Abstract
Viruses have evolved strategies to ensure efficient translation using host cell ribosomes and translation factors. In addition to cleaving translation initiation factors required for host cell translation, poliovirus (PV) uses an internal ribosome entry site (IRES). Recent studies suggest that viruses exploit specific ribosomal proteins to enhance translation of their viral proteins. The ribosomal protein receptor for activated C kinase 1 (RACK1), a protein of the 40S ribosomal subunit, was previously shown to mediate translation from the 5′ cricket paralysis virus and hepatitis C virus IRESs. Here we found that translation of a PV dual-luciferase reporter shows a moderate dependence on RACK1. However, in the context of a viral infection we observed significantly reduced poliovirus plaque size and titers and delayed host cell translational shut-off. Our findings further illustrate the involvement of the cellular translational machinery during PV infection and how viruses usurp the function of specific ribosomal proteins.
Introduction
Since viruses rely on cellular translation factors and ribosomes for the synthesis of viral proteins, viruses battle with host cells for these critical resources. Viral double-stranded RNA intermediates activate interferon-induced, double-stranded RNA-activated protein kinase (PKR), which phosphorylates translation initiation factor eIF2α leading to inhibition of viral and cellular translation (Pakos-Zebrucka et al., 2016; Wek, 2018; Wek et al., 2006). To prevent eIF2α phosphorylation and translational shut-off, viruses target PKR. Some viral proteins directly bind to PKR to prevent its activity, other viruses degrade PKR or alter its subcellular localization (Black et al., 1989; Clarke et al., 1991; Ghadge et al., 1991; Hebner et al., 2006; Langland et al., 1994; Sharp et al., 1993). To efficiently compete for ribosomes, many viruses use translation initiation mechanisms distinct from cellular cap-dependent mRNA translation initiation. All cellular mRNAs are transcribed in the nucleus, where they are also capped and polyadenylated. After export into the cytoplasm, the 5′ m7GpppN cap is bound by the cap binding protein eukaryotic initiation factor 4E (eIF4E) and the polyA-tail is bound by the polyA binding protein (PABP). Through binding of the scaffolding protein eIF4G to eIF4E and PABP, the mRNA is circularized. With help of the RNA helicase eIF4A, the 40S ribosomal subunit in complex with eIF2 and eIF3 scans the 5′ untranslated region (UTR) in an ATP-dependent manner until the start codon is reached and recognized. After 60S ribosomal subunit joining and GTP hydrolysis by eIF5B elongation can proceed. To prevent cap-dependent translation, poliovirus (PV) and other viruses of the Picornaviridae family target these eIFs. Specifically, PV proteases 2A and 3C cleave eIF4G, and PABP and eIF5B, respectively (Bonderoff et al., 2008; de Breyne et al., 2008; Kuyumcu-Martinez et al., 2004, 2002; Svitkin et al., 1999; Ventoso et al., 1998; White et al., 2011). Cleavage of these essential translation factors shuts off host cell translation, while PV uses an internal ribosome entry site (IRES), which does not rely on these translation factors, for synthesis of the viral polyprotein (Pelletier and Sonenberg, 1988; White et al., 2011). Hence, viruses employ strategies that specifically decrease translation of cellular mRNAs but allow for translation of viral RNAs.
In addition to targeting translation initiation factors, several viruses have shown direct usage of ribosomal proteins to increase translation of their viral mRNAs. Lee et al. performed an siRNA screen and identified eight ribosomal proteins including eL40, that are not required for cell viability, but negatively affect Vesicular Stomatitis Virus (VSV) and other related viruses (Lee et al., 2013). Ribosomal protein eL40 was dispensable for viruses that use IRES-mediated translation, but regulated a subset of cellular mRNAs with diverse functions (Lee et al., 2013). In contrast to eL40, eS25, a protein located near the head of the 40S ribosomal subunit, mediates translation of viral mRNAs that initiate translation using IRESs (Hertz et al., 2013; Landry et al., 2009). eS25 directly interacts with the hepatitis C virus (HCV) and the intergenic (IGR) cricket paralysis virus (CrPV) IRESs in cryo-EM structures (Muhs et al., 2011; Nishiyama et al., 2007; Spahn et al., 2001; Yamamoto et al., 2015) and is required for high-affinity binding of the 40S ribosomal subunit to the CrPV IRES (Landry et al., 2009). Further, eS25 not only facilitates translation of other IRESs-containing RNAs such as encephalomyocarditis virus (EMCV) and PV, but also regulates translation of cellular IRES-containing mRNAs (Hertz et al., 2013). More recently, another ribosomal protein, receptor for activated C kinase 1 (RACK1) has been shown to be exploited by different viruses.
