Abstract
Patients with chronic kidney disease (CKD) have elevated circulating levels of trimethylamine N-oxide (TMAO), a metabolite derived from gut microbes and associated with cardiovascular diseases. High circulating levels of TMAO and its dietary precursor, choline, predict increased risk for development of CKD in apparently healthy subjects, and studies in mice fed TMAO or choline suggest that TMAO can contribute to kidney impairment and renal fibrosis. Here we examined the interactions between TMAO, kidney disease, and cardiovascular disease in mouse models. We observed that while female hyperlipidemic apoE KO mice fed a 0.2% adenine diet for 14 weeks developed CKD with elevated plasma levels of TMAO, provision of a non-lethal inhibitor of gut microbial trimethylamine (TMA) production, iodomethylcholine (IMC), significantly reduced multiple markers of renal injury (plasma creatinine, cystatin C, FGF23, and TMAO), reduced histopathologic evidence of fibrosis, and markedly attenuated development of microalbuminuria. In addition, while the adenine-induced CKD model significantly increased heart weight, a surrogate marker for myocardial hypertrophy, this was largely prevented by IMC supplementation. Surprisingly, adenine feeding did not increase atherosclerosis and significantly decreased the expression of inflammatory genes in the aorta compared to the control groups, effects unrelated to TMAO levels. Our data demonstrate that inhibition of TMAO production attenuated CKD development and cardiac hypertrophy in mice, suggesting that TMAO reduction may be a novel strategy in treating CKD and its cardiovascular disease complications.
Subject terms: Cell biology, Molecular biology, Cardiology, Diseases, Nephrology
Introduction
Previous studies by our groups identified a novel dietary/meta-organismal pathway for cardiovascular disease (CVD) whereby bacterial metabolism of dietary choline and L-carnitine in the intestine leads to an intermediate metabolite, trimethylamine (TMA), which is absorbed from the gut and subsequently oxidized by hepatic flavin-containing monooxygenases (FMO) to generate trimethylamine N-oxide (TMAO)1–3. Romano et al. identified eight bacterial species from human intestine representing two different phyla (Firmicutes and Proteobacteria), which can convert choline to TMA in vitro4. A more recent study quantified and characterized bacterial genes encoding enzymes responsible for TMA production, including choline-TMA lyase (CutC), carnitine oxygenase (CntA) and betaine reductase (GrdH) in 89 fecal samples derived from various mammals5. The study showed that bacteria harboring CutC and GrdH are predominately affiliated with various taxa within Firmicutes, whereas CntA comprised sequences primarily linked to Escherichia5. In humans, plasma TMAO levels are elevated in patients with CVD compared to healthy controls, and exhibit a dose-dependent relationship with severity of atherosclerotic disease, and are associated with a ~ twofold increased risk for major adverse cardiac events (MACE), including death, myocardial infarction (MI), and stroke, independent of traditional CVD risk factors, renal function, and medications6–8. Multiple studies have replicated the association of TMAO with CVD and mortality8,9, including in patients with chronic heart failure8,10, diabetes mellitus11 and renal disease12,13. Multiple meta analyses confirm an overall positive association between circulating TMAO levels and CVD and mortality risks8,14, though not all studies have reported the same positive relationships9. Feeding mice with choline or TMAO leads to significant increases in plasma TMAO levels, thereby promoting atherosclerosis and thrombosis2,15. Non-lethal inhibition of gut microbial TMA production has recently been shown to decrease atherosclerosis16 and thrombosis potential17, and benefit tissue remodeling changes in preclinical models of heart failure18 and isoproterenol infusion-driven cardiorenal disease in mice19. Thus, there is growing potential for the use of inhibitors that target the gut microbiome as a novel therapeutic strategy for the treatment of cardiovascular and metabolic diseases.
Chronic kidney disease (CKD) is a serious public health problem that affects 15% (37 million) of United States (US) adults (Center for Disease Control and Prevention. Chronic Kidney Disease Surveillance System—US 2020. Available from: http://www.cdc.gov/ckd) and is associated with premature death, primarily due to CVD20,21. TMAO levels in patients with CKD are elevated12,13,22–24, likely due to impaired renal excretion of TMAO12,23 and possibly due to altered gut microbiome22. In addition, mice fed with choline or TMAO demonstrate significantly increased renal fibrosis and impaired renal function12, suggesting that TMAO is not merely a marker of reduced renal function, but a potential cause of CKD. Furthermore, choline and TMAO feeding in mice is associated with increased phosphorylation of SMAD family member 3 (Smad3) in the kidney12, which is an important regulator of the profibrotic transforming growth factor β (TGF-β)/Smad3 signaling pathway during fibrotic kidney disease25. In a previous study we showed that choline feeding increases TMAO levels and promotes inflammatory responses in arterial vascular cells through the activation of the nuclear factor-kappa B (NF-κB) signaling pathway26. In addition, TMAO has been suggested to promote CVD through several other pro-atherogenic mechanisms, including foam cell formation, reverse cholesterol transport, platelet responsiveness/thrombosis, activation of the NLR family pyrin domain containing 3 (NLRP3) inflammasome, and changes in tissue cholesterol, sterol, and bile acid metabolism2,3,15,27–29. These studies support the notion that the increased CVD risk associated with CKD may arise in part from elevated TMAO levels.
Here, we investigate how inhibition of gut microbiota-dependent TMA production affects the development of adenine-induced CKD and CVD traits in hyperlipidemic apolipoprotein E (apoE) knockout (KO) mice. Our results indicate that TMAO promotes CKD, and that in the context of CKD this contributes to cardiac hypertrophy.
