Abstract
Nowadays, increasing extended-spectrum β-lactamase (ESBL)-producing bacteria have become a global concern because of inducing resistance toward most of the antimicrobial classes and making the treatment difficult. In order to achieve an appropriate treatment option, identification of the prevalent species which generate ESBL as well as their antibiotic susceptibility pattern is essential worldwide. Hence, this study aimed to investigate the prevalence of ESBL-producing bacteria and assess their drug susceptibility in Fardis Town, Iran. A total of 21,604 urine samples collected from patients suspected to have urinary tract infection (UTI) were processed in the current study. The antimicrobial susceptibility of the isolates was tested by the disk diffusion method. The ESBL producing bacteria were determined by Double Disc Synergy Test (DDST) procedure. Bacterial growth was detected in 1408 (6.52%) cases. The most common bacterial strains causing UTI were found E. coli (72.16%), followed by K. pneumoniae (10.3%) and S. agalactiae (5.7%). Overall, 398 (28.26%) were ESBL producer. The highest ESBL production was observed in E. coli, followed by Klebsiella species. ESBL producers revealed a higher level of antibiotic resistance compared with non-ESBLs. In conclusion, ESBL production in uropathogens was relatively high. Carbapenems and Aminoglycosides were confirmed as the most effective treatment options for these bacteria.
Subject terms: Microbiology, Diseases, Infectious diseases, Bacterial infection
Introduction
For decades, antibiotics have been used for the treatment of bacterial infections successfully; however, over the past few years, the abuse of antibiotics has led to the emergence of antimicrobial resistance around the world and has become a serious global threat to the public health1,2. Recently, it has been reported that approximately 700,000 people worldwide die annually from antimicrobial resistance (AMR) infections and it has been predicted that this number would reach 10 million by 20503.
At present, β-lactam drugs are a key factor in the treatment of bacterial infections worldwide and account for almost 65% of antibiotic usage4. They have been classified into six main groups based on the chemical structure of the β-lactam ring which includes Penicillins, Cephalosporins, Cephamycins, Carbapenems, Monobactams, and β-lactamase inhibitors. These drugs block cell wall synthesis by preventing accurate working of the Penicillin-binding protein (PBP), which has a principal role in the synthesis of the bacterial cell wall, and finally leads to cellular death. Nevertheless, it is unfortunate that, in recent years, resistance to this important class of antibiotics is also increasing globally5.
Resistance to b-lactams can occur through different mechanisms such as the generation of efflux pumps, changes in the production of outer membrane porins, alterations of PBPs, and the production of β-lactamase for inactivating antibiotics. Of these mechanisms, the production of B-lactamases is the most prevalent source of resistance to β-lactam antibiotics which are produced by both Gram-positive (extracellularly) and Gram-negative (in the Periplasmic space) bacteria. These enzymes are able to make the β-lactam antibiotics inactive by binding covalently to their carbonyl section and hydrolyzing the b-lactam ring thus enabling β-lactam resistance6,7.
To date, various β-lactamases have been reported to be generated by diverse microorganisms, including Penicillinases, Extended-spectrum β-lactamases (ESBLs), Cephalosporinases (AmpC), Metallo-β-lactamases (MBLs), and Carbapenemases (KPCs). Among these, ESBL-producing bacteria are very important and have attracted the attention of the scientific community8. ESBLs are β-lactamases enzymes with the capability to hydrolyze β-lactam antibiotics containing Penicillins, Aztreonam, as well as the first-, second-, third- and fourth-generation Cephalosporins, while sparing Cephamycins, Moxalactam, and Carbapenems. Further, they are inhibited by β-lactamase inhibitors, such as clavulanic acid, Tazobactam, and Sulbactam8–10. ESBLs producing organisms may also induce resistance to some of the none β-lactam antibiotics including Aminoglycosides, Quinolones, and Trimethoprimsulfamath-oxazoles11.
Today, the outbreak of infections caused by ESBL producing pathogens is becoming increasingly frequent and has become a world health threat12. The plasmid location of ESBL genes contributes to their spread through the horizontal gene transfer among the same and different species of bacteria13. The prevalence of ESBL-producing isolates depends on some factors including species, geographic region, hospital/ward, group of patients and type of infection, and extensive overuse of antibiotics14,15.
ESBLs are mostly produced by Gram-negative bacilli, especially Enterobacteriaceae family8. ESBL-producing Enterobacteriaceae cause a variety of hospital and community-acquired infections such as bloodstream, wound infections, respiratory tract, and urinary tract infections16. Urinary tract infections (UTIs) are very common infectious diseases that occur in a high proportion of the population and are a serious concern in the healthcare system17. At present, Carbapenems are selective drugs for the effective treatment of infections caused by ESBL-producing organisms. However, increasing Carbapenem resistance bacteria has also been associated with the use of Carbapenems18,19.
Due to expanding antibiotic resistance among bacteria and the high distribution of ESBL producing isolates, the recognition of the prevalent species that produce this enzyme as well as their antibiotic susceptibility pattern is necessary for each community to select the most effective treatment options. Thus, the aim of this study was to determine the prevalence of ESBL-producing bacteria and their antibiotic susceptibility pattern among uropathogens isolated from patients referring to the central laboratory of Fardis Town in Alborz province, Iran.
Materials and methods
Study population and samples processing
In the current descriptive cross-sectional study, 21,604 urine samples were aseptically collected from patients suspected to have UTI who were referred to Fardis Town laboratory located in Alborz province, Iran during one year (2018–2019). Positive bacterial growth was detected in urine samples of 1408 (6.52%) patients. The specimens were cultivated on Blood Agar and Eosin Methylene Blue Agar (EMB) medium (Merck, Germany) and incubated at 37 °C for 24 h. Initially, the colonies were counted. In cultures with bacterial counts of > 104 cfu/ml, the specimens were considered as positive, and gram-staining technique was performed. Then, bacterial genus and species were determined by standard biochemical tests.
Antimicrobial susceptibility testing
Antimicrobial susceptibility of the isolates was performed by the standard Kirby–Bauer disk diffusion method on the Mueller–Hinton agar media (Merck, Germany) using commercially available antibiotic disks (Mast, UK). The diameter of inhibition zone was measured for each antibiotic disk, and the results were defined in accordance with the CLSI guidelines20.
Phenotypic identification of ESBL-producing strains
Detection of ESBL-producing organisms was performed by Double Disc Synergy Test (DDST) method following CLSI recommendations. In this method, first, a suspension was prepared for each pure bacterial isolate according to the 0.5 McFarland turbidity standard and cultured on Mueller–Hinton agar. Fifteen minutes after bacterial cultures, pairs of antibiotic disks containing Ceftazidime (30 μg) with Ceftazidime/Clavulanic acid (30/10 μg), and Cefotaxime (30 μg) with Cefotaxime/Clavulanic acid (30/10 μg) were placed on Mueller–Hinton agar medium center to center, at a distance of 20 mm apart from each other. The plates were incubated for 24 h at 37 °C. Then, the diameter of inhibition zone was measured. According to CLSI guidelines, an increase of ≥ 5 mm in the zone diameter around the clavulanic acid combination disks versus the same disks alone confirmed the presence of ESBL producer strains20.
