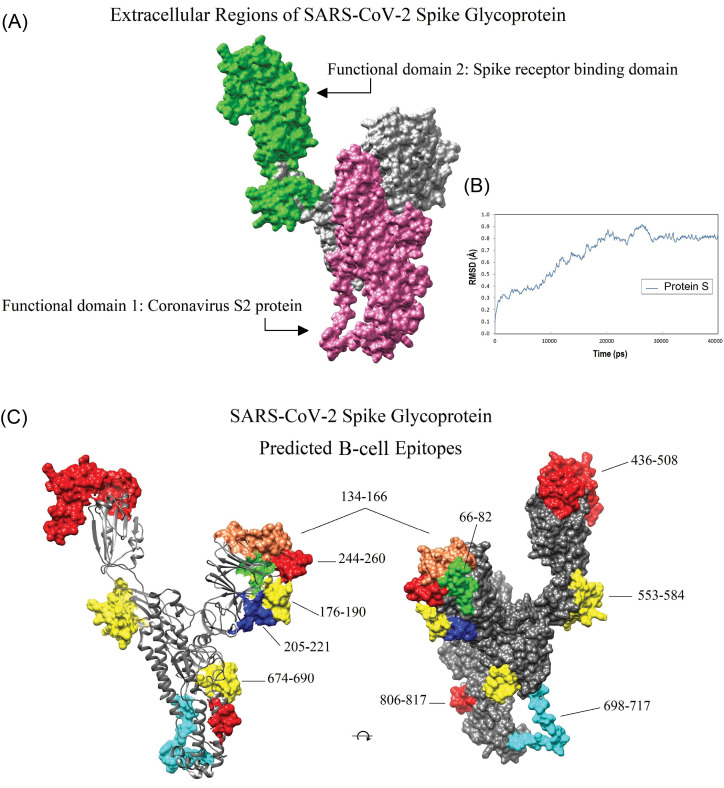
Fig. 4.
Location of the conserved domains and the immunodominant B-cell epitopes of SARS-CoV-2 spike glycoprotein on the homology modeled structure. A) The receptor-binding domain (green), and S2 subunit (pink) of SARS-CoV-2 S glycoprotein. B) The dynamic root-mean-square deviation (RMSD) graph corresponding to all atoms of the modeled spike glycoprotein shows that the simulation time (40000 ps) was long enough to achieve convergence (or stability) for the protein. C) The position of the ten dominant predicted B-cell epitopes. Owing to the lack of crystallized template protein, some residues in the beginning and end of the 3D model (1-27, and 1021-1273, respectively) are missed. The 3D structures were visualized by UCSF Chimera v1.14 software.72