Abstract
COVID-19 raised tension both within China and internationally. Here, we used mathematical modeling to predict the trend of patient diagnosis outside China in future, with the aim of easing anxiety regarding the emergent situation. According to all diagnosis number from WHO website and combining with the transmission mode of infectious diseases, the mathematical model was fitted to predict future trend of outbreak. Daily diagnosis numbers from countries outside China were downloaded from WHO situation reports. The data used for this analysis were collected from January 21, 2020 and currently end at February 28, 2020. A simple regression model was developed based on these numbers, as follows: , where is the total diagnosed patient till the i-th day and t=1 at February 1, 2020. Based on this model, we estimate that there were approximately 34 undetected founder patients at the beginning of the spread of COVID-19 outside China. The global trend was approximately exponential, with an increase rate of 10-fold every 19 days. Through establishment of this model, we call for worldwide strong public health actions, with reference to the experiences learned from China and Singapore.
Keywords: disease management
Significance of this study.
What is already known about this subject?
A novel coronavirus was verified and identified as the seventh member of the enveloped RNA coronaviruses as the cause of the disease, which is referred to as COVID-19.
COVID-19 raised tension both within China and internationally.
Considering the complexity of the real-life situation, a simple model is expected to be more accurate for describing the spread of the virus.
What are the new findings?
A “log-plus” model has been established to predict the situation, which only requires daily number of total diagnoses outside China.
There have been about 34 unobserved founder patients of COVID-19 at the beginning of spread outside China.
The global trend is approximately exponential, at a rate of 10-fold every 19 days.
How might these results change the focus of research or clinical practice?
As a 10-fold increase in patient numbers of COVID-19 every 19 days has been estimated, we call for strong public health actions worldwide.
Introduction
In early December of 2019, pneumonia cases of unknown cause emerged in Wuhan, the capital of Hubei province, China.1 A novel coronavirus (now named SARS-CoV-2) was verified and identified as the seventh member of the enveloped RNA coronaviruses (subgenus, Sarbecovirus; subfamily, Orthocoronavirinae) using high-throughput sequencing2–4 as the cause of the disease, which is referred to as COVID-19. Based on the evidence from early transmission dynamics, human-to-human transmission in hospital and family settings had been accumulating5–7 and occurred among close contacts since the middle of December 2019.8 According to WHO statistics, the accumulated number of diagnosed patients in China on August 08, 2020 was 89,057.9
COVID-19 raised tension both within China and internationally. Since the first case of COVID-19 pneumonia was reported from Wuhan, COVID-19 was rapidly diagnosed in patients in other Chinese cities and in neighboring countries, including Thailand, South Korea, Japan, and even a few Western countries.10–12 On January 13, 2020, the Ministry of Public Health of Thailand reported the first imported case of laboratory-confirmed novel coronavirus (COVID-19).13 After that, surges in cases of COVID-19 in Italy, Japan, and Iran also heightened fears that the world is on the brink of a pandemic. Therefore, on February 28, the WHO increased the assessment of the risk of spread and impact of COVID-19 to very high at the global level. Approximately 19 187 943 reported cases and 716 075 deaths of COVID-2019 have been reported to date August 08, 2020.9 The USA, Brazil, and India are currently the three most affected countries.14
Recently, considerable research resource has been devoted to conducting detailed analysis of the spread of the COVID-19 epidemic.15 16 Several parallel studies have reported that the estimated reproductive number (R0) of COVID-19 is higher than that of SARS, based on different models.17–19 Considering the superspreaders (P), hospitalized (H), and fatality class (F), an ad hoc compartmental mathematical model of the COVID-19 has been established to describe the reality of the Wuhan outbreak and predict the daily number of the confirmed cases.20 Several studies used deep learning to forecast COVID-19 infections.21 22 The disease transmission model predicted the gravity of COVID-19 in Canada using the long short-term memory (LSTM) networks.23 Data-driven estimation methods like LSTM and curve fitting were also used to evaluate the number of COVID-19 cases in India for the next 30 days and the effect of preventive measures.24
Given the limited number of data points and the complexity of the real-life situation, a simple model is expected to be more accurate for describing the spread of the virus (see Discussion section). In this study, we propose a “log-plus” model to predict the situation, which only requires daily number of total diagnoses outside China. This model assumes that there were some unobserved founder patients at the beginning of viral spread outside China and subsequent exponential growth. Despite the simplicity of our model, it fits the data well (R2=0.991). This prediction has potential practical and socially applicable significance and provides evidence that can enhance public health interventions to avoid severe outbreaks.