Ethical considerations
All ethical aspects of this research have been completely observed by the authors. It was approved by Research and Ethics committee of the Iran University of Medical Sciences. All experiments were performed in accordance with relevant guidelines and regulations in Iran. Informed consent was obtained from all participants or their legal guardians before the study. The patient's demographic characteristics were recorded in a questionnaire and their information remained confidential.
Data analysis
Data were analyzed using descriptive statistics (frequency and frequency percent, average and standard deviation), Chi square statistical test, and Fisher's exact test, hierarchical clustering analysis on SPSS software version 22. The confidence limits for statistical tests were considered as 0.95.
Results
Demographic information of the participants
Of a total of 21,604 participants surveyed in the present study, 15,408 (71.3%) were female and 6196 (28.7%) were male. The mean age of subjects was 23.21 ± 33.82 years and their age range was 3 months to 98 years (Table 1).
Table 1.
Test result age group.
| Result | Sex | Age group | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Age ≤ 1 | 2–5 | 6–15 | 16–30 | 31–45 | 46–60 | 61–75 | 75 < Age | |||
| No bacteria growth | 6012 | 13,837 | 1659 | 2047 | 1834 | 3257 | 4923 | 3228 | 2299 | 602 | 19,849 | |
| < 10,000 | 26 | 98 | 58 | 12 | 2 | 19 | 15 | 10 | 3 | 5 | 124 | |
| Candida albicans and non-albicans | 5 | 218 | 1 | 0 | 5 | 73 | 89 | 23 | 18 | 14 | 223 | |
| Positive culture | ||||||||||||
| Enterobacteriaceae | ||||||||||||
| Escherichia coli | 94 | 922 | 125 | 56 | 83 | 101 | 161 | 196 | 204 | 90 | 1016 | |
| Klebsiella pneumoniae | 15 | 129 | 25 | 3 | 2 | 23 | 32 | 26 | 21 | 12 | 144 | |
| Citrobacter diversus | 2 | 21 | 7 | 0 | 0 | 0 | 1 | 3 | 6 | 6 | 23 | |
| Klebsiella oxytoca | 3 | 10 | 2 | 1 | 0 | 3 | 0 | 1 | 3 | 3 | 13 | |
| Proteus mirabilis | 2 | 9 | 6 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 11 | |
| Proteus vulgaris | 1 | 6 | 0 | 4 | 0 | 0 | 2 | 1 | 0 | 0 | 7 | |
| Citrobacter freundii | 3 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 3 | 0 | 5 | |
| Enterobacter aerogenes | 0 | 4 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 4 | |
| Enterobacter agglomerans | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 3 | |
| Streptococcaceae | ||||||||||||
| Streptococcus agalactiae | 2 | 78 | 0 | 0 | 4 | 19 | 24 | 13 | 14 | 6 | 80 | |
| Enterococcus faecalis | 20 | 31 | 7 | 1 | 4 | 2 | 5 | 12 | 13 | 7 | 51 | |
| Non-Enterococcus bovis | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | |
| Staphylococcaceae | ||||||||||||
| Cn Staphylococci | 4 | 7 | 1 | 0 | 0 | 2 | 3 | 1 | 3 | 1 | 11 | |
| Staphylococcus aureus | 1 | 8 | 0 | 0 | 1 | 6 | 1 | 0 | 1 | 0 | 9 | |
| Staphylococcus saprophyticus | 1 | 6 | 0 | 0 | 0 | 3 | 3 | 0 | 0 | 1 | 7 | |
| Pseudomonadaceae | ||||||||||||
| Pseudomonas aeruginosa | 4 | 12 | 2 | 1 | 2 | 0 | 1 | 3 | 2 | 5 | 16 | |
| Other | 0 | 6 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 2 | 6 | |
| Total | 6196 | 15,408 | 1894 | 2127 | 1939 | 3514 | 5263 | 3518 | 2593 | 756 | 21,604 | |
The results of urine culture
Positive bacterial growth was detected in urine samples of 1408 (6.52%) patients. Among uropathogens, E. coli (1016 cases, 72.16%) was the most commonly isolated species, followed by K. pneumoniae (144 cases, 10.3%) and S. agalactiae (80 cases, 5.7%). In addition, fungal infection was found in 223 cases. Out of the 1408 positive bacterial cultures, 1255 (89.13%) cases were related to females. As indicated in Table 1, the patients mostly belonged to the age groups of 60–75 years old (283 cases, 20.11%) and 45–60 years old (254 cases, 18%). In both genders, the main infectious strain was E. coli, while the proportion of E. faecalis infection in males was far higher than in females (13.07–2.47%) (Table 1).
Minority bacteria
Out of the 1408 culture positive cases, the minimum numbers of isolates were related to, Proteus mirabilis (11 cases, 1.6%), Proteus vulgaris (7 cases, 0.5%), Citrobacter freundii (5 cases, 0.35%), Enterobacter aerogenes (4 cases, 0.28%), Enterobacter agglomerans (3 cases, 0.2%), Non-Enterococcus bovis (2 cases, 0.14%), Cn Staphylococci (11 cases, 1.6%), Staphylococcus aureus (9 cases, 0.64%), Staphylococcus saprophyticus strains (7 cases, 0.5%) (Table 1). There isn't any strain producing ESBL among these bacteria.
Antimicrobial susceptibility profile and ESBL production
E. coli
Table 2 indicates the overall antimicrobial susceptibility pattern of E. coli for antibiotics tested. It shows that the highest sensitivity was observed with Imipenem (99.2%), Amikacin (97.9%), Meropenem (97.2%), and Nitrofurantoin (92.8%), respectively. Further, the least sensitivity was to Piperacillin (17%) and Ampicillin (19.1%). Out of 1016 E. coli isolated, 359 (35.7%) were found to be ESBL producer. With the exception of Amikacin, Imipenem, Meropenem, and Nitrofurantoin, ESBL expression had a significant effect on E. coli resistance to other antibiotics tested (Table 2).
Table 2.
Antibiogram result of E. coli.
| Antibiotics | Antibiogram result | Statistical significance | ||||||
|---|---|---|---|---|---|---|---|---|
| Non-ESBL E. coli | ESBL E. coli | Total | Pearson chi square/Fisher's exact test | |||||
| Sensitive | Intermediate | Resistant | Sensitive | Intermediate | Resistant | |||
| Ciprofloxacin | 346 | 3 | 99 | 96 | 3 | 165 | 712 | P value = 0.00 |
| Norfloxacin | 146 | 1 | 41 | 27 | 0 | 72 | 287 | P value = 0.00 |
| Amikacin | 635 | 1 | 7 | 352 | 2 | 11 | 1008 | P value = 0.046 |
| Ceftazidime | 570 | 2 | 60 | 6 | 0 | 351 | 989 | P value = 0.00 |
| Cefotaxime | 485 | 5 | 35 | 7 | 1 | 301 | 834 | P value = 0.00 |
| Ceftriaxone | 88 | 0 | 22 | 2 | 0 | 53 | 165 | P value = 0.00 |
| Gentamycin | 591 | 5 | 45 | 228 | 3 | 131 | 1003 | P value = 0.00 |
| Imipenem | 582 | 0 | 4 | 332 | 0 | 3 | 921 | P value = 0.720 |
| Meropenem | 47 | 0 | 1 | 29 | 0 | 1 | 78 | P value = 0.734 |
| Nitrofurantoin | 610 | 15 | 24 | 330 | 12 | 22 | 1013 | P value = 0.138 |
| Cefalexin | 480 | 16 | 151 | 5 | 1 | 359 | 1012 | P value = 0.00 |
| Ampicillin | 191 | 3 | 449 | 2 | 0 | 364 | 1009 | P value = 0.00 |
| Nalidixic-acid | 333 | 14 | 301 | 56 | 7 | 302 | 1013 | P value = 0.00 |
| Co-Trimoxazole | 321 | 10 | 311 | 48 | 2 | 313 | 1005 | P value = 0.00 |
| Azithromycin | 2 | 0 | 0 | 0 | 0 | 1 | 3 | * |
| Colistin | 2 | 0 | 1 | 1 | 0 | 0 | 4 | |
| Doxycycline | 1 | 0 | 1 | 1 | 1 | 0 | 4 | |
| Chloramphenicol | 2 | 0 | 0 | 1 | 0 | 2 | 5 | |
| Piperacillin | 1 | 0 | 2 | 0 | 0 | 3 | 6 | |
| Tetracycline | 1 | 0 | 1 | 0 | 0 | 1 | 3 | |
| Tobramycin | 0 | 0 | 0 | 3 | 0 | 2 | 5 | |
*No significant statistical test was performed on these antibiotics, due to cases fewer than 6.