Results
Inhibition of TMAO production attenuates chronic kidney disease induced by adenine feeding
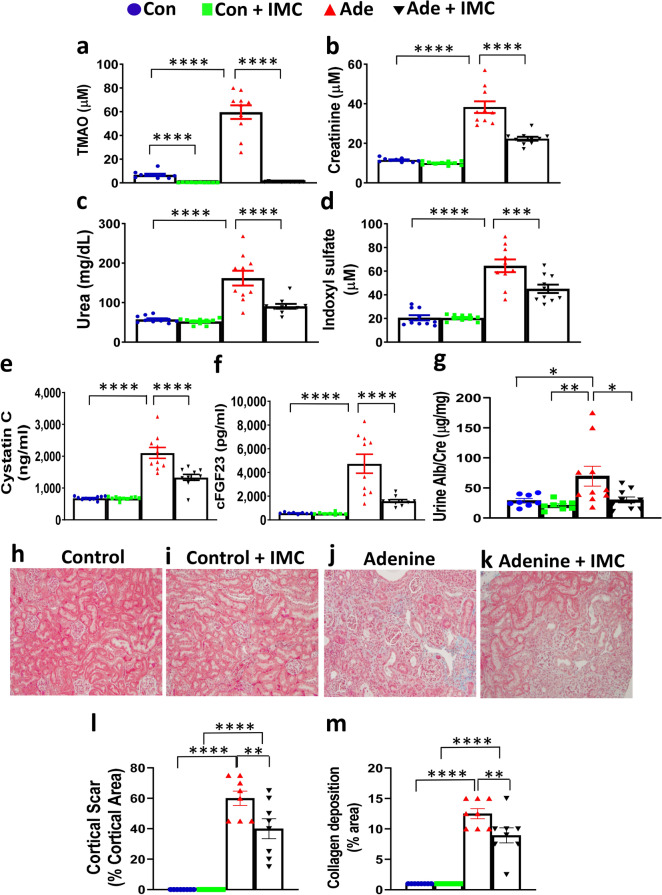
ApoE KO mice, instead of wild-type mice, were chosen for this study since we aimed to study atherosclerosis and these mice develop hypercholesterolemia and atherosclerosis spontaneously without dietary intervention30. Furthermore, CKD induced by 5/6 nephrectomy (5/6Nx) has been shown to increase atherosclerosis in apoE KO mice31–33. Female mice were chosen for the study instead of males since our preliminary data showed that male mice fed the adenine diet for 8 weeks lost a very substantial amount of body weight (19%) and appeared unhealthy for further characterization. It appears that sex hormones may be the cause of greater susceptibility of male kidneys to progressive renal injury in mice34,35. Therefore, one-month old female apoE KO mice were fed standard chow (control; Con) or adenine (Ade) diets with or without iodomethylcholine (IMC; see Methods) for 14 weeks to examine the effects of reduced TMAO on CKD and atherosclerosis. At the end of the 14-week feeding period, the body weights of mice receiving either Ade or Ade + IMC were significantly decreased compared to either Con or Con + IMC groups, whereas there were no significant differences in body weight between Con and Con + IMC or between Ade and Ade + IMC groups (Supplemental Fig. S1). The decreased body weights observed in the Ade and Ade + IMC groups compared to the control groups were not due to decreased food consumption since there were no significant differences in food consumption among the 4 groups of mice (Supplemental Fig. S1). Similar food consumption between the Ade and Ade + IMC mice suggested that similar amounts of adenine were ingested by these two groups of mice. Plasma TMA levels of the Con + IMC group showed a trend of a decrease compared to the Con group (0.08 μM vs. 0.15 μM, p = 0.15, Supplemental Fig. S1). Also, there was a trend of a decrease in TMA levels in the Ade + IMC group compared to the Ade group (0.08 μM vs. 0.14 μM, p = 0.15). Plasma TMAO levels of the Ade group were 8.8-fold higher than the Con group (60 μM vs. 6.8 μM, p < 0.0001, Fig. 1a). IMC dramatically decreased TMAO levels (0.8 μM) in Ade + IMC group compared to the Ade group (p < 0.0001, Fig. 1a). In addition, TMAO levels of Con + IMC group were significantly decreased compared to the Con group (0.4 μM vs. 6.8 μM, p < 0.0001, Fig. 1a). These data demonstrated the effectiveness of IMC in blocking TMAO production in mice fed either a control or an adenine diet.
Figure 1.
Inhibition of TMAO production attenuates chronic kidney development induced by adenine feeding. Female apoE KO mice (n = 10 per group) were fed control (Con), control + IMC (Con + IMC), adenine (Ade), or adenine + IMC (Ade + IMC) diets for 14 weeks before measurements of traits. Plasma levels of (a) TMAO, (b) creatinine, (c) Urea, (d) Indoxyl sulfate, (e) Cystatin C, and (f) c-terminal FGF23 (cFGF23) are shown. (g) Urine albumin (Alb) to creatinine (Cre) ratios are shown. Representative kidney sections stained with Masson’s trichrome are shown in (h–k). Quantification of kidney pathology, (l) cortical scar, and (m) collagen deposition, in the kidney sections of all 4 groups are shown. Individual values, means, and standard errors are shown. Sample sizes were n = 10/group for (a–f), n = 8–10/group for (g), and n = 8 kidneys/group for (l,m). For (a–f,l,m), post hoc multiple comparisons analysis was performed after significant two-way ANOVA. Symbols: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
We next monitored multiple circulating markers of kidney function, including creatinine, urea, cystatin C, and fibroblast growth factor 23 (FGF23). Notably, all markers were significantly elevated in the Ade group compared to the Con group, confirming renal injury and reduced function in the adenine fed mice. Importantly, mice in the Ade + IMC group showed significantly reduced circulating levels of all renal injury markers compared to the Ade group: 41% decrease in creatinine (p < 0.0001), 41% decrease in urea (p < 0.0001), 37% decrease in cystatin C (p < 0.0001), and 64% decrease in FGF23 (p < 0.0001) (Fig. 1b,c,e,f), demonstrating that IMC attenuated CKD induced by adenine feeding. In additional studies, circulating levels of a uremic toxin, indoxyl sulfate, were found to be increased threefold in the Ade group compared to the Con group (Fig. 1d). However, when the gut microbiota targeting inhibitor IMC was provided, there was a significant 31% reduction in indoxyl sulfate levels in the Ade + IMC group versus Ade group (p < 0.001, Fig. 1d). The adenine diet-induced CKD mouse model simulates multiple features of CKD including development of microalbuminuria. We therefore examined urine albumin-to-creatinine ratio (UACR) in the mouse groups and noted a significant increase in the Ade group (compared to the Con and Con + IMC groups; Fig. 1g), indicating impaired kidney function in the former. The Ade + IMC group exhibited significantly decreased UACR (57% decrease, p = 0.03) compared to the Ade group, demonstrating significant improvement in kidney function in the former (Fig. 1g). In fact, there were no significant differences in UACR among the Con, Con + IMC, and Ade + IMC groups (Fig. 1g).
To examine tissue remodeling during CKD, histological sections of kidney were stained with Masson’s trichrome and quantitated for cortical scar area and collagen deposition. Kidney sections of both Con and Con + IMC groups appeared normal (Fig. 1h,i), whereas those of the Ade and Ade + IMC groups showed cortical scar and collagen deposition (Fig. 1j,k). Quantification of the pathological changes revealed no cortical scar and less than 1% collagen deposition in the Con and Con + IMC groups (Fig. 1l,m). There were significant decreases in cortical scar (by 33%, p = 0.003) and collagen deposition (by 28%, p = 0.004) in the Ade + IMC group compared to the Ade group (Fig. 1l,m). There was no global glomerulosclerosis, vascular, or perivascular fibrosis in any group. These data demonstrated that IMC not only significantly decreased circulating levels of kidney disease markers but also decreased renal pathological changes associated with adenine feeding and improved kidney function.