Methods
Data
Daily numbers of COVID-19 diagnoses in countries outside China were downloaded from WHO situation reports (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports). The data used in this analysis start on January 21, 2020 and end at February 28, 2020.
Model
Data were first explored by plotting log-transformed daily case numbers. A linear trend was observed in more recent data, while the fit was relatively poor for earlier time points. The presence of some undetected founder patients at the early time points were considered. Based on exploratory analysis and mathematical intuition, we proposed the following model:
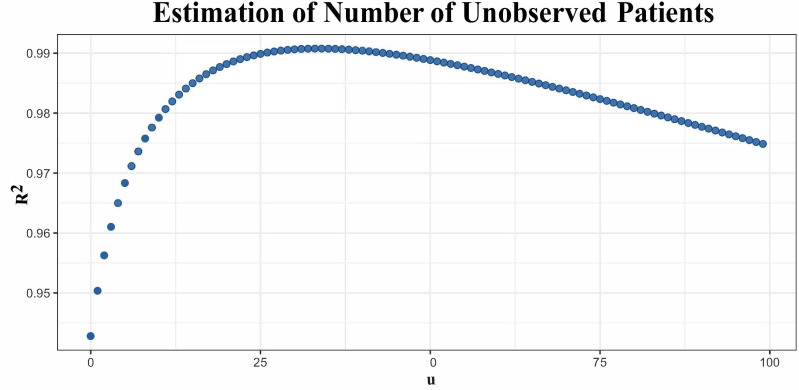
where Nt is the number of patients diagnosed outside China, according to WHO, on the t-th day, t=1 on February 1; u is the number of unobserved founder patients at the beginning of spread outside China; and a and b are simple linear regression parameters. We enumerated u from 0 to 100, with step size 1. For each u, we calculated Pearson’s correlation (R2) between t and , and selected the that maximized R2 and estimated corresponding and , using a simple linear regression between t and .
Availability of source code
The source code of the model is available at: https://github.com/wangyi-fudan/COVID-19_Global_Model
Results
Data table
The WHO daily count of numbers of diagnoses outside China and ‘log-plus’ transformed data, as well as model fit data, are presented in table 1.
Table 1.
WHO daily diagnosis number outside China
| Date | t | Nt | log10(Nt+34) | Model fit |
| January 21 | −10 | 4 | 1.580 | 1.559 |
| January 22 | −9 | 4 | 1.580 | 1.611 |
| January 23 | −8 | 7 | 1.613 | 1.662 |
| January 24 | −7 | 11 | 1.653 | 1.714 |
| January 25 | −6 | 23 | 1.756 | 1.765 |
| January 26 | −5 | 29 | 1.799 | 1.817 |
| January 27 | −4 | 37 | 1.851 | 1.868 |
| January 28 | −3 | 56 | 1.954 | 1.920 |
| January 29 | −2 | 68 | 2.009 | 1.971 |
| January 30 | −1 | 82 | 2.064 | 2.023 |
| January 31 | 0 | 106 | 2.146 | 2.075 |
| February 1 | 1 | 132 | 2.220 | 2.126 |
| February 2 | 2 | 146 | 2.255 | 2.178 |
| February 3 | 3 | 153 | 2.272 | 2.229 |
| February 4 | 4 | 159 | 2.286 | 2.281 |
| February 5 | 5 | 191 | 2.352 | 2.332 |
| February 6 | 6 | 216 | 2.398 | 2.384 |
| February 7 | 7 | 270 | 2.483 | 2.435 |
| February 8 | 8 | 288 | 2.508 | 2.487 |
| February 9 | 9 | 307 | 2.533 | 2.538 |
| February 10 | 10 | 319 | 2.548 | 2.590 |
| February 11 | 11 | 395 | 2.632 | 2.641 |
| February 12 | 12 | 441 | 2.677 | 2.693 |
| February 13 | 13 | 447 | 2.682 | 2.744 |
| February 14 | 14 | 505 | 2.732 | 2.796 |
| February 15 | 15 | 526 | 2.748 | 2.847 |
| February 16 | 16 | 683 | 2.856 | 2.899 |
| February 17 | 17 | 794 | 2.918 | 2.950 |
| February 18 | 18 | 804 | 2.923 | 3.002 |
| February 19 | 19 | 924 | 2.981 | 3.053 |
| February 20 | 20 | 1073 | 3.044 | 3.105 |
| February 21 | 21 | 1200 | 3.091 | 3.156 |
| February 22 | 22 | 1402 | 3.157 | 3.208 |
| February 23 | 23 | 1769 | 3.256 | 3.259 |
| February 24 | 24 | 2069 | 3.323 | 3.311 |
| February 25 | 25 | 2459 | 3.397 | 3.362 |
| February 26 | 26 | 2918 | 3.470 | 3.414 |
| February 27 | 27 | 3664 | 3.568 | 3.466 |
| February 28 | 28 | 4691 | 3.674 | 3.517 |
Parameter estimation
According to February 28 data, , , and were estimated as 34, 0.0515, and 2.075, respectively (figure 1).