K. pneumoniae
According to Table 3, K. pneumoniae showed the highest rate of sensitivity towards Imipenem (99.3%), followed by Amikacin (95.8%), Meropenem (90%), and Gentamycin (90%), and the least sensitivity to tetracycline (0%), Tobramycin (0%), and Ampicillin (19.1%). From 144 K. pneumoniae isolates, 33 (22.9%) were confirmed as ESBL producer. The expression of ESBL resulted in developing resistance to the antibiotics Ceftazidime, Cefotaxime, Ceftriaxone, Gentamycin, Cefalexin, Nalidixic-acid, and Co-trimoxazole.
Table 3.
Antibiogram result of K. pneumoniae.
| Antibiotics | Antibiogram result | Statistical significance | ||||||
|---|---|---|---|---|---|---|---|---|
| Non-ESBL K. pneumoniae | ESBL K. pneumoniae | Total | Pearson chi square/Fisher's exact test | |||||
| Sensitive | Intermediate | Resistant | Sensitive | Intermediate | Resistant | |||
| Ciprofloxacin | 73 | 0 | 9 | 15 | 0 | 8 | 105 | P value = 0.011 |
| Norfloxacin | 24 | 1 | 4 | 5 | 0 | 5 | 39 | P value = 0.060 |
| Amikacin | 109 | 0 | 2 | 28 | 0 | 4 | 143 | P value = 0.023 |
| Ceftazidime | 100 | 0 | 8 | 1 | 0 | 32 | 141 | P value = 0.00 |
| Cefotaxime | 73 | 0 | 6 | 0 | 0 | 26 | 105 | P value = 0.00 |
| Ceftriaxone | 28 | 0 | 3 | 1 | 0 | 5 | 37 | P value = 0.001 |
| Gentamycin | 104 | 0 | 3 | 22 | 1 | 10 | 140 | P value = 0.00 |
| Imipenem | 103 | 0 | 0 | 30 | 0 | 1 | 134 | P value = 0.231 |
| Meropenem | 8 | 0 | 0 | 1 | 0 | 1 | 10 | P value = 0.200 |
| Nitrofurantoin | 50 | 39 | 22 | 17 | 6 | 10 | 144 | P value = 0.148 |
| Cefalexin | 94 | 1 | 16 | 1 | 0 | 32 | 144 | P value = 0.00 |
| Ampicillin | 5 | 0 | 105 | 0 | 0 | 32 | 142 | P value = 0.273 |
| Nalidixic-acid | 88 | 2 | 21 | 13 | 1 | 19 | 144 | P value = 0.00 |
| Co-Trimoxazole | 68 | 2 | 41 | 4 | 0 | 29 | 144 | P value = 0.00 |
| Colistin | 1 | 0 | 0 | 1 | 0 | 1 | 3 | * |
| Chloramphenicol | 0 | 0 | 1 | 1 | 0 | 0 | 2 | |
| Piperacillin | 1 | 0 | 0 | 1 | 0 | 1 | 3 | |
| Tetracycline | 0 | 0 | 1 | 0 | 0 | 1 | 2 | |
| Tobramycin | 0 | 0 | 0 | 0 | 0 | 2 | 2 | |
*Due to fewer than 5 cases, no statistical test was conducted for these antibiotics.
K. oxytoca
The information presented in Table 4 shows that all K. oxytoca isolates were reactive towards Imipenem with a sensitivity rate of 100% followed by Amikacin (92.3%). However, none of the isolates was sensitive to Ampicillin (0%). Of the 23 positive urine cultures of K. oxytoca, 5 (38.46%) were confirmed as ESBL positive. ESBL-producing isolates were highly resistant to Ceftazidime, Cefotaxime, Norfloxacin, Cefalexin, and Co-trimoxazole.
Table 4.
Antibiogram result of K. oxytoca.
| Antibiotics | Antibiogram result | Statistical significance | ||||||
|---|---|---|---|---|---|---|---|---|
| Non-ESBL K. oxytoca | ESBL K. oxytoca | Total | Pearson chi square/Fisher's exact test | |||||
| Sensitive | Intermediate | Resistant | Sensitive | Intermediate | Resistant | |||
| Norfloxacin | 6 | 1 | 1 | 1 | 0 | 4 | 13 | P value = 0.050 |
| Amikacin | 8 | 0 | 0 | 4 | 0 | 1 | 13 | P value = 0.385 |
| Ceftazidime | 8 | 0 | 0 | 0 | 0 | 5 | 13 | P value = 0.001 |
| Cefotaxime | 7 | 0 | 1 | 0 | 0 | 5 | 13 | P value = 0.005 |
| Gentamycin | 8 | 0 | 0 | 3 | 0 | 2 | 13 | P value = 0.128 |
| Imipenem | 8 | 0 | 5 | 0 | 0 | 13 | * | |
| Nitrofurantoin | 6 | 1 | 1 | 2 | 3 | 0 | 13 | P value = 0.174 |
| Cefalexin | 7 | 0 | 1 | 0 | 0 | 5 | 13 | P value = 0.005 |
| Ampicillin | 0 | 0 | 8 | 0 | 0 | 5 | 13 | * |
| Nalidixic-acid | 6 | 0 | 2 | 4 | 1 | 0 | 13 | P value = 0.025 |
| Co-Trimoxazole | 6 | 1 | 1 | 1 | 0 | 4 | 13 | P value = 0.050 |
*No statistical test was conducted for these antibiotics.
S. agalactiae
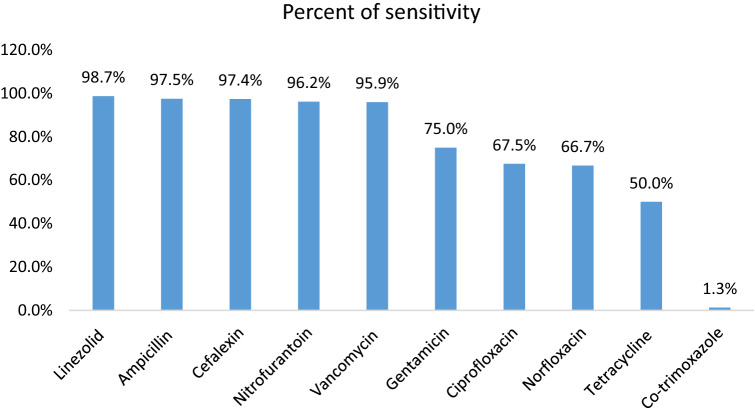
The antibiotic susceptibility pattern of S. agalactiae revealed that the majority of the isolates were sensitive to Linezolid (98.7%), Ampicillin (97.5%), Cefalexin (97.4%), and Nitrofurantoin (96.2%), and the least susceptibility was to Co-trimoxazole (1.3%) (Fig. 1).