IMC supplementation results in a kidney gene expression profile consistent with decreased inflammation and fibrosis following adenine feeding
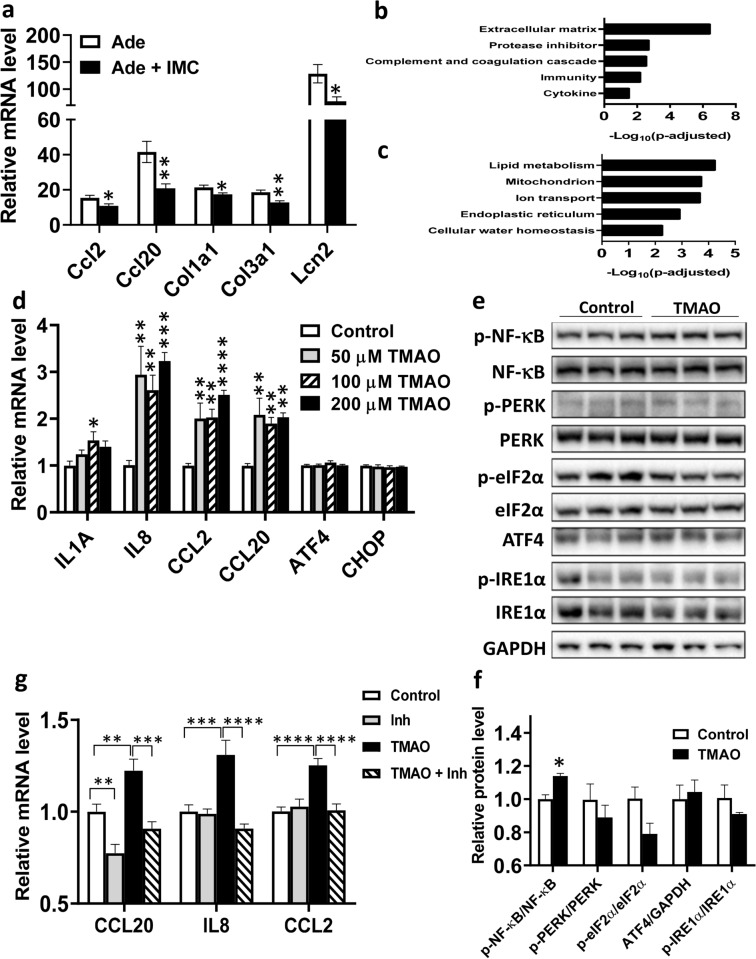
qPCR analysis revealed that adenine treatment significantly increased the mRNA levels of inflammatory and fibrosis genes in the kidney by 15- to 100-fold compared to the Con group (Fig. 2a). IMC supplementation significantly decreased renal expression of inflammatory genes, C–C motif chemokine ligand 2 (Ccl2, p < 0.05), C–C motif chemokine ligand 20 (Ccl20, p < 0.01), and lipocalin-2 (Lcn2, p < 0.05), and fibrosis genes, collagen type I alpha 1 chain (Col1a1, p < 0.05), and collagen type III alpha 1 chain (Col3a1, p < 0.01), caused by adenine feeding (Fig. 2a). RNA-seq analysis revealed 627 down-regulated and 584 up-regulated genes in the kidneys of Ade + IMC group compared to the Ade group (data not shown). Pathway analysis of the downregulated genes showed the enrichment of extracellular matrix, protease inhibitor, complement and coagulation cascade, immunity, and cytokine clusters (Fig. 2b and Supplemental Table S1). The protease inhibitor functional cluster includes: WAP 4-disulfide core domain-2 (Wfdc2), plasminogen activatorinhibitor-1 (PAI-1, encoded by Serpine1), tissue inhibitor of metalloproteinase 1 (Timp1) and 10 other genes (Supplemental Table S1). Overall, our data suggested that IMC supplementation attenuated the pathological processes, such as fibrosis and inflammation, induced by adenine feeding. The up-regulated functional clusters include: lipid metabolism, mitochondria, ion transport, endoplasmic reticulum, and cellular water homeostasis (Fig. 2c and Supplemental Table S2), indicating an improved metabolic state associated with IMC supplementation.
Figure 2.
Inhibition of TMAO generation decreases inflammation and fibrosis in the kidneys of adenine diet-induced CKD. Gene expression analysis of kidney samples from the same mice described in Fig. 1 were performed using qPCR (a), n = 8 to 9 per group, and RNA-seq (b,c). Based on the RNA-seq data, pathway enrichment analysis was performed to identify down-regulated (b), and up-regulated (c) functional clusters in the kidneys of Ade + IMC group compared to the Ade group (n = 6 per group). Gene expression (d, n = 4–5/group) and immunoblotting (e,f, n = 3/group) analysis of MDCK II kidney epithelial cells treated with control, and various doses of TMAO (d) or 200 μM of TMAO (e,f) for 48 h. (g) MDCKII cells were treated with control media, 100 nM NF-κB inhibitor IV (Inh), 200 μM TMAO (TMAO), or 200 μM TMAO + 100 nM NF-κB inhibitor IV (TMAO + Inh) for 48 h before gene expression analysis (n = 9 for each group). For (a,f), two tailed Student’s t-test was performed. Symbols: *p < 0.05, and **p < 0.01. For (d) Post hoc Tukey’s multiple comparisons test was performed after significant one-way ANOVA. Symbols: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 compared to the control group. For (g) Post hoc analysis was performed after significant two-way ANOVA. Symbols: **p < 0.01, ***p < 0.001, and ****p < 0.0001.
TMAO treatment induced inflammation via NF-κB activation in kidney epithelial cells
To better understand the mechanism of the action of TMAO in the kidney, we quantified the expression of inflammatory and unfolded protein response (UPR) genes in MDCK II kidney epithelial cells in response to various doses of TMAO after 48 h. We observed that TMAO (as low as 50 μM up to 200 μM) significantly increased the mRNA levels of the inflammatory genes: interleukin 8 (IL8), CCL2, and CCL20 by two to threefold compared to the control group, whereas the mRNA levels of genes involved in UPR, activating transcription factor 4 (ATF4) and CHOP (DNA damage inducible transcript 3, DDIT3) were similar between the TMAO treated and control cells (Fig. 2d). Immunoblotting analysis revealed that the NF-κB pathway is activated by TMAO treatment, as evidenced by significant increases in both the phosphorylation of the NF-κB p65 subunit (Fig. 2e) and the p-NF-κB p65 to total NF-κB p65 ratio (Fig. 2f). In contrast, under the conditions examined, TMAO treatment did not activate proteins in the UPR pathways, including protein kinase R-like endoplasmic reticulum kinase (PERK), eukaryotic initiation factor 2 alpha (eIF2α), ATF4, and inositol-requiring enzyme 1 alpha (IRE1α), in MDCK II cells (all p > 0.09; Fig. 2e,f). The presence of an NF-κB inhibitor, NF-κB inhibitor IV, completely abolished the stimulatory effect of TMAO on the expression of CCL20 (p < 0.001), IL8 (p < 0.0001), and CCL2 (p < 0.0001) (Fig. 2g), suggesting the importance of NF-κB in mediating the inflammatory effect of TMAO. TMAO treatment also significantly induced the expression of the inflammatory molecules, CCL2 and tumor necrosis factor-alpha (TNF-α), in normal human primary renal proximal tubule epithelial cells (RPTEC) (Supplemental Fig. S2).
