natural product biosynthesis, such as the need for key metabolic precursors and the coordinated activity of multiple enzymes either within or separate from their native production hosts.
Fig. 1.
Biosynthetic pathways for the natural product antibiotics penicillin (a) and erythromycin (b). Highlighted are the multi-domain biosynthetic enzymes for nonribosomal peptide (penicillin) and polyketide (erythromycin) biosynthesis. A: adenylation; T: thiolation; C: condensation; E: epimerization; TE; thioesterase; DEBS: Deoxyerythronolide B Synthase; AT: acyl transferase; ACP: acyl carrier protein; KS: β-keto-acyl synthase; KR: β-keto reductase; KR*: non-functional KR; DH: dehydratase; ER: enoyl reductase.
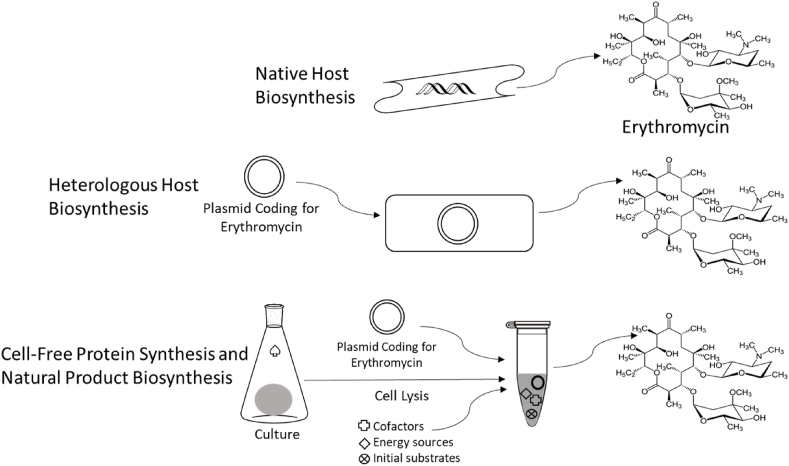
The penicillin and erythromycin examples also serve as key case studies in natural product bio-manufacturing, as they will straddle the approaches and methodologies outlined in the rest of this article. Production efforts are driven by the societally valuable properties of natural products and the desire for more ready access to such products. Scaled production attempts also parallel the timeline of advances in molecular biology. For example, key advances in the discovery and development of penicillin occurred prior to the completely elucidated structure of DNA, with such circumstances prompting production efforts focused on the native system. Alternatively, efforts in the heterologous production of erythromycin depended on advances in molecular biology to enable reconstitution in a surrogate host system capable of providing advantages relative to the native host [[25], [26], [27]]. More recently, elements of both molecular biology and microbiology are considered in efforts to establish biosynthesis in a cell-free format, completely unconstrained from traditional cellular confines. Fig. 2 provides a schematic representation of the native, heterologous, and cell free production systems to be covered, with accompanying commentary connecting production efforts to evolving technical advances. Finally, the chapter also offers a tribute to the central figure of this particular special issue, Dr. Arnold Demain, who made invaluable contributions to established and ongoing efforts in natural product bio-production [[28], [29], [30], [31], [32], [33]].
Fig. 2.
Natural product production options. Featured is the natural product polyketide antibiotic erythromycin, produced natively from Saccharopolyspora erythraea. Heterologous production of erythromycin has been completed using Streptomyces spp. and Escherichia coli as surrogate hosts; whereas, cell-free biosynthesis removes the need for a dedicated host (except for systems that rely on expression machinery provided via cellular lysis, as depicted).
2. Natural product production from native hosts
As introduced, natural products derive from environmental locations [13,34]. Generally, soil and aquatic environs have been fruitful sources of natural products [10,12,[35], [36], [37], [38], [39], [40]]. Within these environs, commonly identified production hosts include plants and microorganisms with natural products possessing biological functions spanning chemical communication to a weaponized form of competition for resources [[41], [42], [43]]. In the latter functionality, compounds like anti-fungals or anti-bacterials would be produced as a form of competitive advantage in warding off microbial rivals for environmental nutrients. Regardless, the bioactivity of natural products served as a key motivator in subsequent efforts to scale production and distribution.
Once a bioactive natural product was identified from an environmental cellular host, a key challenge was to then sufficiently access the active compound [31,32,44]. One option was to generate the same compound using a chemical synthetic approach [45]. The primary obstacle to this approach was the chemical complexity of natural products (featuring numerous functionalities and chiral centers). Thus, though impressive synthetic efforts resulted in natural product generation, doing so in an economic and scalable fashion remained challenging.
As a result, scaled production options then turned to the native hosts responsible for the bioactive natural product [[46], [47], [48]]. Conceptually, this approach resembled the long history of Eastern herbal therapies reliant on nature. The key difference in the cases highlighted in the middle portion of the 20th century (with penicillin as the prime example) was the focus on industrial scale [18,19,49]. From this standpoint, the influence of the Industrial Revolution was available to aid production efforts. Also coinciding with such efforts were the World Wars that painfully emphasized the damage infectious disease inflicted under such circumstances, thus, propelling efforts to overproduce penicillin for battlefield hospitals.
Native sources, again highlighted via herbal medicines, include many plant species [[50], [51], [52]]. However, these particular hosts are less amenable to scaled production relative to single-cell microorganisms (as often observed for fungi and bacteria), and discovery efforts underway were successfully isolating the microbial species (largely from soil samples) demonstrating bioactive natural product formation [35,53]. Thus, bio-production or bio-manufacturing evolved as a combination of the industrialization of microbial growth and natural product production [54].
Such efforts are best illustrated in the case of penicillin, which has been covered extensively in previous reviews [18,19,48]. Importantly, the template for scaled penicillin production was generally adopted in efforts to isolate, identify, and produce subsequent bioactive natural products in their so-called Golden Age (ca. 1940–70) [47,[55], [56], [57]]. During this period, it is important to emphasize the nascent development of the field that would be called molecular biology. Again citing the penicillin example, a fully scaled bio-manufacturing process was established prior to the complete structural elucidation of DNA and, thus, prior to the molecular biology tools available to current engineering efforts [58]. Table 1 outlines efforts to bio-manufacture several historically-relevant (based upon health and economic impact) natural products from native production hosts. In several cases, key production milestones (processes and production levels) were reached prior to the establishment of molecular biology in the 1970s. Industrial microbiology is a field that best describes efforts in this time period. Early in the process, there was a need to identify environmental bioactivity, the responsible microbial source, and microbiological details about the source. In tandem, efforts commenced to both isolate and culture the identified microbe for advanced study, including the need to confirm natural product production and to elucidate the chemical structure of the natural product. A key scientific element missing from such efforts was the genetic and biochemical analysis of the biosynthetic process, mainly due to emerging understandings of such intracellular workings. As such, efforts to scale production of confirmed bioactive natural products from isolated microbes relied upon both physical and biological techniques available at the time.