Figure 1.
Antibiogram result of S. agalactiae.
E. faecalis
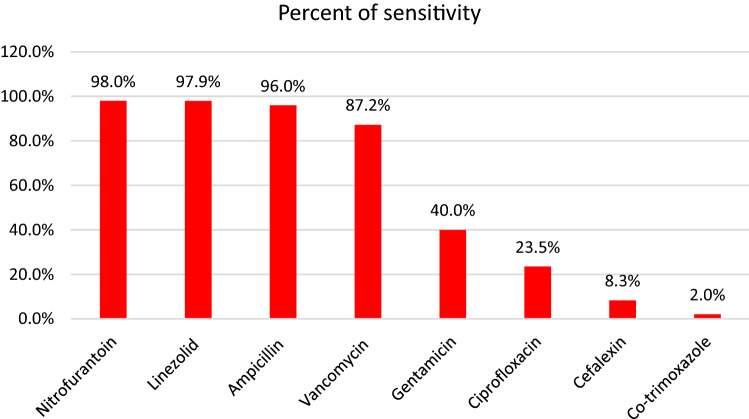
According to the results of antibiogram for E. faecalis, Nitrofurantoin (98%), Linezolid (97.9%), and Ampicillin (96%) revealed a superior sensibility for this bacterium. On the other hand, low level of susceptibility was observed in Co-trimoxazole (2%) and Cefalexin (8.3%) (Fig. 2).
Figure 2.
Antibiogram result of E. faecalis.
C. diversus
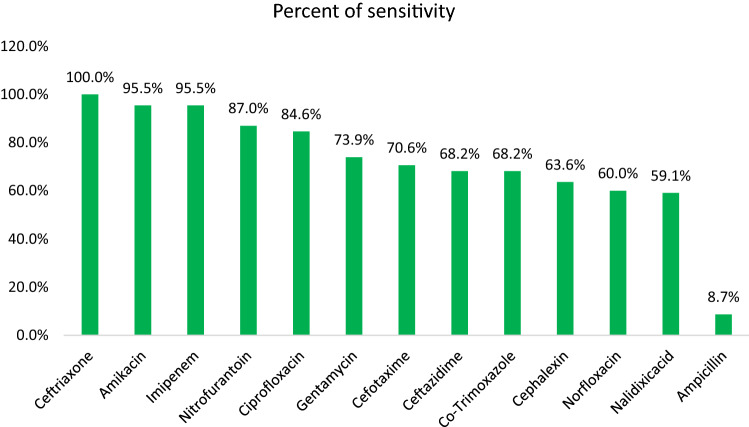
In the case of C. diversus, Ceftriaxone (100%), Amikacin (95.5%), and Imipenem (95.5%) were found to be most effective antibiotics. However, Ampicillin (8.7%) and Nalidixic-acid (59.1%) showed the least susceptibility rate (Fig. 3). Among 23 C. diversus isolates screened for ESBL, one of them was positive for ESBL production which was resistant to antibiotics tested, except of Amikacin and Imipenem.
Figure 3.
Antibiogram result of C. diversus.
P. aeruginosa
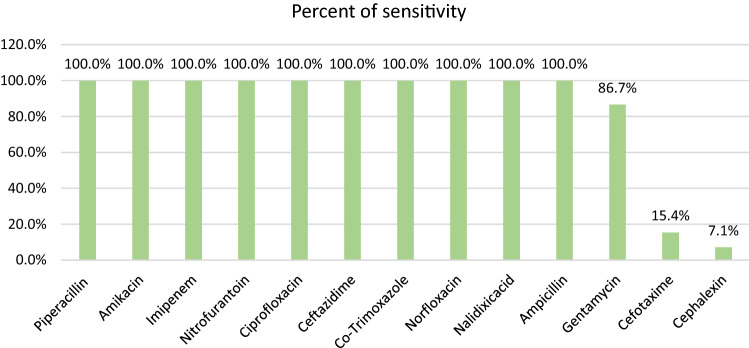
As depicted in Fig. 4, the least sensitivity rate for P. aeruginosa isolates was against Cefalexin (7.1%) and Cefotaxime (15.4%).
Figure 4.
Antibiogram result of P. aeruginosa.
Hierarchical clustering analysis
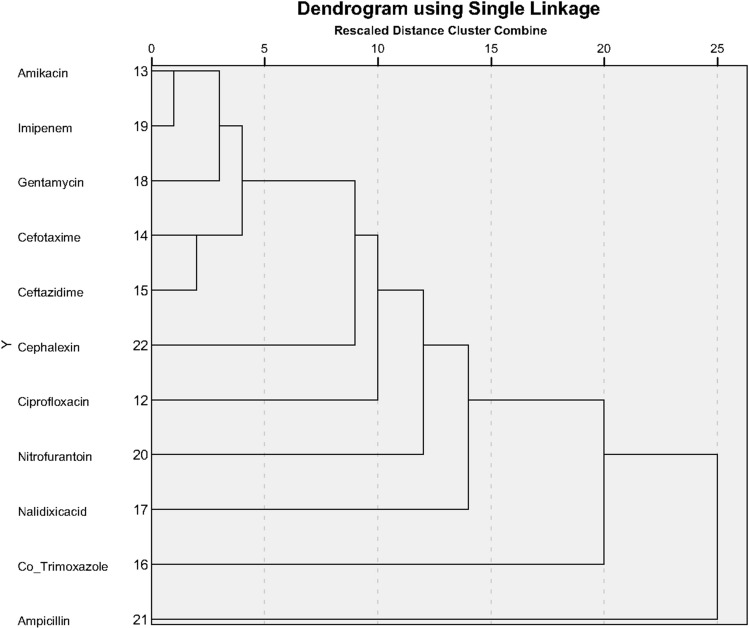
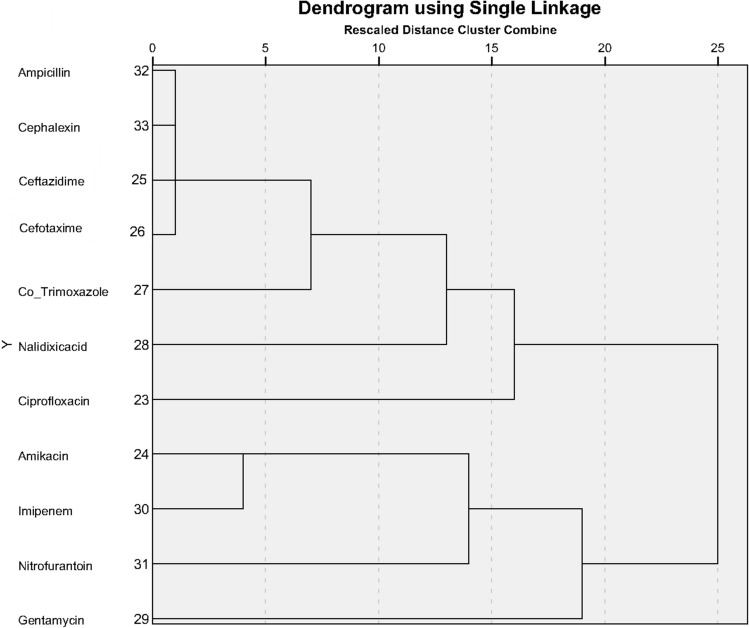
Using Hierarchical cluster analysis for pattern of antibiotic effect for ESBL-negative/positive bacterial strains, we plotted 11 commonly used antibiotics for three of the bacterial strains producing ESBL (E. coli, K. pneumoniae and K. oxytoca). The goal was to cluster antibiotics with similar efficacy. Clustering was done by the nearest neighbor method and Minkowski measure. Figures 5 and 6 (Dendrogram plots) show the clustering of antibiotics with similar effects for ESBL-negative and ESBL-positive bacterial strains.