Effects of IMC on cardiovascular disease related traits in mice fed the adenine diet
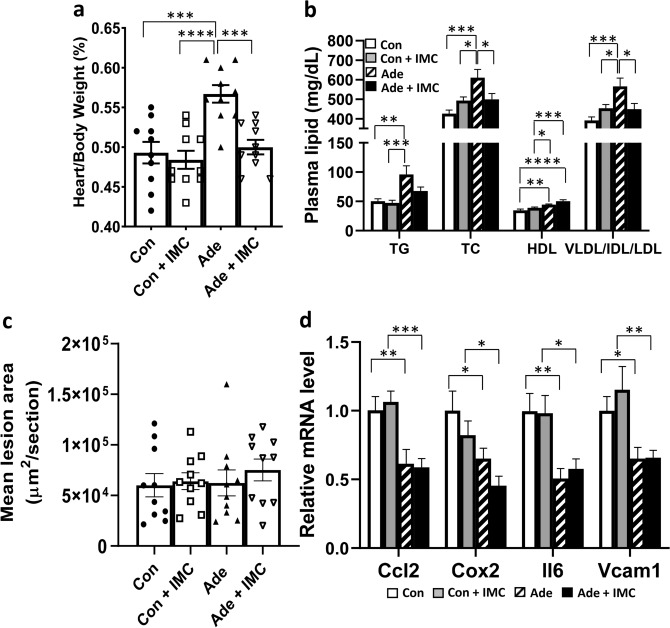
Adenine-fed mice exhibited significantly increased heart weight/body weight ratio (a surrogate marker for myocardial hypertrophy36) compared to Con, Con + IMC, and Ade + IMC groups (Fig. 3a), whereas no differences in heart weight/body weight ratio were observed among the Con, Con + IMC, and Ade + IMC groups (Fig. 3a). These data suggested that adenine feeding induced heart hypertrophy in mice—likely as a result of CKD—and cardiac hypertrophy was blocked by IMC treatment.
Figure 3.
IMC supplementation protects against myocardial hypertrophy and dyslipidemia associated with adenine diet-induced CKD. Data obtained from tissue samples of the same animals described in Fig. 1 are presented in this figure, n = 10 per group. (a) Heart weight/body weight ratios, (b) Plasma levels of triglyceride (TG), total cholesterol (TC), HDL cholesterol (HDL), and VLDL/IDL/LDL cholesterol (VLDL/IDL/LDL), (c) mean atherosclerotic lesion area at the aortic root, and (d) aortic gene expression analysis by qPCR are shown. For data presented in all panels, post hoc multiple comparisons analysis was performed after significant two-way ANOVA. Symbols: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Adenine diet also worsened the hyperlipidemia phenotypes in apoE KO mice with increased plasma triglyceride, total cholesterol, and VLDL/IDL/LDL cholesterol levels compared to Con and Con + IMC groups (Fig. 3b). In contrast, the Ade + IMC group exhibited significant decreases in plasma total cholesterol (by 18%, p < 0.05) and VLDL/IDL/LDL cholesterol (by 21%, p < 0.05) levels compared to the Ade group, suggesting improved plasma lipoprotein profile (Fig. 3b). Surprisingly, both the Ade and Ade + IMC groups had significantly increased HDL cholesterol levels compared to the Con and Con + IMC group (Fig. 3b). Unexpectedly, there were no significant differences in mean atherosclerotic lesion area at the aortic root region among the 4 groups of mice (Fig. 3c). Gene expression analysis of the aorta showed significantly reduced mRNA levels of Ccl2, Cox2 (prostaglandin-endoperoxide synthase 2, Ptgs2), interleukin 6 (Il6), and vascular cell adhesion molecule 1 (Vcam1) in the Ade and Ade + IMC groups compared to those of the Con and Con + IMC groups (Fig. 3d), suggesting decreased inflammation in the aortas of mice of the former groups, despite induction of CKD.
Discussion
In this study, we demonstrated that targeted inhibition of bacterial choline TMA lyase activity dramatically decreased circulating TMAO levels and multiple indices of renal function and adverse remodeling in an adenine-induced CKD mouse model. In the adenine-fed apoE KO model where renal function declined, multiple markers of injury were increased and adverse histopathological changes occurred. Provision of IMC blocked these adverse effects, normalizing renal function and histology, and improving the many CKD markers including creatinine, urea, cystatin C, FGF23, TMAO, and UACR. Of importance, we observed suppression of TMAO with the non-lethal inhibitor significantly decreased kidney cortical scar and fibrosis area, and also decreased expression of multiple inflammatory and fibrosis genes. RNA-seq analysis in the kidneys of Ade + IMC group compared to the Ade group revealed downregulation of pathways involved in extracellular matrix, immunity, and cytokine, and upregulation of pathways involved in lipid metabolism, mitochondria, endoplasmic reticulum, ion transport and water homeostasis. Therefore, gut microbiota dependent inhibition of TMA generation using IMC attenuated CKD development in the host and preserved kidney function. Recent studies demonstrated halomethylcholine based microbial choline TMA lyase inhibitors block TMAO generation in the host and elicit protective effects on atherosclerosis, thrombosis, and heart failure16–18. In agreement with our findings, in male wild-type C57BL/6 mice, IMC has been shown to inhibit TMA and TMAO production and prevent choline diet-induced renal functional decline and adverse remodeling in a chronic sympathetic-driven (isoproterenol infusion) model of CKD19. Our study provides further evidence that inhibition of a gut microbe pathway (the TMAO metaorganismal pathway) can have a profound effect on the host—this time demonstrating marked inhibition in progression of CKD in an adenine feeding model.