Table 1.
Natural product production from native microbial sources.
| Natural Product (Activity) | Native Host | Enhanced Production Methods | Production Metrics | References |
|---|---|---|---|---|
| Penicillin (antibiotic) | Penicillium notatum |
|
Titer: 5.5 g/L Productivity: 34 mg/L/h (160 h) |
[[59], [60], [61]] |
| Vancomycin (antibiotic) | Streptomyces orientalis |
|
|
[[62], [63], [64]] |
| Erythromycin (antibiotic) | Saccharopolyspora erythraea |
|
|
[[65], [66], [67]] |
| Lovastatin (cholesterol lowering) | Aspergillus terreus |
|
|
[[68], [69], [70]] |
| Cyclosporin A (immunosuppressive) | Tolypocladium inflatum |
|
|
[71] |
| Rapamycin (immunosuppressive) | Streptomyces hygroscopicus |
|
|
[72,73] |
| Actinomycin D (anticancer) | Streptomyces antibioticus |
|
|
[74,75] |
| Doxorubicin (anticancer) | Streptomyces peucetius |
|
|
[76] |
| Artemisinin (antimalarial) | Artemisia annua |
|
|
[77,78] |
| Tetracycline (antibiotic) | Streptomyces aureofaciens |
|
|
[79,80] |
| Tobramycin (antibiotic) | Streptomyces tenebrarius |
|
|
[81,82] |
| Daptomycin (antibiotic) | Streptomyces roseosporus |
|
|
[[83], [84], [85]] |
From a physical perspective, optimization of culture parameters (medium components, process controls [temperature, aeration, pH, etc.]) offered a readily available means of improving production titers. Likewise, the refinement of culture conditions that embraced process control and unit operations (in particular, the bioreactor and its formats across batch, fed-batch, and continuous operations [[86], [87], [88]]) further contributed to improved and scalable production. Biological approaches leveraged current and emerging data about cellular biology. In particular, the concept of mutation and screening allowed improvements in titer by effectively treating the cellular system as a black box in which a perturbation was applied (in the way of a mutational event) followed by laborious screening of single cell mutants demonstrating enhanced production. Though less directed than genetic engineering alternatives that would emerge over the last 50 years, such approaches proved ultimately effective in establishing both scalable natural product production and an industry built around this approach.
3. Natural product production from heterologous hosts
Though native host production proved a viable method for access to a series of impactful natural products, the approach also had limitations. First, the process could only begin upon the successful isolation of a native natural host. The natural environments hold an immeasurable amount of biological content [42,89], and only a small fraction of natural product producing microbes have been isolated through conventional culturing methods [43], which was required for eventual scaled native host production. Because the same conventional methods of source organism culture were used repeatedly for natural product discovery, the issue of replication emerged, where researchers would continually isolate the same production hosts producing the same compounds. Once genotyping technology emerged [[90], [91], [92]], it became obvious that there was a massive gap between those microbes that exist in nature and those that could be successfully isolated and cultured using traditional microbiology [42,89]. Thus, access to a potentially massive repository of novel natural products was being neglected.
In addition, those organisms that were successfully isolated and culturable often possessed less than ideal innate biological properties, such as growth rate, cellular morphology, and the potential for precise genetic manipulation. From a practical standpoint, such innate properties complicated production efforts due to the challenges in culturing the native hosts, long time scales, and the potential for contamination. Moreover, even as genomic analysis emerged and a better understand of the genetic and biochemical workings of these microbes became clear, the lack of molecular engineering tools limited any directed efforts to improve cellular level production metrics.
These issues associated with native production hosts were in sharp contrast to workhorse microbes gaining traction in recombinant DNA practices that emerged in the 1970s. In contrast, these organisms had rapid growth rates (compared to most native natural product producers), single cell morphologies that supported scaled culture, and rapidly emerging genetic manipulation protocols. Example microbes included Escherichia coli and Saccharomyces cerevisiae. In addition, those working more closely within the natural products community were developing similar tools in hosts such as Streptomyces spp., noting that this bacterial group possessed native metabolism able to support complex natural product biosynthesis [53,[93], [94], [95], [96]]. Doing so enabled the first efforts at heterologous natural product biosynthesis, in which the genetic content responsible for a natural product was transferred from the original host to a host like Streptomyces coelicolor [97,98]. In so doing, the heterologous host enables the application of molecular biology manipulation of both the heterologous host and the implanted natural product pathway [[99], [100], [101]].
Successful heterologous natural product biosynthesis in Streptomyces spp. helped to validate the approach and led to a wave of impactful studies that deftly combined the new recombinant tools for these hosts with the unique biosynthetic pathways of complex natural products. A primary result was the directed production of unique compounds [102,103]. This outcome spurred an ongoing movement towards the application and refinement of heterologous natural product biosynthesis.
However, there was a key remaining drawback to the Streptomyces spp. used for heterologous natural product biosynthesis. Namely, they still retained nonideal growth properties relative to an alternative bacterial option such as E. coli. Unlike Streptomyces hosts, however, E. coli did not possess the native metabolism capable of supporting complex natural product biosynthesis. Hence, effort was required to both prep the cellular background in anticipation of complex natural product biosynthesis and modify established molecular biology tools, such as expression plasmids, to account for the size and number of heterologous genes needed to be transferred [104,105]. After overcoming a series of such technical challenges, E. coli was demonstrated capable of supporting complex natural product biosynthesis, with the erythromycin compound successfully produced from this host [106]. Table 2 highlights other successful examples of heterologous biosynthesis applied in the context of natural product production. Of note, the options of hosts have expanded across the bacterial and fungal species with a general heuristic being that the heterologous host match the type of native host (e.g., transfer from a eukaryotic native host to a eukaryotic heterologous host).
Table 2.
Natural product production from heterologous microbial sources.