Figure 5.
Clustering pattern of antibiotic effect for ESBL-negative bacterial strains.
Figure 6.
Clustering pattern of antibiotic effect for ESBL-positive bacterial strains.
As depicted in Fig. 5, the two antibiotics Amikacin and Imipenem were most similar in their effect on ESBL-negative bacterial strains.
As depicted in Fig. 6, the antibiotics Ampicillin, Cephalexin, Ceftazidime, and Cefotaxime had the most similarity in their effect on ESBL-positive bacterial strains.
Discussion
Urinary tract infection (UTI) is the second common infectious disease throughout the world caused by a wide range of microbial pathogens21. In the present work, from 21,604 suspected UTI patients, 1408 (6.52%) uropathogenic bacteria containing different species of Gram-negative and Gram-positive bacteria were isolated. The majority of the isolates were obtained from females 1255 (89.13%). This finding is supported by other studies reporting a higher rate of UTI prevalence in female patients compared to males14,22. These studies suggest that females are more at risk of developing infection by uropathogens which is due to their anatomical structure23. In terms of age, it was found that the most frequently uropathogens were related to the age groups of 60–75 (20.11%) and 45–60 (18%) years old. These outcomes agree with previous studies in which the incidence of UTIs was higher among elderly patients24,25.
The members of Enterobacteriaceae family especially E. coli and Klebsiella spp. are identified as vital causative agents of UTIs as they possess a number of factors including adhesion, pilli, fimbrae, and P1 blood group genotype receptor, which contribute to the attachment of bacteria to the urothelium26. In this regard, here the predominant urinary isolates were E. coli 1016 (72.16%) followed by K. pneumoniae 144 (10.3%). These results are in line with the earlier studies conducted by other researchers27–29. In addition, we found S. agalactiae 80 (5.7%) as the most frequently isolated Gram-positive bacterium involved in UTI.
At present, increasing Enterobacteriaceae producing ESBLs is a global healthcare concern because of high antibiotic resistance and the restricted treatment options30. In this survey, ESBL production was found in 28.26% (398/1408). In line with this result, some researchers reported the rate of ESBL production as 29% and 30.23%31,32. In contrast, an earlier study revealed that 11.75% of uropathogens were ESBL positive33 which is lower than our result. Elsewhere, the researcher reported 55.4% for ESBL production rate which is larger than ours34. These differences may be due to geographical area, time, and the diagnostic technique used32.
Among the ESBL producers, the highest rate was observed in E. coli (35.7%), followed by Klebsiella spp. (22.7%) and C. diversus (4.34%). This is in accordance with other studies in which among various ESBL isolates, E. coli species was the most prominent isolates followed by Klebsiella spp. On the other hand, the minimum ESBL isolates were related to Citrobacter spp.35–37. In contrast, several studies reported the highest ESBL production among K. pneumoniae followed by E. coli which do not match our results38–40, since we found E. coli as the predominant ESBL producer.
The antibiotic susceptibility testing by commonly prescribed antibiotics was accomplished for the most frequent pathogens found in our study. In determining the antibiotic susceptibility patterns of E. coli, a high level of sensitivity to Imipenem (99.2%), Amikacin (97.9%), Meropenem (97.2%), and Nitrofurantoin (92.8%) was observed, while the least sensitivity was related to Piperacillin (17%) and Ampicillin (19.1%). Most studies on the antibiotic susceptibility of urinary pathogens around the world have found similar results. For example, in Ahmed et al.’s study, E. coli showed high resistance to Ampicillin and Piperacillin, and low resistance rates against Meropenem, Amikacin, and Nitrofurantoin41. In another study by Mohammed et al., isolated E. coli was highly sensitive to Amikacin, Imipenem, and Meropenem, and extremely resistant to Ampicillin42. Shakibaie et al. exhibited that uropathogenic E. coli were 100% sensitive to Imipenem and Meropenem, and 94.4% to Amikacin, while all of them were resistant to Ampicillin43. Another study conducted by Koshesh et al. revealed a low rate of resistance to Imipenem for E. coli uropathogens44. In Khan et al.'s study, E. coli showed 96.2% susceptibility to Imipenem, 85.1% to Amikacin, and 72.6% to Nitrofurantoin45.
In the case of P. aeruginosa, we found it 100% sensitive to 10 antibiotics from 13 antibiotics tested. Its minimum sensitivity was observed to Cefalexin (7.1%) and Cefotaxime (15.4%). A similar study by Shah et al. reported that Imipenem, Piperacillin/Tazobactam, and Amikacin with a minimum resistance rate were the most effective antibiotics against P. aeruginosa in UTI patients46. In contrast, a study by Abdollahi et al. showed that P. aeruginosa isolated from UTI patients were highly resistant to Ampicillin (100%), Gentamicin (66.7%), and Nalidixic-acid (66.7%)47. On the other hand, we found no resistance to Ampicillin and Nalidixic-acid, while resistance to Gentamicin was very low (13.3%).
S. agalactiae isolates in our study were highly sensitive to Linezolid, Ampicillin, Cefalexin, and Nitrofurantoin. In agreement with our findings, Shayanfar et al. found high susceptibility to Ampicillin (96%), Nitrofurantoin (95.5%), Vancomycin (95%), and Norfloxacin (96.5%). They also found the most resistance to Tetracycline (81.6%) and Co-trimoxazole (68.9%)48, while in our study the most resistance was seen to Co-trimoxazole (98.7%) followed by Tetracycline (50%). Another study conducted by Tayebi et al. showed that all S. agalactiae urinary isolates were sensitive to Linezolid and Vancomycin, 99.6% to Ampicillin and nitrofurantoin, which is in line with our results. Nevertheless, they showed 12% sensitivity to Tetracycline which was lower than ours (50%)49.
Regarding E. faecalis isolates, we found Nitrofurantoin (98%), Linezolid (97.9%), Ampicillin (96%), and Vancomycin (87.2%) as the most effective antibiotics. Similar to our results, Goel et al. reported that E. faecalis isolated from UTI patients was highly sensitive to Linezolid (100%), Nitrofurantoin (86%), Vancomycin (77.1%), Gentamicin (67.3%), Ampicillin (63.9%), and Ciprofloxacin (31.2%)50. Their Gentamicin susceptibility was more than ours. Further, Akhter et al. demonstrated considerable sensitivity to Vancomycin (100%), Nitrofurantoin (85.72%), and Ciprofloxacin (23.81%), which is similar to this study's findings. They also showed 28.58 sensitivity rate for Gentamicin and Co-trimoxazole, which is different with our results51.