Chronic kidney disease is associated with altered gut microbiome in humans and animals22,37,38. Also, CKD patients tend to have higher percentages of opportunistic pathogens and decreased percentages of beneficial microbes in the gut22. Furthermore, CKD patients had increased plasma TMAO levels and significantly increased abundance of bacterial genes related to TMA production in the intestine compared to healthy controls22. A fecal microbiota transplantation study showed that antibiotic-treated mice receiving fecal samples from CKD patients had significantly higher plasma TMAO levels and different composition of gut microbiota than did mice receiving fecal samples from healthy subjects22. These data suggest that elevated plasma TMAO level associated with CKD is caused at least in part by increased abundance of TMA producing bacteria in the gut. Therefore, our strategy of non-lethal inhibition of gut microbial TMA production is likely to be beneficial to CKD patients in slowing the progression of CKD and decreasing CVD risk.
Our cell culture studies demonstrated that TMAO treatment induced the expression of inflammatory genes by activation of NF-κB in kidney epithelial cells, similar to our previous observation in human aortic endothelial and smooth muscle cells26. Therefore, we postulate that TMAO exacerbates the development of CKD, at least in part via activation of NF-κB pathway in relevant cell types in the kidney. Unlike the data previously reported in primary hepatocytes39, we failed to observe activation of PERK or other UPR pathways in TMAO-treated kidney epithelial cells. This may have resulted from differences in cell types, experimental conditions, or other factors. Whether PERK plays a role in recognition of TMAO as a receptor involved in phenotypes monitored is unclear, and merits further investigation. Thus far, only glucose related phenotypes linked to TMAO have been implicated in being transmitted in part via the involvement of PERK and other UPR genes39.
Increased NF-κB activation has been observed in the kidneys of patients with type 2 diabetic nephropathy40. Inhibition of NF-κB signaling through feeding of a chemical inhibitor, pyrrolidine dithiocarbamate, has been shown to decrease inflammation and CKD development in various animal models, including: type 1 diabetes41, adenine overload42, and 5/6 nephrectomy43. Our findings that TMAO activated NF-κB, leading to increased expression of inflammatory genes in kidney epithelial cells, suggest NF-κB activation as one of the mechanisms by which TMAO may promote CKD. We postulate that activation of NF-κB by TMAO leads to increased inflammation which is a major driver of fibrosis44,45. In fact we observed significantly decreased expression of genes belonged to the extracellular matrix pathway in the kidneys of Ade + IMC group compared to those of Ade group (Fig. 2b and Supplemental Table S1), supporting our hypothesis.
We observed significantly decreased expression of Wfdc2, PAI-1/Serpine1, Timp1 and 10 other genes of the protease inhibitor functional cluster in the kidneys of Ade + IMC group compared to those of the Ade group (Supplemental Table S1). Wfdc2 is a serine protease inhibitor that is upregulated in human and mouse fibrotic kidneys and suppresses the degradation of type I collagen. Administration of Wfdc2 neutralizing antibodies accelerated collagen I degradation and inhibited fibrosis in various mouse models of renal disease, demonstrating a profibrotic role for Wfdc246. PAI-1/Serpine1, a serine protease inhibitor, is thought to be profibrotic by impairing the turnover and degradation of extracellular matrix proteins. Furthermore, PAI-1 plays an important role in the interstitial recruitment of inflammatory macrophages and myofibroblasts in response to chronic kidney injury47,48. Timp1 inhibits the activities of matrix metalloproteinases. Timp1 deficiency in mice is associated with significantly reduced myocardial fibrosis in models of cardiomyopathy in vivo49. Timp1 also mediates an association between CD63 (cell surface receptor for TIMP1) and integrin β1 on fibroblasts, initiates activation and nuclear translocation of Smad2/3 and β-catenin, leading to de novo collagen synthesis49. Therefore, the decreased expression of these protease inhibitor genes in the kidneys of the Ade + IMC mice compared to those of the Ade mice may explain, in part, the decreased collagen deposition in kidneys of the former (Fig. 1m).
We observed significantly decreased heart weight/body weight ratios in the Ade + IMC group as compared to the Ade group, suggesting decreased cardiac hypertrophy in the former. FGF23 is a bone derived hormone with a central role in the regulation of phosphate homeostasis50. CKD patients exhibit elevated circulating levels of FGF23, which has been shown to induce left ventricular hypertrophy51 that can lead to heart failure. We observed significantly increased circulating FGF23 levels in the adenine-fed mice compared to the control mice, whereas IMC supplementation significantly decreased FGF23 levels induced by adenine feeding. Therefore, the decrease in heart weight/body weight ratios in the Ade + IMC group compared to the Ade group could be attributed in part to decreased FGF23 levels.
CKD induced by 5/6 nephrectomy (5/6Nx) has been shown to increase atherosclerosis in apoE KO mice31–33. Adenine feeding, a non-surgical approach to induce CKD, induces tubulointerstitial nephropathy in mice52. It also induces cardiac hypertrophy and decreases cardiac function in mice53. Surprisingly, atherosclerotic lesion areas at the aortic root of the Ade group were not increased compared to the Con or Con + IMC groups. In fact, gene expression analysis of the aortas revealed that expression of inflammatory genes was decreased in the Ade and Ade + IMC groups compared to the Con and Con + IMC groups, suggesting that, by unknown mechanisms, adenine feeding decreased inflammation locally in the aorta despite induction of CKD. Adenine is the precursor of adenosine that is known to play a role in the control of inflammation54. One of the adenosine receptors, A2B adenosine receptor, is expressed in the vasculature and macrophages and protects against inflammation and excessive vascular adhesion55. We hypothesize that adenine feeding may lead to increased adenosine level in the vasculature, leading to increased activation of A2B adenosine receptor, decreased vascular inflammation and atherosclerosis. Therefore, our data suggest that although adenine feeding is suitable for studying CKD development and CKD-induced cardiac hypertrophy and dysfunction, the model did not permit investigation of the relationship between CKD and atherosclerosis.
In summary, our study demonstrated that inhibition of TMAO production in an adenine-induced model of CKD significantly attenuated the development of CKD and cardiac hypertrophy. Our cell culture studies support the concept that TMAO promotes development of CKD by activation of NF-κB, leading to aggravated inflammation. Inhibition of TMAO production provides a novel strategy in the treatment CKD and reno-cardiac syndrome.
Materials and methods
Animal studies
All animal experiments were approved by the UCLA Animal Care and Use Committees, in accordance with PHS guidelines. One-month-old female apoE KO mice on C57BL/6J background (stock number: 002052, Jackson laboratory, Bar Harbor, ME), n = 10/group, were fed ab libitum the following diets from Envigo (Madison, WI) for 14 weeks before tissue collection: (1) chow diet (Con, TD.110846), (2) chow diet + 0.06% iodomethylcholine (IMC) (Con + IMC, TD.170932), (3) chow diet + 0.2% adenine (Ade, TD.170076), and (4) chow diet + 0.2% adenine + 0.06% IMC (Ade + IMC, TD.170093). At the 12th week of diet feeding, mice were individually placed in metabolic cages for collection of urine for 24 h. At the end of the diet feeding period, mice were fasted for 4 h before blood and tissue collection.