| Natural Product Class | Representative Examples (Activity) | Native Host | Heterologous Host | Enhanced Production Methods | Production Metrics | References |
|---|---|---|---|---|---|---|
| Polyketide | ||||||
| Type I | Salinomycin (anticancer) | Streptomyces albus | Streptomyces coelicolor |
|
|
• [107] |
| Lovastatin (cholesterol lowering) | Aspergillus terreus | Pichia pastoris |
|
|
• [108] | |
| Type II | Chlorotetracycline (antibiotic) | Streptomyces aureofaciens | Streptomyces rimosus |
|
|
• [109] |
| Mithramycin A (anticancer) | Streptomyces argillaceus | Streptomyces lividans |
|
|
• [110] | |
| Nonribosomal peptide | Epothilone (anticancer) | Sorangium cellulosum | Schlegelella brevitalea |
|
|
• [111] |
| Daptomycin (antibiotic) | Streptomyces roseosporus | Streptomyces coelicolor |
|
|
• [112] | |
| Isoprenoid | Dihydroxy artemisinic acid (precursor to artemisin- antimalarial) | Artemisia annua | Saccharomyces cerevisiae |
|
|
• [113] |
| Naringenin (anti-inflammatory) | Arabidopsis thaliana/Rhodotorula glutinis | Yarrowia lipolytica |
|
|
• [114] | |
| Curcumin (anti-allergy and anticancer) | Arabidopsis thaliana | Escherichia coli |
|
|
[115] | |
| Guaia-6,10 (14)-diene (Precursor to Englerin A-anticancer) | Phyllanthus engleri | Saccharomyces cerevisiae |
|
|
[116] | |
| RiPP | Telomestatin (anticancer) | Streptomyces annulatus | Streptomyces avermitilis |
|
|
[117] |
Table 2 also highlights the evolution of the research fields of metabolic engineering and synthetic biology as they relate to heterologous natural product biosynthesis [[118], [119], [120]]. Technically, both fields exists separate from heterologous biosynthesis, but this particular approach offers an excellent means of implementing each of these fields. Before being formally conceptualized, the approaches used to design and transfer natural product pathways from native to heterologous hosts employed now common tenants of synthetic biology. All of the detail that must be considered in gene and pathway expression design were features of the heterologous biosynthetic process to later be merged with the explosion in tools and concepts associated with synthetic biology (itself closely tied to breakthroughs in gene sequencing and synthesis). Similarly, metabolic engineering emerged as a field dedicated to understanding and directing intracellular metabolism for applied purposes. This approach directly resonates with the cellular level optimization available to natural product heterologous biosynthetic efforts. Both of these fields require host systems with amenable genetic manipulation tools. Hence, the progression of heterologous biosynthesis through genetically-amenable systems like bacteria and fungi served to tighten the relationships to metabolic engineering and synthetic biology.
4. Cell free complex natural product biosynthesis
With the proficiency of sequencing technology developing over the last 20 years, there has been a concomitant emergence of natural product pathways (either confirmed or putative) [[121], [122], [123], [124], [125]]. This information has greatly impacted prospects for heterologous biosynthesis as key blueprint is now available to seed reconstitution efforts. Furthermore, with the continuing emergence of DNA synthesis technology, natural product genetic pathways designed for optimal expression within a target heterologous host is becoming a realistic option [25,126].
While the above technical developments support continued efforts at heterologous biosynthesis, there are other emerging paradigms for the rapid reconstitution of target natural product pathways. One such concept is cell free biosynthesis. In this approach, which stems from cell free protein synthesis [127], one would completely eliminate the cellular framework.
Instead, a more controlled, fabricated reaction would be assembled from the key components of the biosynthetic process. Genetic code would be input to such a system, together with the transcription and translation machinery required for protein formation. In this case, however, the resulting proteins would be expected to convert additional metabolic building blocks into a final natural product.
While heterologous biosynthesis leverages the positive traits of a host system, there is still extensive background metabolism that at best is divorced from the biosynthetic process. However, other times, there is unwanted cross-talk between native and heterologous metabolism that detracts from final heterologous biosynthetic success. There are also post-translational steps, including protein folding [106,128], that will be strongly influenced by the native cellular environment. Whereas, in a cell free setting, the environment could be more finely controlled towards such ends as improved protein solubility and activity. Finally, the heterologous process is also constrained by associated host properties such as growth rate and mutational potential, issues that can be eliminated in a cell free format.
A cell free system, however, would require a degree of complexity to mimic the gene expression process that happens intracellularly. Many ongoing commercial and academic approaches utilize cellular components or extracts from popular heterologous host options, most notably, E. coli. This of course dictates genetic pathway designs that are congruent with the transcription and translation mechanics associated with the host from which they have been isolated.
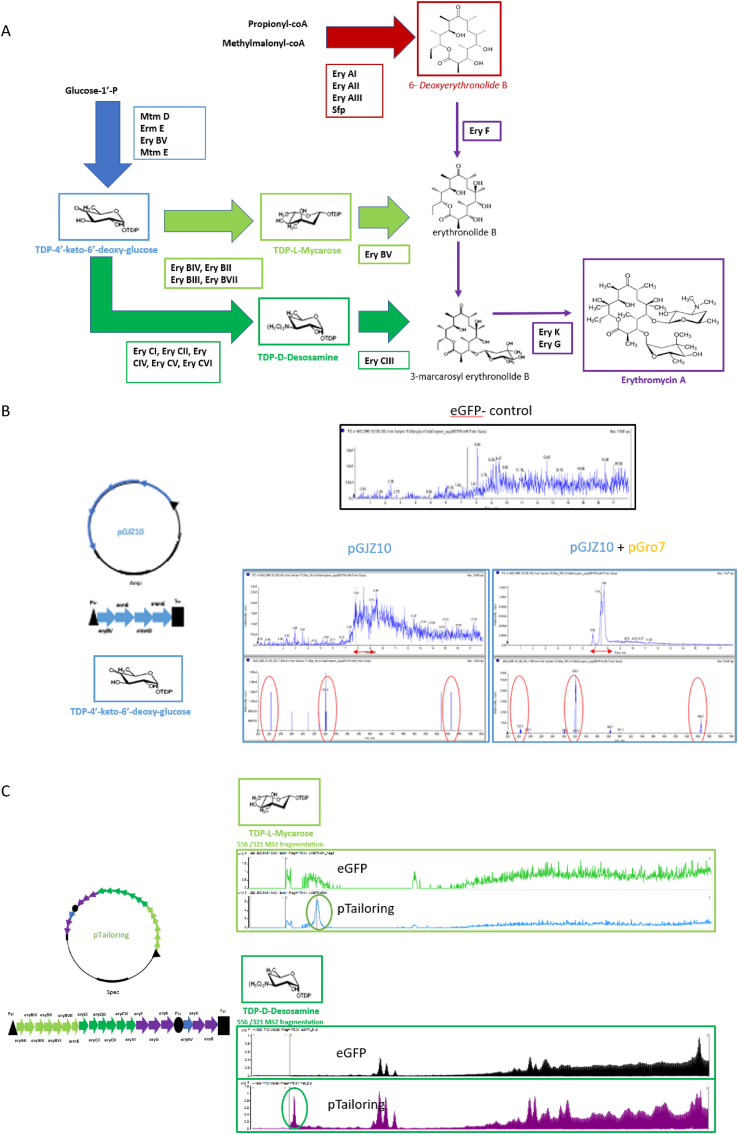
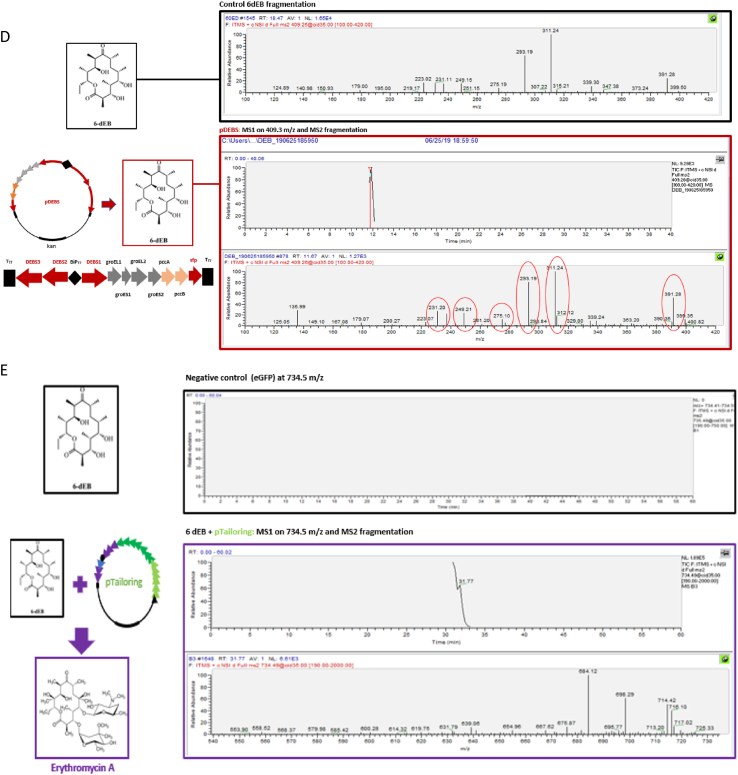
Table 3 highlights ongoing efforts to generate complex natural products using cell free platforms. Further, Fig. 3 highlights our group's efforts to generate the erythromycin product using a cell free approach. These examples and initial data show promise for the approach. Future considerations must include the ultimate end goal of cell free biosynthesis. Challenges will be encountered in efforts to scale cell free production efforts (just as they exist for production from native and heterologous hosts). However, the platform may better serve to support screening efforts of natural product discovery from newly identified sequence information as a way to confirm biosynthesis of a new compound.