Concerning C. diversus isolates, they were highly sensitive to Ceftriaxone (100%), Amikacin (95.5%), and Imipenem (95.5%). Sami et al. found that Citrobacter species isolated from UTI patients were sensitive to Imipenem (100), Amikacin (85.2%), and Gentamicin (77.4%), which is consistent with our findings. In addition, they found sensitivity to Nitrofurantoin (66.1%), Ciprofloxacin (56.2%), Ceftriaxone (50.9%), and cefotaxime (43.3%) which are lower than ours52.
Considering the results of antibiogram test for K. oxytoca isolates, the greatest sensitivity was related to Imipenem (100%) and Amikacin (92.3%). This agrees with Razzaque et al.'s study which found Imipenem (94.7%) and Amikacin (92.3%) as the most effective antibiotics for K. oxytoca urinary isolates. They also found sensitivity to Nitrofurantoin (84.6%) and Gentamicin (65.5%), which are different to our results53.
Depending on the ESBL-positive or negative bacteria, this study suggested that ESBL-producing isolates had higher resistance to some antibiotics tested compared to non-ESBL producers. For example, ESBL-positive isolates of E. coli were more resistant to antibiotics tested except Amikacin, Imipenem, Meropenem, and Nitrofurantoin. ESBL-producing K. pneumoniae presented a higher resistance rate to Ceftazidime, Cefotaxime, Ceftriaxone, Gentamycin, Cefalexin, Nalidixic-acid, and Co-trimoxazole. Also, in C. diversus isolates, ESBL production caused resistance to antimicrobial agents except for Amikacin and Imipenem. Regarding K. oxytoca, it led to resistance to Ceftazidime, Cefotaxime, Norfloxacin, Cefalexin, and Co-trimoxazole.
In line with these outcomes, a study carried out by Poovendran et al. indicated that resistance to Tetracycline, Amikacin, Ampicillin, Tobramycin, and Norfloxacin was comparatively higher among uropathogenic E. coli ESBL-producer than non-ESBL producer. Nevertheless, both groups of isolates were 100% sensitive to Imipenem54. Furthermore, Albu et al. found that ESBL-producing E. coli and K. pneumoniae isolates were more resistant to most of the antibiotics tested compared to non-ESBLs, except for Amikacin for E. coli and Imipenem for K. pneumoniae, which had lower resistance rates than non-ESBLs55. Further, another study by Abayneh et al. indicated that ESBL-positive bacteria had higher resistance to most of the antimicrobial agents tested, while in both ESBL-producing and non-ESBL-producing isolates, no resistance was observed toward Imipenem and resistance to Amikacin was low56. Additionally, we used Hierarchical clustering analysis based on the antibiogram pattern of ESBL-negative/positive E. coli, K. pneumoniae and K. oxytoca strains for clustering antibiotics with similar effects. In the case of ESBL positive strains, the results showed that the effectiveness of Cephalexin, Cefotaxime, and Ceftazidime was similar to that of Ampicillin, an antibiotic which had poor effect, and about the ESBL-negative strains, Amikacin and Imipenem demonstrated the most similarity in their efficacy.
In conclusion, the current study indicated a significant rate of infection with ESBL-producing Gram-negative bacilli among UTI patients. The ESBL production was found predominantly among E. coli followed by Klebsiella spp. An intensifying level of resistance to various classes of antimicrobial agents was observed among ESBL producers compared with non-ESBLs.
Hierarchical cluster analysis on the antibiogram pattern showed that in ESBL-positive bacterial strains, the efficacy of Cephalosporins; Cephalexin, Cefotaxime, and Ceftazidime was similar to that of Ampicillin, an antibiotic that had very little effect. With respect to the results of this research and similar investigations worldwide, Carbapenems and aminoglycosides were confirmed as the best options for antibiotic therapy against the ESBL-producing isolates. On the other hand, Penicillins and Co-trimoxazole are not recommended in the treatment of these bacteria; also, the administration of Cephalosporins should be limited. In conclusion, given the rise of antibiotic resistance and the high prevalence of ESBL production in Gram-negative bacteria, plus the importance of this issue in the field of treatment and public health and the costs associated with it, precise infection control and careful monitoring of antibiotic administration is crucial. Thus, routine screening of ESBL-producing isolates before prescribing antibiotics is recommended to prevent prolonged and inappropriate use of antibiotics and therapeutic failures.
Acknowledgements
The authors would like to thank the technical staff of Fardis Central Laboratory particularly for their valuable assistance in collecting and processing of specimens. Also, we are grateful to the research staff of the Department of Medical Laboratory Sciences, Faculty of Allied Medicine, Iran University of Medical Sciences, Tehran, Iran, for contribution to improving the quality of this research.
Author contributions
M.J.G. and N.R. conceived the study and designed experiments. P.R.A., M.S. and Z.Y. conducted the experiment. J.Z. analysed the data. P.R.A. drafted the first manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chiang C-Y, Uzoma I, Moore RT, Gilbert M, Duplantier AJ, Panchal RG. Mitigating the impact of antibacterial drug resistance through host-directed therapies: Current progress, outlook, and challenges. mBio. 2018;9:e01932-1917. doi: 10.1128/mBio.01932-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saipriya JB, Shubha DS, Sudhindra KS, Sumantha A, Madhuri KR. Clinical importance of emerging ESKAPE pathogens and antimicrobial susceptibility profile from a tertiary care centre. Int. J. Curr. Microbiol. Appl. Sci. 2018;7:2881–2891. doi: 10.20546/ijcmas.2018.705.336. [DOI] [Google Scholar]
- 3.O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Review on Antimicrobial Resistance. London: Wellcome Trust; 2016. [Google Scholar]
- 4.Patel MP, et al. Synergistic effects of functionally distinct substitutions in β-lactamase variants shed light on the evolution of bacterial drug resistance. J. Biol. Chem. 2018;293:17971–17984. doi: 10.1074/jbc.RA118.003792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ur Rahman S, Ali T, Ali I, Khan NA, Han B, Gao J. The growing genetic and functional diversity of extended spectrum beta-lactamases. Biomed. Res. Int. 2018;2018:9519718. doi: 10.1155/2018/9519718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eiamphungporn W, Schaduangrat N, Malik AA, Nantasenamat C. Tackling the antibiotic resistance caused by class A β-lactamases through the use of β-lactamase inhibitory protein. Int. J. Mol. Sci. 2018;19:2222. doi: 10.3390/ijms19082222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tewari R, et al. Prevalence of extended spectrum β-lactamase, AmpC β-lactamase and metallo β-lactamase mediated resistance in Escherichia coli from diagnostic and tertiary healthcare centers in south Bangalore, India. Int. J. Res. Med. Sci. 2018;6:1308–1313. doi: 10.18203/2320-6012.ijrms20181288. [DOI] [Google Scholar]
- 8.Ye Q, et al. Characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae from retail food in China. Front. Microbiol. 2018;9:1709. doi: 10.3389/fmicb.2018.01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madhi F, et al. Febrile urinary-tract infection due to extended-spectrum beta-lactamase-producing Enterobacteriaceae in children: A French prospective multicenter study. PLoS ONE. 2018;13:e0190910. doi: 10.1371/journal.pone.0190910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajivgandhi, G., Maruthupandy, M., Ramachandran, G., Priyanga, M. & Manoharan, N. Detection of ESBL genes from ciprofloxacin resistant Gram negative bacteria isolated from urinary tract infections (UTIs). Front. Lab. Med.2, 5–13. http://creativecommons.org/licenses/by-ncnd/4.0/ (2018).