RACK1 is a core ribosomal protein (Gerbasi et al., 2004) that belongs to the tryptophan-aspartate repeat (WD-repeat) protein family. The seven-bladed β-propeller structure of RACK1 is located near the mRNA exit tunnel where it makes contacts with the ribosomal RNA through lysine and arginine residues and neighboring ribosomal proteins (Anger et al., 2013; Coyle et al., 2009; Sengupta et al., 2004). RACK1 is often termed a scaffolding protein and has been implicated in a variety of biological functions on and off the ribosome. In addition to binding to its eponym protein kinase C βII (PKCβII) and being involved in cellular signaling via Src protein-tyrosine kinase (Chang et al., 2001; Dobrikov et al., 2018a, 2018b), RACK1 has been shown to interact with the microRNA machinery (Jannot et al., 2011), bind eIF6 to regulate the 60S ribosomal subunit (Ceci et al., 2003) and regulate ribosome-associated quality control (Matsuda et al., 2014; Sundaramoorthy et al., 2017). At the level of tissues and organisms, RACK1 regulates axonal growth (Kershner and Welshhans, 2017), neural tube closure in Xenopus laevis (Wehner et al., 2011), and is essential for development in mice (Volta et al., 2013), Drosophila melanogaster (Kadrmas et al., 2007) and Arabidopsis thaliana (Guo et al., 2011; Guo and Chen, 2008), but appears to be dispensable in single cell organisms such as yeast (Coyle et al., 2009). Directly and/or indirectly, RACK1 also influences translation of cellular mRNAs. The Saccharomyces cerevisiae RACK1 homolog, Asc1, facilitates efficient translation of mRNAs containing a short open reading frame (Thompson et al., 2016), while in mammalian cells, RACK1 appears to stimulate cell proliferation in a PKCβII-dependent manner (Dobrikov et al., 2018a; Grosso et al., 2008; Romano et al., 2019).
At the intersection of cellular signaling and translational regulation, RACK1 represents a critical regulatory target for many viruses. Vaccinia virus, which belongs to the poxviruses and contains a dsDNA genome, but replicates exclusively in the cytoplasm, and encodes a kinase that phosphorylates a flexible loop in RACK1 (Jha et al., 2017). Through phosphorylation, this RACK1 loop becomes negatively charged, which allows for translation of the poxvirus polyA-leader containing mRNAs (Jha et al., 2017). In plants, where polyA-leader sequences are commonly found, this RACK1 loop contains several glutamic acid residues, hence poxvirus evolution likely rediscovered efficient translation of polyA-leaders through phosphorylation of RACK1. Viruses from the Dicistroviridae family encode two polyproteins, and translation of each polyprotein is mediated by an IRES (Hertz and Thompson, 2011). In contrast to eS25, RACK1 is dispensable for translation mediated by the CrPV IGR IRES, but its loss inhibits translation initiation from both the 5′ IRES of CrPV as well as the HCV IRES (Majzoub et al., 2014).
The finding that RACK1 facilitates efficient HCV IRES-mediated translation prompted us to explore if the need for RACK1 is more broadly conserved. Using a RACK1 knockout cell line generated by CRISPR-Cas9 genome editing (Jha et al., 2017), we first tested translation efficiency using dual-luciferase constructs. We found that HCV, EMCV and PV IRES-mediated translation are all reduced in cells lacking RACK1. Although the effect on PV IRES-mediated translation in the context of the dual-luciferase reporter is moderate, loss of RACK1 causes a significant decrease in the PV plaque size and virion release. This decrease is due to attenuated translation prior to and post translational shut-off, possibly lengthening the virus life cycle in cells lacking RACK1.