Table 3.
Natural product production using cell free biosynthesis.
| Natural Product | Natural Product Class | Production Methods | Production Metrics | References |
|---|---|---|---|---|
| Diketopiperazine | NRPS | E. coli BL21 Star (DE3) extract | Titer: 12 mg/L Productivity: 0.6 mg/L/h (20 h) Yield: 50 mg/g phenylalanine |
[129] |
| Z-5-Bromo-3 (2-isocyanovinyl)-1H-indole | RiPP | New England Biolabs PURExpress system | Titer 6.6 g/L Productivity: 330 mg/L/h (20 h) |
[130] |
| Limonene | Terpene | E. coli BL21 Star (DE3) extract | Titer: 610 mg/L Productivity: 25.4 mg/L/h (24 h) Yield: 17 mg/g glucose |
[131] |
| Lanthipeptide (NisinZ) | RiPP | Multiple E. coli strains | Titer: 4–5 mg/L Productivity: 0.83 mg/L/h (6 h s) |
[132] |
Fig. 3.
Cell free biosynthesis for erythromycin A. The erythromycin A biosynthetic pathway (a) color-coded for steps leading to complete compound formation. Cell free biosynthesis for TDP-4′-keto-6′-deoxy glucose using plasmids pGJZ10 (indicated [27]) and pGro7 (providing the GroEL/ES chaperonin genes), using a plasmid expressing eGFP as a negative control and compared to samples with pGJZ10 and pGJZ10+pGro7 as analyzed by LC-MS (b). Cell free production for TDP-l-mycarose and TDP-d-desosamine using pTailoring previously described [25] (c). Cell free biosynthesis of 6-deoxyerythronolide B (6dEB) using plasmids pDEBS [25] assessed by LC-MS (d). Combining the pTailoring plasmid with the 6dEB molecule indicated erythromycin A formation by LC-MS (e). Cell free extract was prepared as follows: 10 mL of overnight-cultured E. coli BL21 Star (DE3) was inoculated in 1 L of 2 × YTPG medium (10 g/L yeast extract, 16 g/L tryptone, 3 g/L K2HPO4, 7 g/L KH2PO4, 5 g/L NaCl, and 18 g/L glucose) and grown at 37 °C with 250 rpm shaking. The culture was induced with 0.5 mM IPTG at an OD600nm of 0.6–0.8, and the cells were grown until OD600nm 3.5. The cells were harvested by centrifuge (8000 rpm, 10 min) and washed thrice with S30 Buffer (10 mM Tris-acetate, 14 mM Mg(OAc)2, 60 mM KOAc, 5 mM DTT) and stored at −80 °C after flash-freeze using liquid nitrogen. The cells were resuspended in S30 Buffer (1.2 g/mL) and aliquoted to 1.5 mL. Aliquots were sonicated with a FB50 (Thermo Fisher) sonifier with a 3 mm microtip (10 s on/10 s off) 5 times. The extract was prepared by collecting supernatant after 12,000 rpm of centrifugation. Cell free natural product biosynthesis was performed by using eGFP plasmid DNA as a control and a reaction buffer with the following contents: 1.2 mM ATP; 0.85 mM each of GTP, UTP and CTP; 34 μg/mL folinic acid; 171 μg/mL T7 RNA polymerase; 2 mM each of the 20 translatable amino acids, 0.33 mM nicotinamide adenine dinucleotide (NAD), 0.26 mM coenzyme A (CoA), 33 mM PEP, 130 mM potassium glutamate, 10 mM ammonium glutamate, 12 mM magnesium glutamate, 1.5 mM spermidine, 1 mM putrescine, 57 mM HEPES, 4 mM sodium oxalate, and 1 μL of 20 mg/mL Sfp [133]. To the reaction buffer, 0.25 volume of cell extract (described above) and 20 μg/mL plasmid DNA were added to reach a final volume of 25 μL. The reaction was held for 20 h at 30 °C and the required natural product metabolic substrates were added: gluocse-1-phosphate (10 mM) and dTTP (10 mM) for tailoring sugar biosynthesis, and propionyl-CoA (1 mM) and methyl malonyl-CoA (6 mM) for 6dEB biosynthesis. After 20 h, the samples were moved to −20 °C prior to SDS-PAGE and LC-MS analysis. Samples were prepared for analysis by diluting three times in LC-MS grade methanol and centrifuging at 13,000 rpm for 15 min with supernatant used for analysis. Erythromycin and 6dEB samples were applied to a Thermo Scientific Orbitrap XL with a C-18 analytical column. TDP-deoxysugar samples (mycarose, desosamine) were measured by Agilent G6545A quadrupole-time-of-flight (Q-TOF) using an XBridge Shield RP18 3.5 μm, 3.0 mm × 150 mm column from Waters. All MS analyses were conducted in positive ion mode. A linear gradient of 80% buffer A (95% water/5% acetonitrile/0.1% formic acid) to 100% buffer B (5% water/95% acetonitrile/0.1% formic acid) was used at a flow rate of 10 μL/min for the LC.