- 11.Taghizadeh S, Haghdoost M, Owaysee Osquee H, Pourjafar H, Pashapour K, Ansari F. Epidemiology of extended spectrum Β-lactamase producing gram negative bacilli of community acquired urinary tract infection in Tabriz, Iran. J. Res. Med. Dent. Sci. 2018;6:199–204. doi: 10.24896/jrmds.20186231. [DOI] [Google Scholar]
- 12.Baziboroun M, Bayani M, Poormontaseri Z, Shokri M, Biazar T. Prevalence and antibiotic susceptibility pattern of extended spectrum beta lactamases producing Escherichia coli isolated from outpatients with urinary tract infections in Babol, Northern of Iran. Curr. Issues Pharm. Med. Sci. 2018;31:61–64. doi: 10.1515/cipms-2018-0013. [DOI] [Google Scholar]
- 13.Stadler T, et al. Transmission of ESBL-producing Enterobacteriaceae and their mobile genetic elements—Identification of sources by whole genome sequencing: Study protocol for an observational study in Switzerland. BMJ Open. 2018;8:e021823. doi: 10.1136/bmjopen-2018-021823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shakya P, Shrestha D, Maharjan E, Sharma VK, Paudyal R. ESBL production among E. coli and Klebsiella spp. causing urinary tract infection: A hospital based study. Open Microbiol. J. 2017;11:23–30. doi: 10.2174/1874285801711010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNulty CAM, et al. CTX-M ESBL-producing Enterobacteriaceae: Estimated prevalence in adults in England in 2014. J. Antimicrob. Chemother. 2018;73:1368–1388. doi: 10.1093/jac/dky007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, Tullu KD. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control. 2019;8:39. doi: 10.1186/s13756-019-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moustafa A, et al. Microbial metagenome of urinary tract infection. Sci. Rep. 2018;8:4333. doi: 10.1038/s41598-018-22660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giwa, F. J., Ige, O. T., Haruna, D. M., Yaqub, Y., Lamido, T. Z. & Usman, S. Y. Extended-spectrum beta-lactamase production and antimicrobial susceptibility pattern of uropathogens in a Tertiary Hospital in Northwestern Nigeria. Ann. Trop. Pathol. 9, 11–6. http://www.atpjournal.org/text.asp?2018/9/1/11/234155 (2018).
- 19.Meije Y, et al. Non-intravenous carbapenem-sparing antibiotics for definitive treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum β-lactamase (ESBL) or AmpC β-lactamase: A propensity score study. Int. J. Antimicrob. Agents. 2019;54:189–196. doi: 10.1101/304246. [DOI] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing. 28th edn. CLSI supplement M100 (CLSI, Wayne, 2018).
- 21.Behzadi, P., Behzadi, E. & Pawlak-Adamska, E. A. Urinary tract infections (UTIs) or genital tract infections (GTIs)? It's the diagnostics that count. GMS Hyg. Infect. Control.14, Doc14. http://www.egms.de/en/journals/dgkh/2019-14/dgkh000320 (2019). [DOI] [PMC free article] [PubMed]
- 22.Yadav K, Prakash S. Screening of ESBL producing multidrug resistant E. coli from urinary tract infection suspected cases in Southern Terai of Nepal. J. Infect. Dis. Diagn. 2017;2:116. doi: 10.4172/2576-389X.1000116. [DOI] [Google Scholar]
- 23.Alqasim A, Abu Jaffal A, Alyousef AA. Prevalence of multidrug resistance and extended-spectrum β-lactamase carriage of clinical uropathogenic Escherichia coli isolates in Riyadh, Saudi Arabia. Int. J. Microbiol. 2018;2018:3026851. doi: 10.1155/2018/3026851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akhtar N, Rahman R, Sultana S, Rahman MR. Antimicrobial sensitivity pattern of bacterial pathogens associated with urinary tract infection. Delta. Med. Coll. J. 2017;5:57–62. doi: 10.3329/dmcj.v5i2.33342. [DOI] [Google Scholar]
- 25.Raja NS. Oral treatment options for patients with urinary tract infections caused by extended spectrum βeta-lactamase (ESBL) producing Enterobacteriaceae. J. Infect. Public Health. 2019;12:843–846. doi: 10.1016/j.jiph.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Shrestha A, Manandhar S, Pokharel P, Panthi P, Chaudhary DK. Prevalence of extended spectrum beta-lactamase (ESBL) producing multidrug resistance gram-negative isolates causing urinary tract infection. EC Microbiol. 2016;4:749–755. [Google Scholar]
- 27.Kengne M, Dounia AT, Nwobegahay JM. Bacteriological profile and antimicrobial susceptibility patterns of urine culture isolates from patients in Ndjamena, Chad. Pan Afr. Med. J. 2017;28:258. doi: 10.11604/pamj.2017.28.258.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rimal U, Thapa S, Maharjan R. Prevalence of extended spectrum beta-lactamase producing Escherichia coliand Klebsiellaspecies from urinary specimens of children attending Friendship International Children’s Hospital. Nepal J. Biotechnol. 2017;5:32–38. doi: 10.3126/njb.v5i1.18868. [DOI] [Google Scholar]
- 29.Al Yousef SA, Younis S, Farrag E, Moussa HSh, Bayoumi FS, Ali AM. Clinical and laboratory profile of urinary tract infections associated with extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae. Ann. Clin. Lab. Sci. 2016;46:393–400. [PubMed] [Google Scholar]
- 30.Falodun OI, Morakinyo YM, Fagade OE. Determination of water quality and detection of extended beta-lactamase producing gram-negative bacteria in selected rivers located in Ibadan, Nigeria. Jord. J. Biol. Sci. 2018;11:107–112. [Google Scholar]
- 31.Fatima S, Muhammad IN, Usman S, Jamil S, Khan MN, Khan SI. Incidence of multidrug resistance and extended-spectrum beta-lactamase expression in community-acquired urinary tract infection among different age groups of patients. Indian J. Pharmacol. 2018;50:69–74. doi: 10.4103/ijp.IJP_200_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nanoty VV, Agrawal GN, Tankhiwale SS. Evaluation of antibiotic resistance and β-lactamase production in clinical isolates from a tertiary care Hospital in Central India. J. Clin. Basic Res. 2018;2:1–5. doi: 10.29252/jcbr.2.1.1. [DOI] [Google Scholar]
- 33.Kargar, M., Kargar, M., Jahromi, M. Z., Najafi, A. & Ghorbani-Dalini, S. Molecular detection of ESBLs production and antibiotic resistance patterns in Gram negative bacilli isolated from urinary tract infections. Indian J. Pathol. Microbiol. 57, 244–248. http://www.ijpmonline.org/text.asp?2014/57/2/244/134688 (2014). [DOI] [PubMed]