Figure 1.
Estimation of u parameter by enumeration.
Global trend model
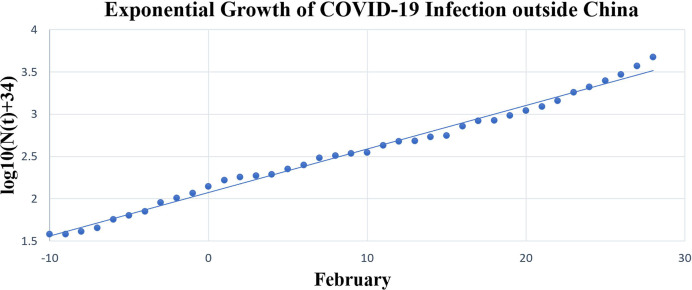
Next, we plotted against time to visualize model fitting (figure 2). The R2 value for the model was 0.991, indicating an excellent fit.
Figure 2.
Exponential growth of COVID-19 infection outside China.
Future number prediction
The number of COVID-19 diagnoses as of February 28 was 4691. Our model predicts that the number of diagnoses outside China will expand exponentially at a rate of 10-fold every 19 days in the absence of strong public health interventions.
Discussion
In this report, only the total number of diagnoses outside China was analyzed. Country-scale data are also available, but is less complete than the total numbers; hence, we limited our analysis to capture the global trend.
This model is a minimal extension of the “default” exponential growth model, using an estimate of 34 undetected founder patients outside China. An almost perfect model fit (R2=0.991) indicates that the spread of disease does follow our model.
A simple and straightforward linear model has some advantages: (1) it works for small sample sizes, due to limited observation or somewhat imperfect data; (2) it is relatively robust in complex situations, and the virus spreading pattern is complex and varies across the world, hence a simple model can provide coarse-grained trend estimation; and (3) a linear model easier to extrapolate than more complex models (eg, neural networks).
The existence of 34 undetected founder patients is not surprising. Actually, founder patients are those patients who are not reported at the beginning (January 22) of WHO reports. Thus, most of them are not under control and continually contribute to the pandemic. These individuals may have had mild symptoms and thus did not attend hospital; however, we do not preclude that they were already present before, or parallel with, the outbreak in Wuhan.
Based on this model, we estimate that there were approximately 34 undetected founder patients at the beginning of the spread of COVID-19 outside China. This suggests that the disease stably followed an approximate exponential growth model at the very beginning. This situation is dangerous, as we expect a 10-fold increase in patient numbers every 19 days, in the absence of strong intervention. We call for strong public health actions worldwide, referring to the experiences learned from China and Singapore.
The manuscript has been preprinted on the medRxiv (doi: https://doi.org/10.1101/2020.03.01.20029819). It is our pleasure that many researchers and social media care more about the outbreak trend outside China through our manuscript. The results of this article have been read more than 9000 times, picked by seven news outlets, and cited more than 10 times.25–29 We reproduced the disease’s initial spread to the world, which would impose a positive impact on other countries to pay attention to the development of COVID-19 and take powerful measures in time.
Acknowledgments
We thank the Fudan University High-End Computing Center for supporting computations involved in this study.
Footnotes
YL and ML contributed equally.
Contributors: YW conceived the idea and wrote the source code. YW, YL, ML, and LJ contributed to the data analysis, generating of tables and figures, and manuscript writing. YL, ML, XY, XL, MH, ZH, YW, and LJ contributed to the theoretical analysis and manuscript revision. All authors contributed to the final revision of the manuscript.
Funding: Data in this study are publicly available and were downloaded from the WHO Website. Our research was supported by the Postdoctoral Science Foundation of China (2018M640333) and Shanghai Municipal Science and Technology Major Project (2017SHZDZX01).
Competing interests: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as supplementary information. The source code of the model is available at https://github.com/wangyi-fudan/COVID-19_Global_Model.