5. Conclusions
The production methods for complex natural products outlined in this article span native production hosts to cell free platforms, each possessing various advantages and drawbacks, as summarized in Table 4. Over time, the transition from native natural product production hosts to heterologous host options to cell free systems has been spurred by technical advancements in parallel with ongoing engineering efforts to address production challenges. These same technical advances have produced stand-alone academic fields of metabolic engineering and synthetic biology, both widely utilized across the natural product production platforms outlined herein. Moving forward, technical advances will continue to influence the choice of natural product biosynthetic system, with researchers having the option of native host, heterologous host, or cell free systems. The ultimate choice will be dictated by the economical and sustainable discovery, production, and distribution of a target natural product for societal benefit.
Table 4.
Summary and comparison of natural product production options.
| Pros | Cons | ||
|---|---|---|---|
| Native Host Biosynthesis |
|
|
|
| Heterologous Host Biosynthesis | Prokaryotic |
|
|
| Eukaryotic |
|
|
|
| Cell-Free Protein Synthesis and Natural Product Biosynthesis |
|
|
|
Declaration of competing interest
The Authors Declare No Conflicts.
Acknowledgement
The authors recognize support from the University at Buffalo Blue Sky Initiative for funding related to natural product heterologous biosynthesis and the NIH (AI126367).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Baker D.D. The value of natural products to future pharmaceutical discovery. Nat Prod Rep. 2007;24(6):1225–1244. doi: 10.1039/b602241n. [DOI] [PubMed] [Google Scholar]
- 2.Cragg G.M., Newman D.J. Natural products: a continuing source of novel drug leads. Biochim Biophys Acta. 2013;1830(6):3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grushkin D. Natural products emergent. Nat Med. 2013;19(4):390–392. doi: 10.1038/nm0413-390. [DOI] [PubMed] [Google Scholar]
- 4.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair S.K., Jez J.M. Natural product biosynthesis: what's next? An introduction to the JBC Reviews Thematic Series. J Biol Chem. 2020;295(2):335–336. doi: 10.1074/jbc.REV119.011586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staunton J., Weissman K.J. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18(4):380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 7.Walsh C.T. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303(5665):1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 8.Weissman K.J., Leadlay P.F. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3(12):925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 9.Park D., Swayambhu G., Pfeifer B.A. Heterologous biosynthesis as a platform for producing new generation natural products. Curr Opin Biotechnol. 2020;66:123–130. doi: 10.1016/j.copbio.2020.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Konig G.M. Natural products from marine organisms and their associated microbes. Chembiochem. 2006;7(2):229–238. doi: 10.1002/cbic.200500087. [DOI] [PubMed] [Google Scholar]
- 11.Molinari G. Natural products in drug discovery: present status and perspectives. Adv Exp Med Biol. 2009;655:13–27. doi: 10.1007/978-1-4419-1132-2_2. [DOI] [PubMed] [Google Scholar]
- 12.Newman D.J., Cragg G.M. Marine natural products and related compounds in clinical and advanced preclinical trials. J Nat Prod. 2004;67(8):1216–1238. doi: 10.1021/np040031y. [DOI] [PubMed] [Google Scholar]
- 13.Rateb M.E. Diverse metabolic profiles of a Streptomyces strain isolated from a hyper-arid environment. J Nat Prod. 2011;74(9):1965–1971. doi: 10.1021/np200470u. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H. Methods and options for the heterologous production of complex natural products. Nat Prod Rep. 2011;28(1):125–151. doi: 10.1039/c0np00037j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCaskill D., Croteau R. Prospects for the bioengineering of isoprenoid biosynthesis. Adv Biochem Eng Biotechnol. 1997;55:107–146. doi: 10.1007/BFb0102064. [DOI] [PubMed] [Google Scholar]
- 16.Bergman M.E., Davis B., Phillips M.A. Medically useful plant terpenoids: biosynthesis, occurrence, and mechanism of action. Molecules. 2019;24(21) doi: 10.3390/molecules24213961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming A. Classics in infectious diseases: on the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae by Alexander Fleming, Reprinted from the British Journal of Experimental Pathology 10:226-236, 1929. Rev Infect Dis. 1980;2(1):129–139. [PubMed] [Google Scholar]
- 18.Demain A.L., Elander R.P. The beta-lactam antibiotics: past, present, and future. Antonie Leeuwenhoek. 1999;75(1–2):5–19. doi: 10.1023/a:1001738823146. [DOI] [PubMed] [Google Scholar]
- 19.Kardos N., Demain A.L. Penicillin: the medicine with the greatest impact on therapeutic outcomes. Appl Microbiol Biotechnol. 2011;92(4):677–687. doi: 10.1007/s00253-011-3587-6. [DOI] [PubMed] [Google Scholar]
- 20.Haight T.H., Finland M. The antibacterial action of erythromycin. Proc Soc Exp Biol Med. 1952;81(1):175–183. doi: 10.3181/00379727-81-19815. [DOI] [PubMed] [Google Scholar]
- 21.Cortes J. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature. 1990;348(6297):176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 22.Reeves A.R. Transcriptional organization of the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea. J Bacteriol. 1999;181(22):7098–7106. doi: 10.1128/jb.181.22.7098-7106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.W M. Production of erythromycin with Saccharopolyspora erythraea. In: JL B., editor. Microbial processes and products. Humana Press: Methods in Biotechnology; 2005. [Google Scholar]
- 24.Weber J.M., Wierman C.K., Hutchinson C.R. Genetic analysis of erythromycin production in Streptomyces erythreus. J Bacteriol. 1985;164(1):425–433. doi: 10.1128/jb.164.1.425-433.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang L. Heterologous erythromycin production across strain and plasmid construction. Biotechnol Prog. 2018;34(1):271–276. doi: 10.1002/btpr.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang L. Broadened glycosylation patterning of heterologously produced erythromycin. Biotechnol Bioeng. 2018;115(11):2771–2777. doi: 10.1002/bit.26735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang G. Tailoring pathway modularity in the biosynthesis of erythromycin analogs heterologously engineered in E. coli. Sci Adv. 2015;1(4) doi: 10.1126/sciadv.1500077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adrio J.L., Demain A.L. Genetic improvement of processes yielding microbial products. FEMS Microbiol Rev. 2006;30(2):187–214. doi: 10.1111/j.1574-6976.2005.00009.x. [DOI] [PubMed] [Google Scholar]