Results
RACK1-FLAG is incorporated into polysomes
To investigate the function of mammalian RACK1 in translation, we established a functional rescue by expressing RACK1-FLAG in HAP1-derived CRISPR genome edited RACK1 knockout cell line RACK1 KO #1 described by Jha et al. (Jha et al., 2017) using lentiviral transduction (Johnson et al., 2019). HAP1 cells are a near-haploid human cell line derived from chronic myelogenous leukemia KBM-7 cells (Carette et al., 2011). RACK1 was undetectable in RACK1 KO #1 and RACK1 KO #2 cell lines. Following lentiviral transduction of RACK1-FLAG into RACK1 KO #1 cells, RACK1 levels were partially restored (figure 1A). To examine incorporation of FLAG-tagged RACK1 into translating ribosomes rather than other high molecular weight cytosolic complexes, we performed polysome analysis by sucrose gradient ultracentrifugation. When cell lysate is treated with the translation elongation inhibitor cycloheximide, translation will be stalled. Upon sucrose gradient ultracentrifugation, the translating ribosomes, polysomes, are separated from the ribosomal subunits. When sucrose gradient analysis was performed on wildtype HAP1 and RACK1-FLAG expressing RACK1 KO #1 cells, no major differences in the overall polysome profile and in the distribution of RACK1 within the sucrose gradient were detected (figures 1B and 1C). Cells lacking RACK1 still had significant polysome levels, but the heaviest polysomes were decreased, causing a marked shift in the overall profile (supplementary figure 1). Polysome distribution was rescued in RACK1-FLAG expressing RACK1 KO #1 cells, while polysome levels were partially rescued. Since we did not observe changes in translation by metabolic labeling (Jha et al., 2017, figure 4A), this result suggests that RACK1 alters the translation of a subset of mRNAs. We found that wildtype RACK1 and RACK1-FLAG co-sedimented in the polysomal fractions 9-14 (figure 1B and figure 1C, left panel) with eS25 and uL13, ribosomal proteins of the 40S and 60S ribosomal subunits, respectively. Although this result suggested that RACK1-FLAG was incorporated into polysomes, we could not exclude that it sedimented in heavy sucrose fractions because it formed aggregates. To further validate that RACK1-FLAG indeed was incorporated into polysomes, we treated cell lysate with puromycin. Puromycin is a tRNA analog, which stalls translation elongation and releases the growing peptide chain. When cell lysate treated with puromycin is heated to 37°C, the two ribosomal subunits are separated, and the mRNA is released (Blobel and Sabatini, 1971). Puromycin treatment alters the sedimentation pattern for ribosomal proteins, which now sediment in lighter sucrose fractions where the ribosomal protein subunits are found. Following puromycin treatment, RACK1-FLAG now sedimented in the lighter sucrose gradient fractions 3 and 4 (figure 1C, right panel), where it again co-sedimented with eS25. Taken together, these results indicate that RACK1-FLAG is incorporated into translating ribosomes and likely fully functional.
Figure 1: RACK1-FLAG is incorporated into polysomes.
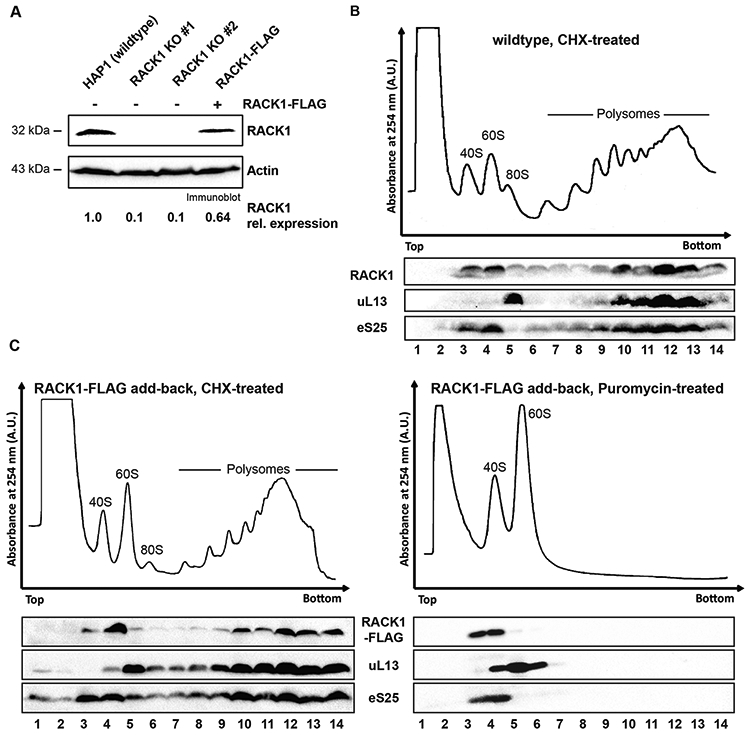
(A) RACK1 levels can be partially restored in RACK1 KO cells by expression of RACK1-FLAG. RACK1 levels were quantified by immunoblot analysis of RACK1 and the loading control actin. (B) Polysome trace of HAP1 wildtype cells and distribution of the ribosomal proteins RACK1, eS25 and uL13 in the gradients. Cell lysate was separated in 10-50% sucrose gradient and absorbance at 254 nm was measured. Proteins from all gradient fractions were analyzed by immunoblotting All ribosomal proteins sediment in fractions according to the distribution of ribosomal subunits and polysomal fractions in the trace. (C) Sucrose gradient analysis of FLAG-tagged RACK1 protein. In cells treated with the translation elongation inhibitor cycloheximide, RACK1-FLAG, detected by immunoblotting using an anti-FLAG antibody, co-sediments with polysomes (fractions 9-14). Upon treatment of cell lysate with the translation elongation puromycin, which separates actively translating ribosomes into the ribosomal subunits, RACK1-FLAG co-sediments with 40S ribosomal subunits in lighter sucrose gradient fractions (fractions 3-4). Immunoblot analysis for eS25 and ul13 are used as indicators for sedimentation of protein components of the small and large ribosomal subunits, respectively.