- 34.Yousefipour M, et al. Bacteria producing extended spectrum β-lactamases (ESBLs) in hospitalized patients: Prevalence, antimicrobial resistance pattern and its main determinants. Iran. J. Pathol. 2019;14:61–67. doi: 10.30699/ijp.14.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eltai NO, et al. Molecular characterization of extended spectrum β-lactamases enterobacteriaceae causing lower urinary tract infection among pediatric population. Antimicrob. Resist. Infect. Control. 2018;7:90. doi: 10.1186/s13756-018-0381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bharara T, Sharma A, Gur R, Duggal SD, Jena PP, Kumar A. Comparative analysis of extended-spectrum beta-lactamases producing uropathogens in outpatient and inpatient departments. Int. J. Health Allied Sci. 2018;7:45–50. doi: 10.4103/ijhas.IJHAS_33_17. [DOI] [Google Scholar]
- 37.Kalaivani R, Charles P, Kunigal S, Prashanth K. Incidence of SHV-1 and CTX-M-15 extended spectrum of β-lactamases producing Gram-negative bacterial isolates from antenatal women with asymptomatic bacteriuria. J. Clin. Diagn. Res. 2018;12:1–4. doi: 10.7860/JCDR/2018/35728.11568. [DOI] [Google Scholar]
- 38.Koksal E, et al. Investigation of risk factors for community-acquired urinary tract infections caused by extended-spectrum beta-lactamase Escherichia coli and Klebsiella species. Investig. Clin. Urol. 2019;60:46–53. doi: 10.4111/icu.2019.60.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taha AA, Shtawi A, Jaradat A, Dawabsheh Y. Prevalence and risk factors of extended spectrum beta-lactamase-producing uropathogens among UTI patients in the governmental hospitals of North West Bank: A cross-sectional study. J. Infect. Dis. Preve. Med. 2018;6:183. doi: 10.4172/2329-8731.1000183. [DOI] [Google Scholar]
- 40.Senbayrak S, et al. Antibiotic resistance trends and the ESBL prevalence of Escherichia coli and Klebsiella spp urinary isolates in in-and outpatients in a tertiary care hospital in Istanbul, 2004–2012. Jundishapur J. Microbiol. 2017;10:e13098. doi: 10.5812/jjm.13098. [DOI] [Google Scholar]
- 41.Ahmed SS, Shariq A, Alsalloom AA, Babikir IH, Alhomoud BN. Uropathogens and their antimicrobial resistance patterns: Relationship with urinary tract infections. Int. J. Health Sci. 2019;13:48–55. [PMC free article] [PubMed] [Google Scholar]
- 42.Mohammed MA, Alnour TM, Shakurfo OM, Aburass MM. Prevalence and antimicrobial resistance pattern of bacterial strains isolated from patients with urinary tract infection in Messalata Central Hospital, Libya. Asian Pac. J. Trop. Med. 2016;9:771–776. doi: 10.1016/j.apjtm.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Shakibaie MR, Adeli S, Salehi MH. Antimicrobial susceptibility pattern and ESBL production among uropathogenic Escherichia coli isolated from UTI children in pediatric unit of a hospital in Kerman, Iran. Br. Microbiol. Res. J. 2014;4:262–273. doi: 10.9734/BMRJ/2014/6563. [DOI] [Google Scholar]
- 44.Koshesh M, Mansouri S, Hashemizadeh Z, Kalantar-Neyestanaki D. Identification of extended-spectrum β-lactamase genes and AmpC-β-lactamase in clinical isolates of Escherichia colirecovered from patients with urinary tract infections in Kerman, Iran. Arch. Pediatr. Infect. Dis. 2017;5:e37968. doi: 10.5812/pedinfect.37968. [DOI] [Google Scholar]
- 45.Khan IU, et al. Antimicrobial susceptibility pattern of bacteria isolated from patients with urinary tract infection. J. Coll. Physicians Surg. Pak. 2014;24:840–844. doi: 10.2014/JCPSP.840844. [DOI] [PubMed] [Google Scholar]
- 46.Saha S, et al. Antimicrobial resistance in uropathogen isolates from patients with urinary tract infections. Biomed. Res. Ther. 2015;2:263–269. doi: 10.7603/s40730-015-0011-3. [DOI] [Google Scholar]
- 47.Abdulahi II, Gebre-Selassie S. Common bacterial pathogens and their antimicrobial susceptibility patterns in patients with symptomatic urinary tract infections at Hiwot-Fana and Jugal Hospitals, Harar City, Eastern Ethiopia. Galore Int. J. Health. Sci. Res. 2018;3:33–49. [Google Scholar]
- 48.Shayanfar N, Mohammadpour M, Hashemi-Moghadam SA, Haghi Ashtiani MT, Zare Mirzaie A, Rezaei N. Group B streptococci urine isolates and their antimicrobial susceptibility profiles in a group of Iranian females: Prevalence and seasonal variations. Acta Clin. Croat. 2012;51:623–626. [PubMed] [Google Scholar]
- 49.Tayebi Z, Saderi H, Gholami M, Houri H, Samie S, Boroumandi S. Evaluation of antimicrobial susceptibility of Streptococcus agalactiae isolates from patients with urinary tract infection (UTI) symptoms. Infect. Epidemiol. Med. 2016;2:17–19. doi: 10.18869/modares.iem.2.4.17. [DOI] [Google Scholar]
- 50.Goel V, Kumar D, Kumar R, Mathur P, Singh S. Community acquired enterococcal urinary tract infections and antibiotic resistance profile in North India. J. Lab. Physicians. 2016;8:50–54. doi: 10.4103/0974-2727.176237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akhter J, Ahmed S, Anwar S. Antimicrobial susceptibility patterns of Enterococcus species isolated from urinary tract infections. Bangladesh J. Med. Microbiol. 2014;8:16–20. doi: 10.3329/bjmm.v8i1.31069. [DOI] [Google Scholar]
- 52.Sami H, et al. Citrobacteras a uropathogen, its prevalence and antibiotics susceptibility pattern. CHRISMED J. Health Res. 2017;4:23–26. doi: 10.4103/2348-3334.196037. [DOI] [Google Scholar]
- 53.Razzaque SA, et al. A cross sectional study highlighting the sensitivity patterns and incidence of extended spectrum beta lactamase producing Klebsiella oxytoca in patients with urinary tract infection. Int. J. Pulmonol. Infect. Dis. 2017;1:1–5. doi: 10.15226/2637-6121/1/1/00102. [DOI] [Google Scholar]
- 54.Poovendran P, Vidhya N, Murugan S. Antimicrobial susceptibility pattern of ESBL and non-ESBL producing uropathogenic Escherichia coli (UPEC) and their correlation with biofilm formation. Int. J. Microbiol. Res. 2013;4:56–63. doi: 10.5829/idosi.ijmr.2013.4.1.71235. [DOI] [Google Scholar]
- 55.Albu S, et al. Bacteriuria and asymptomatic infection in chronic patients with indwelling urinary catheter: The incidence of ESBL bacteria. Medicine. 2018;97:e11796. doi: 10.1097/MD.0000000000011796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abayneh M, Tesfaw G, Abdissa A. Isolation of extended-spectrum β-lactamase-(ESBL-)producing Escherichia coli and Klebsiella pneumoniae from patients with community-onset urinary tract infections in Jimma University Specialized Hospital, Southwest Ethiopia. Can. J. Infect. Dis. Med. Microbiol. 2018;2018:4846159. doi: 10.1155/2018/4846159. [DOI] [PMC free article] [PubMed] [Google Scholar]