- 29.Adrio J.L., Demain A.L. Recombinant organisms for production of industrial products. Bioeng Bugs. 2010;1(2):116–131. doi: 10.4161/bbug.1.2.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demain A.L. Mutation and the production of secondary metabolites. Adv Appl Microbiol. 1973;16:177–202. doi: 10.1016/s0065-2164(08)70027-3. [DOI] [PubMed] [Google Scholar]
- 31.Demain A.L. From natural products discovery to commercialization: a success story. J Ind Microbiol Biotechnol. 2006;33(7):486–495. doi: 10.1007/s10295-005-0076-x. [DOI] [PubMed] [Google Scholar]
- 32.Demain A.L., Adrio J.L. Strain improvement for production of pharmaceuticals and other microbial metabolites by fermentation. Prog Drug Res. 2008;65(251):253–289. doi: 10.1007/978-3-7643-8117-2_7. [DOI] [PubMed] [Google Scholar]
- 33.Demain A.L., Vaishnav P. Involvement of nitrogen-containing compounds in beta-lactam biosynthesis and its control. Crit Rev Biotechnol. 2006;26(2):67–82. doi: 10.1080/07388550600671466. [DOI] [PubMed] [Google Scholar]
- 34.Kaeberlein T., Lewis K., Epstein S.S. Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science. 2002;296(5570):1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 35.Torsvik V., Goksoyr J., Daae F.L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56(3):782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torsvik V., Ovreas L. Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol. 2002;5(3):240–245. doi: 10.1016/s1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- 37.Zhou J., Bruns M.A., Tiedje J.M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62(2):316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russo P., Nastrucci C., Cesario A. From the sea to anticancer therapy. Curr Med Chem. 2011;18(23):3551–3562. doi: 10.2174/092986711796642652. [DOI] [PubMed] [Google Scholar]
- 39.Blunt J.W. Marine natural products. Nat Prod Rep. 2013;30(2):237–323. doi: 10.1039/c2np20112g. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L. Exploring novel bioactive compounds from marine microbes. Curr Opin Microbiol. 2005;8(3):276–281. doi: 10.1016/j.mib.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Clardy J., Fischbach M.A., Walsh C.T. New antibiotics from bacterial natural products. Nat Biotechnol. 2006;24(12):1541–1550. doi: 10.1038/nbt1266. [DOI] [PubMed] [Google Scholar]
- 42.Davies J., Ryan K.S. Introducing the parvome: bioactive compounds in the microbial world. ACS Chem Biol. 2012;7(2):252–259. doi: 10.1021/cb200337h. [DOI] [PubMed] [Google Scholar]
- 43.Miao V.D.J. vol. 10. Springer-Verlag; Berlin: 2009. (Metagenomics and antibiotic discovery from bacteria. Uncultivated organisms; microbiology monographs). [Google Scholar]
- 44.Zhuang K.H., Herrgard M.J. Multi-scale exploration of the technical, economic, and environmental dimensions of bio-based chemical production. Metab Eng. 2015;31:1–12. doi: 10.1016/j.ymben.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Nicolaou K.C. Total synthesis of taxol. Nature. 1994;367(6464):630–634. doi: 10.1038/367630a0. [DOI] [PubMed] [Google Scholar]
- 46.Baltz R.H., Hosted T.J. Molecular genetic methods for improving secondary-metabolite production in actinomycetes. Trends Biotechnol. 1996;14(7):245–250. doi: 10.1016/0167-7799(96)10034-2. [DOI] [PubMed] [Google Scholar]
- 47.Katz L., Baltz R.H. Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol. 2016;43(2–3):155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 48.Demain A.L., Sanchez S. Microbial drug discovery: 80 years of progress. J Antibiot (Tokyo) 2009;62(1):5–16. doi: 10.1038/ja.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kardos N., Demain A.L. Ernst Chain: a great man of science. Appl Microbiol Biotechnol. 2013;97(15):6613–6622. doi: 10.1007/s00253-013-5017-4. [DOI] [PubMed] [Google Scholar]
- 50.Kirby J., Keasling J.D. Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu Rev Plant Biol. 2009;60:335–355. doi: 10.1146/annurev.arplant.043008.091955. [DOI] [PubMed] [Google Scholar]
- 51.Song M.C. Microbial biosynthesis of medicinally important plant secondary metabolites. Nat Prod Rep. 2014;31(11):1497–1509. doi: 10.1039/c4np00057a. [DOI] [PubMed] [Google Scholar]
- 52.Sham T.T. A review on the traditional Chinese medicinal herbs and formulae with hypolipidemic effect. BioMed Res Int. 2014;2014:925302. doi: 10.1155/2014/925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hopwood D.A. Soil to genomics: the Streptomyces chromosome. Annu Rev Genet. 2006;40:1–23. doi: 10.1146/annurev.genet.40.110405.090639. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y.P., Sun J., Ma Y. Biomanufacturing: history and perspective. J Ind Microbiol Biotechnol. 2017;44(4–5):773–784. doi: 10.1007/s10295-016-1863-2. [DOI] [PubMed] [Google Scholar]
- 55.Hutchings M.I., Truman A.W., Wilkinson B. Antibiotics: past, present and future. Curr Opin Microbiol. 2019;51:72–80. doi: 10.1016/j.mib.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 56.Lewis K. New approaches to antimicrobial discovery. Biochem Pharmacol. 2017;134:87–98. doi: 10.1016/j.bcp.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 57.Rishton G.M. Natural products as a robust source of new drugs and drug leads: past successes and present day issues. Am J Cardiol. 2008;101(10A):43D–49D. doi: 10.1016/j.amjcard.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 58.Watson J.D., Crick F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 59.Shwartzman G. Enhanced production of penicillin in fluid medium containing cellophane. Science. 1944;100(2600):390–392. doi: 10.1126/science.100.2600.390-a. [DOI] [PubMed] [Google Scholar]
- 60.Pirt S.J., Righelato R.C. Effect of growth rate on the synthesis of penicillin by Penicillium chrysogenum in batch and chemostat cultures. Appl Microbiol. 1967;15(6):1284–1290. doi: 10.1128/am.15.6.1284-1290.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paul G.C., Thomas C.R. A structured model for hyphal differentiation and penicillin production using Penicillium chrysogenum. Biotechnol Bioeng. 1996;51(5):558–572. doi: 10.1002/(SICI)1097-0290(19960905)51:5<558::AID-BIT8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 62.Mertz F.P., Doolin L.E. The effect of inorganic phosphate on the biosynthesis of vancomycin. Can J Microbiol. 1973;19(2):263–270. doi: 10.1139/m73-040. [DOI] [PubMed] [Google Scholar]
- 63.McIntyre J.J., Bull A.T., Bunch A.W. Vancomycin production in batch and continuous culture. Biotechnol Bioeng. 1996;49(4):412–420. doi: 10.1002/(SICI)1097-0290(19960220)49:4<412::AID-BIT8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 64.McCormick M.H., McGuire J.M. Vancomycin and method for its preparation. United States, U.S. Patent No. 1962;3 067,099. [Google Scholar]