References
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet 2020;395:565–74. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WHO WHO Director-General's remarks at the media briefing on 2019-nCoV on 11 February 2020, 2020. Available: https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020
- 4. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med Overseas Ed 2020;382:727–33. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan JF-W, Yuan S, Kok K-H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020;395:514–23. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Phan LT, Nguyen TV, Luong QC, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med 2020;382:872–4. 10.1056/NEJMc2001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. JT W, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modeling study. Lancet 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 2020;382:1199–207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. WHO Coronavirus disease (COVID-19) Situation Report – 201, 2020. Available: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200808-covid-19-sitrep-201.pdf?sfvrsn=121bb855_2
- 10. Kim H. Outbreak of novel coronavirus (COVID-19): what is the role of radiologists? 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shao N, Pan H, Li X, et al. CoVID-19 in Japan: what could happen in the future? MedRxiv 2020. [Google Scholar]
- 12. Ryu S, Ali ST, Lim J, et al. The estimate of infected individuals of the 2019-Novel coronavirus in South Korea by incoming international students from the countries of risk of 2019-Novel coronavirus: a simulation study. MedRxiv 2020. [Google Scholar]
- 13. WHO Coronavirus disease 2019 (COVID-19) situation report – 38. Available: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200227-sitrep-38-covid-19.pdf?sfvrsn=9f98940c_2
- 14. Ilie O-D, Cojocariu R-O, Ciobica A, et al. Forecasting the spreading of COVID-19 across nine countries from Europe, Asia, and the American continents using the ARIMA models. Microorganisms 2020;8:1158. 10.3390/microorganisms8081158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shao N, Cheng J, Chen W. The reproductive number R0 of COVID-19 based on estimate of a statistical time delay dynamical system. MedRxiv 2020. [Google Scholar]
- 16. Zhao Q, Chen Y, Small DS. Analysis of the epidemic growth of the early 2019-nCoV outbreak using internationally confirmed cases. MedRxiv 2020. [Google Scholar]
- 17. Xiong H, Yan H. Simulating the infected population and spread trend of 2019-nCov under different policy by EIR model. SSRN Journal 2020. 10.2139/ssrn.3537083 [DOI] [Google Scholar]
- 18. Mizumoto K, Kagaya K, Chowell G. Early epidemiological assessment of the transmission potential and virulence of 2019 novel coronavirus in Wuhan City: China, 2019-2020. MedRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao S, Lin Q, Ran J, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis 2020;92:214–7. 10.1016/j.ijid.2020.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ndaïrou F, Area I, Nieto JJ, et al. Mathematical modeling of COVID-19 transmission dynamics with a case study of Wuhan. Chaos Solitons Fractals 2020;135:109846. 10.1016/j.chaos.2020.109846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang W, Schotten HD. Deep learning for fading channel prediction. IEEE Open J Commun Soc 2020;1:320–32. 10.1109/OJCOMS.2020.2982513 [DOI] [Google Scholar]
- 22. Huang CJ, Chen YH, Ma Y, et al. Multiple-input deep convolutional neural network model for covid-19 forecasting in China. MedRxiv 2020. [Google Scholar]
- 23. Chimmula VKR, Zhang L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos Solitons Fractals 2020;135:109864. 10.1016/j.chaos.2020.109864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tomar A, Gupta N. Prediction for the spread of COVID-19 in India and effectiveness of preventive measures. Sci Total Environ 2020;728:138762. 10.1016/j.scitotenv.2020.138762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Business Insider The number of coronavirus cases outside China could jump tenfold every 19 days without ‘strong intervention,’ a study says, 2020. Available: https://www.businessinsider.nl/coronavirus-outside-china-tenfold-increase-19-days-strong-intervention-study-2020-3/
- 26. Yahoo News The number of coronavirus cases outside China could jump tenfold every 19 days without 'strong intervention,' a study says, 2020. Available: https://news.yahoo.com/number-coronavirus-cases-outside-china-171856791.html
- 27. Tsiotas D, Magafas L. The effect of Anti-COVID-19 policies on the evolution of the disease: a complex network analysis of the successful case of Greece. Physics 2020;2:325–39. 10.3390/physics2020017 [DOI] [Google Scholar]
- 28. Igor N, Ihor K, Gerhard D. Coronavirus pandemic dynamics in March, 2020. What can we expect in April? Available: https://www.researchgate.net/publication/340385105_Coronavirus_pandemic_dynamics_in_March_2020_What_can_we_expect_in_April
- 29. Rossman H, Keshet A, Shilo S, et al. A framework for identifying regional outbreak and spread of COVID-19 from one-minute population-wide surveys. Nature Medicine 2020:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]