Biochemical assays
Plasma TMA, TMAO and creatinine levels were determined by mass spectrometry as previously described2. Indoxyl sulfate was determined by Shimadzu 8050 LC/mass spectrometer by using a phenyl column (250 × 2 mm, Thermo Scientific) and resolving by a gradient generated between A, 0.2% formic acid in water and B, 0.2% formic acid in methanol with the initial two minutes at 0% B, then linearly rose to 90% B over 7.3 min, then to 100% B over 0.5 min and held for 3 min, then back to 0% B and equilibrium for 3 min. Indoxyl sulfate (Cayman Chemical, Ann Arbor, MI) and its respective internal standard, indoxyl sulfate (ring-d4) were monitored using ESI in positive-ion mode with multiple reaction monitoring (MRM) of precursor and characteristic product ion transitions as: m/z 212 → 80 for indoxyl sulfate, 216 → 80 for indoxyl sulfate (ring-d4). Sample preparation for the measurement of indoxyl sulfate followed the same procedure as the measurement of TMAO with 1 μM indoxyl sulfate (ring-d4) added to the TMAO internal standard mix in methanol2. Plasma total cholesterol, HDL cholesterol, and triglyceride levels were determined as previously described27. Plasma urea levels were determined using a colorimetric assay (Bioassay Systems, Hayward, CA). Plasma FGF23 (Mouse/Rat FGF-23 (C-Term) ELISA kit, Quidel, San Diego, CA) and cystatin C (R&D Systems, Minneapolis, MN) levels were determined by ELISA. Urine albumin and creatinine levels were determined using an ELISA kit (Exocell, Philadelphia, PA) and a colorimetric assay kit (Exocell), respectively for calculation of UACR.
RNA isolation and quantitative RT-PCR analyses
Total RNA was isolated from tissue samples using the miRNA isolation kit (Qiagen, Germantown, MD) according to the protocol provided by the manufacturer. The cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Quantitative PCR was performed using gene-specific primers (Supplemental Table S3) and the Roche SYBR green master mix. Samples were run on a LightCycler 480 II system (Roche, Pleasanton, CA) and analyzed using the Roche LightCycler 1.5.0 software26. All qPCR targets were quantified based on standard curves ran on the sample plate. The mRNA levels of specific genes were normalized to the mRNA levels of housekeeping genes of the same sample. The mRNA levels of housekeeping genes eukaryotic translation initiation factor 2A (Eif2a, Fig. 2a), glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Fig. 2d,g), ribosomal protein L13a (Rpl13a, Fig. 3d), and β2 microglobulin (B2M, Supplemental Fig. S2) were used for normalization.
Kidney histology
Histologic Sections (5 μm) from formalin fixed paraffin embedded mouse kidneys (8 kidneys per group) were stained with Masson’s trichrome and quantitated for cortical scar area and collagen deposition in a blinded fashion. The cortical scarring and collagen deposition were measured over the entire cortical tubulointerstitial area via semi-quantitative visual assessment performed by a renal pathologist.
Atherosclerosis
Atherosclerotic lesions in the proximal aorta were quantitated as described56. Briefly, the heart was flushed with PBS and embedded in OCT. Frozen Sections (10 μM) were stained with Oil Red O and lesion area quantified every 6th section beginning at the aortic valves. The average of the first 10 scored sections was then used as a measure of lesion size.
RNA-seq
Total RNA from kidney was isolated as described above. RNA libraries were prepared using the Illumina TruSeq kits (Illumina, San Diego, CA). Following barcoding, 24 samples per lane were sequenced on a HiSeq4000 using 50 bp single-end protocol. Reads were QC’d using FastQC in batch mode and mapped to the mouse genome (mm10) using STAR aligner version 2.3.1. The count data were normalized using DESeq2′s median of ratios method57. Differential expression analysis was performed using DEseq258 with statistically significant genes called using adjusted p-value cutoffs of less than 0.1. Gene ontology analysis was performed using DAVID 6.8 (https://david.ncifcrf.gov/). The Benjamini–Hochberg method was used to obtain false discovery rates for enriched pathways.
Cell culture studies
The Madin-Darby canine kidney II (MDCK II, from Sigma-Aldrich, St. Louis, MO) epithelial cells were cultured in DMEM (Corning, CA) supplemented with 5% fetal calf serum (Hyclone, Logan, UT) and 100 U/ml penicillin/100 μg/ml streptomycin (Gibco, Carlsbad, CA). For TMAO treatment, cells were plated in 6-well plates for 24 h, followed by treatment with TMAO (0, 50, 100, or 200 μM, Sigma-Aldrich) in the presence or absence of an NF-κB inhibitor (NF-κB inhibitor IV, 100 nM, Millipore, Burlington, MA) for 48 h before gene expression and immunoblotting analyses.
Normal human primary renal proximal tubule epithelial cells (RPTEC, ATCC, Manassas, VA) were grown in renal epithelial cell basal media (ATCC PCS400030) supplemented with renal epithelial cell growth kit (ATCC PCS400040), and 100 U/ml penicillin/100 μg/ml streptomycin (Gibco, Carlsbad, CA) according to the supplier’s protocol. For TMAO treatment, cells were plated in 6-well plates for 24 h, followed by treatment with TMAO (0, 50, 100, 200, or 400 μM, Sigma-Aldrich) for 48 h before gene expression analysis.
Immunoblotting
Immunoblotting was performed using 20 μg of proteins from cell lysate per sample as previously described26. Primary antibodies against phospho-NF-κB p65, NF-κB p65, phospho-PERK, PERK, phospho-eIF2α, eIF2α, phospho-IRE1α, IRE1α, ATF4, and GAPDH (Cell Signaling Technology, Danvers, MA) were used in the experiments. After incubation with secondary antibody and extensive washing, blots were placed in Amersham ECL Prime Western Blotting Detection Reagent (GE Life Sciences). Blots were then imaged using the ChemiDoc MP system (Bio-Rad), and bands were quantified using ImageJ software (National Institutes of Health)26. Band intensities of phospho-NF-κB p65, phospho-PERK, phospho-eIF2α, and phospho-IRE1α were normalized by band intensities of total NF-κB p65, PERK, eIF2α, and IRE1α, respectively, of the same samples on the same blot after blot stripping and re-probing. Band intensities of ATF4 were normalized by band intensities of GAPDH of the same samples on the same blot.