- 65.Cheruy A., Durand A. Optimization of erythromycin biosynthesis by controlling pH and temperature: theoretical aspects and practical application. Biotechnol Bioeng Symp. 1979;(9):303–320. [PubMed] [Google Scholar]
- 66.Minas W. Improved erythromycin production in a genetically engineered industrial strain of Saccharopolyspora erythraea. Biotechnol Prog. 1998;14(4):561–566. doi: 10.1021/bp980055t. [DOI] [PubMed] [Google Scholar]
- 67.Johnson L.R.E. Process for producing erythromycin. United States, U.S. Patent No. 1957;2 809,151. [Google Scholar]
- 68.Novak N., Gerdin S., Berovic M. Increased lovastatin formation by Aspergillus terreus using repeated fed-batch process. Biotechnol Lett. 1997;19(10):947–948. [Google Scholar]
- 69.Szakacs G., Morovjan G., Tengerdy R.P. Production of lovastatin by a wild strain of Aspergillus terreus. Biotechnol Lett. 1998;20(4):411–415. [Google Scholar]
- 70.Kumar M.S. Repeated fed-batch process for improving lovastatin production. Process Biochem. 2000;36(4):363–368. [Google Scholar]
- 71.Kim J.W. Process for preparing cyclosporin A. United States, U.S. Patent No. 1999;5 874,572. [Google Scholar]
- 72.Sehgal S.N., Blazekovic Teodora M., Vezina Claude. Rapamycin and process of preparation. United States, U.S. Patent No. 1975;3 929,992. [Google Scholar]
- 73.Lee M.S., Kojima I., Demain A.L. Effect of nitrogen source on biosynthesis of rapamycin by Streptomyces hygroscopicus. J Ind Microbiol Biotechnol. 1997;19(2):83–86. doi: 10.1038/sj.jim.2900434. [DOI] [PubMed] [Google Scholar]
- 74.Katz E., Goss W.A. Influence of amino-acids of actinomycin biosynthesis. Nature. 1958;182(4650):1668–1669. doi: 10.1038/1821668b0. [DOI] [PubMed] [Google Scholar]
- 75.Katz E. Influence of valine, isoleucine, and related compounds on actinomycin synthesis. J Biol Chem. 1960;235:1090–1094. [PubMed] [Google Scholar]
- 76.Arcamone F. Adriamycin, 14-hydroxydaunomycin, a new antitumor antibiotic from S. peucetius var. caesius. Biotechnol Bioeng. 1969;11(6):1101–1110. doi: 10.1002/bit.260110607. [DOI] [PubMed] [Google Scholar]
- 77.Sa G. Effects of ipt gene expression on the physiological and chemical characteristics of Artemisia annua L. Plant Sci. 2001;160(4):691–698. doi: 10.1016/s0168-9452(00)00453-2. [DOI] [PubMed] [Google Scholar]
- 78.Baldi A., Dixit V.K. Yield enhancement strategies for artemisinin production by suspension cultures of Artemisia annua. Bioresour Technol. 2008;99(11):4609–4614. doi: 10.1016/j.biortech.2007.06.061. [DOI] [PubMed] [Google Scholar]
- 79.Yang S.S., Yuan S.S. Oxytetracycline production byStreptomyces rimosus in solid state fermentation of sweet potato residue. World J Microbiol Biotechnol. 1990;6(3):236–244. doi: 10.1007/BF01201291. [DOI] [PubMed] [Google Scholar]
- 80.Minieri P.P., Firman M.C., Mistretta A.G., Abbey A., Bricker C.E., Rigler N.E., Sokol H. A new broad spectrum antibiotic product of the tetracycline group. Antibiot Annu. 1954;(1953):81–87. [Google Scholar]
- 81.Gladkikh E.G., Korobkova T.P., Motkova M.O. [Effect of culture parameters on tobramycin biosynthesis] Antibiotiki. 1983;28(12):889–893. [PubMed] [Google Scholar]
- 82.Hong W., Yan S. Engineering Streptomyces tenebrarius to synthesize single component of carbamoyl tobramycin. Lett Appl Microbiol. 2012;55(1):33–39. doi: 10.1111/j.1472-765X.2012.03254.x. [DOI] [PubMed] [Google Scholar]
- 83.Lu W. Kinetic analysis and modeling of daptomycin batch fermentation by Streptomyces roseosporus. Appl Biochem Biotechnol. 2011;163(4):453–462. doi: 10.1007/s12010-010-9053-6. [DOI] [PubMed] [Google Scholar]
- 84.Ng I.S. Daptomycin antibiotic production processes in fed-batch fermentation by Streptomyces roseosporus NRRL11379 with precursor effect and medium optimization. Bioproc Biosyst Eng. 2014;37(3):415–423. doi: 10.1007/s00449-013-1007-2. [DOI] [PubMed] [Google Scholar]
- 85.Huber F.M., Pieper R.L., Tietz A.J. The formation of daptomycin by supplying Decanoic acid to streptomyces-roseosporus cultures producing the antibiotic complex A21978c. J Biotechnol. 1988;7(4):283–292. [Google Scholar]
- 86.Gomes J., Menawat A.S. Fed-batch bioproduction of spectinomycin. Adv Biochem Eng Biotechnol. 1998;59:1–46. doi: 10.1007/BFb0102295. [DOI] [PubMed] [Google Scholar]
- 87.Korz D.J. Simple fed-batch technique for high cell density cultivation of Escherichia coli. J Biotechnol. 1995;39(1):59–65. doi: 10.1016/0168-1656(94)00143-z. [DOI] [PubMed] [Google Scholar]
- 88.Yamane T., Shimizu S. Fed-batch techniques in microbial processes. Adv Biochem Eng. 1984;30:147–194. [Google Scholar]
- 89.Davies J. How to discover new antibiotics: harvesting the parvome. Curr Opin Chem Biol. 2011;15(1):5–10. doi: 10.1016/j.cbpa.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 90.Liles M.R. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl Environ Microbiol. 2003;69(5):2684–2691. doi: 10.1128/AEM.69.5.2684-2691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morrow K.M. Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl Environ Microbiol. 2012;78(18):6438–6449. doi: 10.1128/AEM.01162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fierer N. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol. 2007;73(21):7059–7066. doi: 10.1128/AEM.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baltz R.H. Gene expression in recombinant Streptomyces. Bioprocess Technol (N Y) 1995;22:309–381. [PubMed] [Google Scholar]
- 94.Baltz R.H. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol. 2010;37(8):759–772. doi: 10.1007/s10295-010-0730-9. [DOI] [PubMed] [Google Scholar]
- 95.Borodina I., Krabben P., Nielsen J. Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res. 2005;15(6):820–829. doi: 10.1101/gr.3364705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hopwood D.A. Streptomyces genes: from waksman to sanger. J Ind Microbiol Biotechnol. 2003;30(8):468–471. doi: 10.1007/s10295-003-0031-7. [DOI] [PubMed] [Google Scholar]
- 97.Kao C.M., Katz L., Khosla C. Engineered biosynthesis of a complete macrolactone in a heterologous host. Science. 1994;265(5171):509–512. doi: 10.1126/science.8036492. [DOI] [PubMed] [Google Scholar]
- 98.Malpartida F. Cloning and expression in a heterologous host of the complete set of genes for biosynthesis of the Streptomyces coelicolor antibiotic undecylprodigiosin. Gene. 1990;93(1):91–99. doi: 10.1016/0378-1119(90)90141-d. [DOI] [PubMed] [Google Scholar]