Statistical analyses
Two-tailed Student’s t-test was utilized to compare means between 2 groups. One-way ANOVA with Tukey’s multiple comparisons test was performed to analyze data presented in Fig. 2d and Supplemental Fig. S2. For experiments with a two-factorial design (data presented in Figs. 1a–g,l,m, 2g, 3a–d, and Supplemental Fig. S1a and S1b), a two-way ANOVA was performed to establish that not all groups were equal. The Holm–Sidak post hoc analysis was then used for specific between-group comparisons after statistical significance was established by ANOVA. Statistical analyses were performed in GraphPad Prism version 8.
Supplementary Information
Acknowledgements
We thank Jennifer Buffa for excellent technical support. This work is supported by NIH Grants: 1R01 HL148110-01A1, 1R01 HL147883, 1RO1HL130819, and 1R01 HL144651-01, P01 HL147823, R01 HL103866 and UCLA Cardiovascular Theme‐CTSI Team Science Award. ZW and SLH were also supported in part by a Leducq Foundation Award.
Author contributions
W.Z.: carried out part of the experimental work; shared in manuscript writing. A.M.: carried out part of the experimental work. J. Z.: carried out part of the experimental work. X.J.: carried out part of the experimental work; Z.W.: carried out part of the experimental work. S.C.: carried out part of the experimental work. Z.Z.: carried out part of the experimental work. K.E.K.-U.: carried out data analysis. C.M.: carried out part of the experimental work. F.G.: carried out part of the experimental work. M.P.: supervised data analysis. S.L.H.: shared in experimental design. S.N.: shared in experimental design and obtaining funding. A.J.L.: shared in obtaining funding and manuscript writing. D.M.S.: shared in experimental design; carried out part of the experimental work; shared in obtaining funding and manuscript writing. All authors reviewed the manuscript.
Data availability
All data are contained within the manuscript. The RNA-seq data presented in this publication have been deposited in NCBI's Gene Expression Omnibus59 and are accessible through GEO Series accession number GSE163276 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE163276).
Competing interests
SLH and ZW report being named as co-inventors on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics and therapeutics. SLH also reports being a paid consultant for Procter & Gamble, having received research funds from Procter & Gamble, and Roche Diagnostics, and both SLH and ZW report being eligible to receive royalty payments for inventions or discoveries related to cardiovascular diagnostics or therapeutics from Cleveland Heart Lab (a subsidiary of Quest Diagnostics) and P&G. All other authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80063-0.
References
- 1.Bennett BJ, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell. Metab. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio. 2015;6:e02481. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rath S, Rud T, Pieper DH, Vital M. Potential TMA-producing bacteria are ubiquitously found in mammalia. Front. Microbiol. 2019;10:2966. doi: 10.3389/fmicb.2019.02966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li XS, et al. Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes: A prognostic marker for incident cardiovascular events beyond traditional risk factors. Eur. Heart J. 2017;38:814–824. doi: 10.1093/eurheartj/ehw582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiattarella GG, et al. Gut microbe-generated metabolite trimethylamine-N-oxide as cardiovascular risk biomarker: A systematic review and dose-response meta-analysis. Eur. Heart J. 2017;38:2948–2956. doi: 10.1093/eurheartj/ehx342. [DOI] [PubMed] [Google Scholar]
- 9.Qi J, et al. Circulating trimethylamine N-oxide and the risk of cardiovascular diseases: A systematic review and meta-analysis of 11 prospective cohort studies. J. Cell. Mol. Med. 2018;22:185–194. doi: 10.1111/jcmm.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Troseid M, et al. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J. Intern. Med. 2015;277:717–726. doi: 10.1111/joim.12328. [DOI] [PubMed] [Google Scholar]
- 11.Lever M, et al. Betaine and trimethylamine-N-oxide as predictors of cardiovascular outcomes show different patterns in diabetes mellitus: An observational study. PLoS ONE. 2014;9:e114969. doi: 10.1371/journal.pone.0114969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang WH, et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 2015;116:448–455. doi: 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stubbs JR, et al. Serum trimethylamine-N-oxide is elevated in CKD and correlates with coronary atherosclerosis burden. J. Am. Soc. Nephrol. 2016;27:305–313. doi: 10.1681/ASN.2014111063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heianza Y, Ma W, Manson JE, Rexrode KM, Qi L. Gut microbiota metabolites and risk of major adverse cardiovascular disease events and death: A systematic review and meta-analysis of prospective studies. J. Am. Heart Assoc. 2017 doi: 10.1161/JAHA.116.004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu W, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165:111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163:1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts AB, et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 2018;24:1407–1417. doi: 10.1038/s41591-018-0128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Organ CL, et al. Nonlethal inhibition of gut microbial trimethylamine n-oxide production improves cardiac function and remodeling in a murine model of heart failure. J. Am. Heart Assoc. 2020;9:e016223. doi: 10.1161/JAHA.119.016223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta N, et al. Targeted inhibition of gut microbial trimethylamine N-oxide production reduces renal tubulointerstitial fibrosis and functional impairment in a murine model of chronic kidney disease. Arterioscler. Thromb. Vasc. Biol. 2020;40:1239–1255. doi: 10.1161/ATVBAHA.120.314139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gansevoort RT, et al. Chronic kidney disease and cardiovascular risk: Epidemiology, mechanisms, and prevention. Lancet. 2013;382:339–352. doi: 10.1016/S0140-6736(13)60595-4. [DOI] [PubMed] [Google Scholar]
- 21.Go AS, Chertow GM, Fan D, McCulloch CE, Hsu CY. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N. Engl. J. Med. 2004;351:1296–1305. doi: 10.1056/NEJMoa041031. [DOI] [PubMed] [Google Scholar]
- 22.Xu KY, et al. Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients. Sci. Rep. 2017;7:1445. doi: 10.1038/s41598-017-01387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomlinson JAP, Wheeler DC. The role of trimethylamine N-oxide as a mediator of cardiovascular complications in chronic kidney disease. Kidney Int. 2017;92:809–815. doi: 10.1016/j.kint.2017.03.053. [DOI] [PubMed] [Google Scholar]
- 24.Rhee EP, et al. A combined epidemiologic and metabolomic approach improves CKD prediction. J. Am. Soc. Nephrol. 2013;24:1330–1338. doi: 10.1681/ASN.2012101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu X, et al. Regulation of renal fibrosis by Smad3 Thr388 phosphorylation. Am. J. Pathol. 2014;184:944–952. doi: 10.1016/j.ajpath.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Seldin MM, et al. Trimethylamine N-oxide promotes vascular inflammation through signaling of mitogen-activated protein kinase and nuclear factor-kappaB. J. Am. Heart Assoc. 2016 doi: 10.1161/JAHA.115.002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih DM, et al. Flavin containing monooxygenase 3 exerts broad effects on glucose and lipid metabolism and atherosclerosis. J. Lipid Res. 2015;56:22–37. doi: 10.1194/jlr.M051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warrier M, et al. The TMAO-generating enzyme flavin monooxygenase 3 is a central regulator of cholesterol balance. Cell. Rep. 2015;10:326–338. doi: 10.1016/j.celrep.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen ML, et al. Trimethylamine-N-oxide induces vascular inflammation by activating the NLRP3 inflammasome through the SIRT3-SOD2-mtROS signaling pathway. J. Am. Heart Assoc. 2017 doi: 10.1161/JAHA.117.006347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang SH, Reddick RL, Piedrahita JA, Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 1992;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 31.Bro S, et al. Chronic renal failure accelerates atherogenesis in apolipoprotein E-deficient mice. J. Am. Soc. Nephrol. 2003;14:2466–2474. doi: 10.1097/01.asn.0000088024.72216.2e. [DOI] [PubMed] [Google Scholar]
- 32.Buzello M, et al. The apolipoprotein e knockout mouse: A model documenting accelerated atherogenesis in uremia. J. Am. Soc. Nephrol. 2003;14:311–316. doi: 10.1097/01.asn.0000045048.71975.fc. [DOI] [PubMed] [Google Scholar]
- 33.Massy ZA, et al. Uremia accelerates both atherosclerosis and arterial calcification in apolipoprotein E knockout mice. J. Am. Soc. Nephrol. 2005;16:109–116. doi: 10.1681/ASN.2004060495. [DOI] [PubMed] [Google Scholar]
- 34.Elliot SJ, et al. Gender-specific effects of endogenous testosterone: Female alpha-estrogen receptor-deficient C57Bl/6J mice develop glomerulosclerosis. Kidney Int. 2007;72:464–472. doi: 10.1038/sj.ki.5002328. [DOI] [PubMed] [Google Scholar]
- 35.Si H, et al. Human and murine kidneys show gender- and species-specific gene expression differences in response to injury. PLoS ONE. 2009;4:e4802. doi: 10.1371/journal.pone.0004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rambausek M, Ritz E, Mall G, Mehls O, Katus H. Myocardial hypertrophy in rats with renal insufficiency. Kidney Int. 1985;28:775–782. doi: 10.1038/ki.1985.197. [DOI] [PubMed] [Google Scholar]
- 37.Vaziri ND, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83:308–315. doi: 10.1038/ki.2012.345. [DOI] [PubMed] [Google Scholar]
- 38.Chen YY, et al. Microbiome-metabolome reveals the contribution of gut-kidney axis on kidney disease. J. Transl. Med. 2019;17:5. doi: 10.1186/s12967-018-1756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, et al. Trimethylamine N-oxide binds and activates PERK to promote metabolic dysfunction. Cell. Metab. 2019;30:1141–1151. doi: 10.1016/j.cmet.2019.08.021. [DOI] [PubMed] [Google Scholar]
- 40.Verzola D, et al. Enhanced glomerular toll-like receptor 4 expression and signaling in patients with type 2 diabetic nephropathy and microalbuminuria. Kidney Int. 2014;86:1229–1243. doi: 10.1038/ki.2014.116. [DOI] [PubMed] [Google Scholar]
- 41.Lee FT, et al. Interactions between angiotensin II and NF-kappaB-dependent pathways in modulating macrophage infiltration in experimental diabetic nephropathy. J. Am. Soc. Nephrol. 2004;15:2139–2151. doi: 10.1097/01.ASN.0000135055.61833.A8. [DOI] [PubMed] [Google Scholar]
- 42.Okabe C, et al. NF-kappaB activation mediates crystal translocation and interstitial inflammation in adenine overload nephropathy. Am. J. Physiol. Renal Physiol. 2013;305:F155–163. doi: 10.1152/ajprenal.00491.2012. [DOI] [PubMed] [Google Scholar]
- 43.Fujihara CK, et al. Chronic inhibition of nuclear factor-kappaB attenuates renal injury in the 5/6 renal ablation model. Am. J. Physiol. Renal. Physiol. 2007;292:F92–99. doi: 10.1152/ajprenal.00184.2006. [DOI] [PubMed] [Google Scholar]
- 44.Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140:871–882. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- 45.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 46.LeBleu VS, et al. Identification of human epididymis protein-4 as a fibroblast-derived mediator of fibrosis. Nat. Med. 2013;19:227–231. doi: 10.1038/nm.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuo S, et al. Multifunctionality of PAI-1 in fibrogenesis: Evidence from obstructive nephropathy in PAI-1-overexpressing mice. Kidney Int. 2005;67:2221–2238. doi: 10.1111/j.1523-1755.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 48.Oda T, et al. PAI-1 deficiency attenuates the fibrogenic response to ureteral obstruction. Kidney Int. 2001;60:587–596. doi: 10.1046/j.1523-1755.2001.030002587.x. [DOI] [PubMed] [Google Scholar]
- 49.Takawale A, et al. Tissue inhibitor of matrix metalloproteinase-1 promotes myocardial fibrosis by mediating CD63-integrin beta1 Interaction. Hypertension. 2017;69:1092–1103. doi: 10.1161/HYPERTENSIONAHA.117.09045. [DOI] [PubMed] [Google Scholar]
- 50.Vervloet M. Renal and extrarenal effects of fibroblast growth factor 23. Nat. Rev. Nephrol. 2019;15:109–120. doi: 10.1038/s41581-018-0087-2. [DOI] [PubMed] [Google Scholar]
- 51.Faul C, et al. FGF23 induces left ventricular hypertrophy. J. Clin. Invest. 2011;121:4393–4408. doi: 10.1172/JCI46122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia T, et al. A novel model of adenine-induced tubulointerstitial nephropathy in mice. BMC Nephrol. 2013;14:116. doi: 10.1186/1471-2369-14-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kieswich JE, et al. A novel model of reno-cardiac syndrome in the C57BL/6 mouse strain. BMC Nephrol. 2018;19:346. doi: 10.1186/s12882-018-1155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koupenova M, Johnston-Cox H, Ravid K. Regulation of atherosclerosis and associated risk factors by adenosine and adenosine receptors. Curr. Atheroscler. Rep. 2012;14:460–468. doi: 10.1007/s11883-012-0263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang D, et al. The A2B adenosine receptor protects against inflammation and excessive vascular adhesion. J. Clin. Invest. 2006;116:1913–1923. doi: 10.1172/JCI27933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shih DM, et al. Combined serum paraoxonase knockout/apolipoprotein E knockout mice exhibit increased lipoprotein oxidation and atherosclerosis. J. Biol. Chem. 2000;275:17527–17535. doi: 10.1074/jbc.M910376199. [DOI] [PubMed] [Google Scholar]
- 57.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript. The RNA-seq data presented in this publication have been deposited in NCBI's Gene Expression Omnibus59 and are accessible through GEO Series accession number GSE163276 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE163276).