- 99.Katz L., McDaniel R. Novel macrolides through genetic engineering. Med Res Rev. 1999;19(6):543–558. doi: 10.1002/(sici)1098-1128(199911)19:6<543::aid-med5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 100.McDaniel R. Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature. 1995;375(6532):549–554. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 101.Gokhale R.S. Dissecting and exploiting intermodular communication in polyketide synthases. Science. 1999;284(5413):482–485. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- 102.Carreras C. Saccharopolyspora erythraea-catalyzed bioconversion of 6-deoxyerythronolide B analogs for production of novel erythromycins. J Biotechnol. 2002;92(3):217–228. doi: 10.1016/s0168-1656(01)00372-8. [DOI] [PubMed] [Google Scholar]
- 103.McDaniel R. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel "unnatural" natural products. Proc Natl Acad Sci U S A. 1999;96(5):1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pfeifer B.A., Admiraal S.J., Gramajo H., Cane D.E., Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291(5509):1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 105.Pfeifer B.A. Biosynthesis of Yersiniabactin, a complex polyketide-nonribosomal peptide, using Escherichia coli as a heterologous host. Appl Environ Microbiol. 2003;69(11):6698–6702. doi: 10.1128/AEM.69.11.6698-6702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang H. Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host. Chem Biol. 2010;17(11):1232–1240. doi: 10.1016/j.chembiol.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 107.Luhavaya H. Site-specific modification of the anticancer and antituberculosis polyether salinomycin by biosynthetic engineering. Chembiochem. 2014;15(14):2081–2085. doi: 10.1002/cbic.201402300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu Y. Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol. Metab Eng. 2018;45:189–199. doi: 10.1016/j.ymben.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 109.Wang X. Heterologous production of chlortetracycline in an industrial grade Streptomyces rimosus host. Appl Microbiol Biotechnol. 2019;103(16):6645–6655. doi: 10.1007/s00253-019-09970-1. [DOI] [PubMed] [Google Scholar]
- 110.Novakova R. Increased heterologous production of the antitumoral polyketide mithramycin A by engineered Streptomyces lividans TK24 strains. Appl Microbiol Biotechnol. 2018;102(2):857–869. doi: 10.1007/s00253-017-8642-5. [DOI] [PubMed] [Google Scholar]
- 111.Yu Y. Reassembly of the biosynthetic gene cluster enables high epothilone yield in engineered schlegelella brevitalea. ACS Synth Biol. 2020;9(8):2009–2022. doi: 10.1021/acssynbio.0c00100. [DOI] [PubMed] [Google Scholar]
- 112.Choi S. Heterologous expression of daptomycin biosynthetic gene cluster via Streptomyces artificial chromosome vector system. J Microbiol Biotechnol. 2019;29(12):1931–1937. doi: 10.4014/jmb.1909.09022. [DOI] [PubMed] [Google Scholar]
- 113.Tsuruta H. High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PloS One. 2009;4(2):e4489. doi: 10.1371/journal.pone.0004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wei W. Metabolically engineering of Yarrowia lipolytica for the biosynthesis of naringenin from a mixture of glucose and xylose. Bioresour Technol. 2020;314:123726. doi: 10.1016/j.biortech.2020.123726. [DOI] [PubMed] [Google Scholar]
- 115.Rodrigues J.L., Gomes D., Rodrigues L.R. A combinatorial approach to optimize the production of curcuminoids from tyrosine in Escherichia coli. Front Bioeng Biotechnol. 2020;8:59. doi: 10.3389/fbioe.2020.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Siemon T. Semisynthesis of plant-derived englerin A enabled by microbe engineering of guaia-6,10(14)-diene as building block. J Am Chem Soc. 2020;142(6):2760–2765. doi: 10.1021/jacs.9b12940. [DOI] [PubMed] [Google Scholar]
- 117.Amagai K. Identification of a gene cluster for telomestatin biosynthesis and heterologous expression using a specific promoter in a clean host. Sci Rep. 2017;7(1):3382. doi: 10.1038/s41598-017-03308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boyle P.M., Silver P.A. Parts plus pipes: synthetic biology approaches to metabolic engineering. Metab Eng. 2012;14(3):223–232. doi: 10.1016/j.ymben.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mitchell W. Natural products from synthetic biology. Curr Opin Chem Biol. 2011;15(4):505–515. doi: 10.1016/j.cbpa.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 120.Yadav V.G. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab Eng. 2012;14(3):233–241. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Danhorn T., Young C.R., DeLong E.F. Comparison of large-insert, small-insert and pyrosequencing libraries for metagenomic analysis. ISME J. 2012;6(11):2056–2066. doi: 10.1038/ismej.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Erlich Y. DNA Sudoku--harnessing high-throughput sequencing for multiplexed specimen analysis. Genome Res. 2009;19(7):1243–1253. doi: 10.1101/gr.092957.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kamb A. Next-generation sequencing and its potential impact. Chem Res Toxicol. 2011;24(8):1163–1168. doi: 10.1021/tx200121m. [DOI] [PubMed] [Google Scholar]
- 124.Tettelin H., Feldblyum T. Bacterial genome sequencing. Methods Mol Biol. 2009;551:231–247. doi: 10.1007/978-1-60327-999-4_18. [DOI] [PubMed] [Google Scholar]
- 125.Udwary D.W. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci U S A. 2007;104(25):10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kodumal S.J. Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc Natl Acad Sci U S A. 2004;101(44):15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Swartz J. A PURE approach to constructive biology. Nat Biotechnol. 2001;19(8):732–733. doi: 10.1038/90773. [DOI] [PubMed] [Google Scholar]
- 128.Betancor L. Improved catalytic activity of a purified multienzyme from a modular polyketide synthase after coexpression with Streptomyces chaperonins in Escherichia coli. Chembiochem. 2008;9(18):2962–2966. doi: 10.1002/cbic.200800475. [DOI] [PubMed] [Google Scholar]
- 129.Goering A.W. In vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth Biol. 2017;6(1):39–44. doi: 10.1021/acssynbio.6b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khatri Y. Multicomponent microscale biosynthesis of unnatural cyanobacterial indole alkaloids. ACS Synth Biol. 2020;9(6):1349–1360. doi: 10.1021/acssynbio.0c00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dudley Q.M. In vitro prototyping of limonene biosynthesis using cell-free protein synthesis. Metab Eng. 2020;61:251–260. doi: 10.1016/j.ymben.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 132.Liu R. A cell-free platform based on nisin biosynthesis for discovering novel lanthipeptides and guiding their overproduction in vivo. Adv Sci. 2020;7(17):2001616. doi: 10.1002/advs.202001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Quadri L.E. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37(6):1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]