Figure 4: RACK1 aids PV translation prior and post host cell translational shut-off.
(A) 35S metabolic pulse labeling of uninfected (mock) and PV-infected cells. Cells were mock-infected or infected with PV Mahoney at MOI of 1 (MOI of 10 for positive control) and 35S pulse-labeled for 10 min at 185 minutes post infection. Total protein lysates were separated in 10% SDS-PAGE and visualized by exposure to a phosphor-screen. Poliovirus-specific protein products P1, 3CD, and 2C are indicated (Florez et al., 2005). (B) Same experiment as in (A), but at MOI of 10 and labeled at 160 minutes post infection. (C) Immunoblot analysis for expression of poliovirus proteins 3CD and 3D following infection at MOI of 1 and MOI of 10. (D) Expression of a PV-Luc replicon is inefficient in cells lacking RACK1. HAP1, RACK1 KO #1, and RACK-1 FLAG cells were transfected with in vitro transcribed PV-Luc replicon RNA. An aliquot of cells was removed 3, 5, 7, and 9 hours post RNA transfection and Firefly luminescence was measured. Error bars represent the standard error of the mean of at least three independent experiments. Left panel: PV-Luc translation is decreased in cells lacking RACK1 during the replication phase. Middle panel: To limit the observation to the early translation phase, PV-Luc replication was inhibited with 2 mM guanidine hydrochloride immediately after RNA transfection. Right panel: Translation of all PV-Luc reporters is completely inhibited upon treatment with 25 μg/ml cycloheximide. Cells were treated with cycloheximide immediately after RNA transfection.
RACK1 mediates translation of viral IRESs
Loss of RACK1 has been previously shown to inhibit HCV and CrPV 5′ IRES-mediated translation (Majzoub et al., 2014) raising the possibility that RACK1 generally facilitates viral IRES-mediated translation. To test this hypothesis, we used dicistronic luciferase reporters, in which translation of the Renilla luciferase uses canonical cap-dependent translation initiation, while translation of the Firefly luciferase is mediated by a viral IRES (figure 2A). We tested the importance of RACK1 for Firefly luciferase translation from four viral IRESs, specifically PV, EMCV, HCV and CrPV intergenic IRESs. These IRESs represent four major types of viral IRESs and use different mechanisms for translation initiation (Balvay et al., 2009). None of these viral IRESs use the cap-binding function of eIF4E, although a recent study showed that eIF4E stimulates the helicase activity of eIF4A on the PV IRES independent of its cap-binding function (Avanzino et al., 2017). In contrast to PV, neither the EMCV nor the HCV IRES use a scanning mechanism but instead directly recruit the ribosome to the start codon (Kieft, 2008). While the PV and EMCV IRESs require all canonical translation initiation factors except for eIF4E, HCV IRES-mediated translation initiation uses a subset of translation initiation factors. In agreement with previous observations, loss of RACK1 did not alter CrPV intergenic (IGR) IRES-mediated translation, but inhibited HCV IRES-mediated translation (figures 2B and 2C, and (Majzoub et al., 2014)). In addition, we observed that RACK1 also facilitated Firefly luciferase translation from the EMCV and PV IRESs (figures 2D and 2E). Expression of RACK1-FLAG in RACK1 knockout cells partially rescued the defect in IRES-mediated translation (figure 2C-E). Taken together, these data indicate that RACK1 to aids translation from HCV, EMCV and PV IRESs, but not from the CrPV IGR IRES.
Figure 2: RACK1 aids in the translation of viral IRESs.
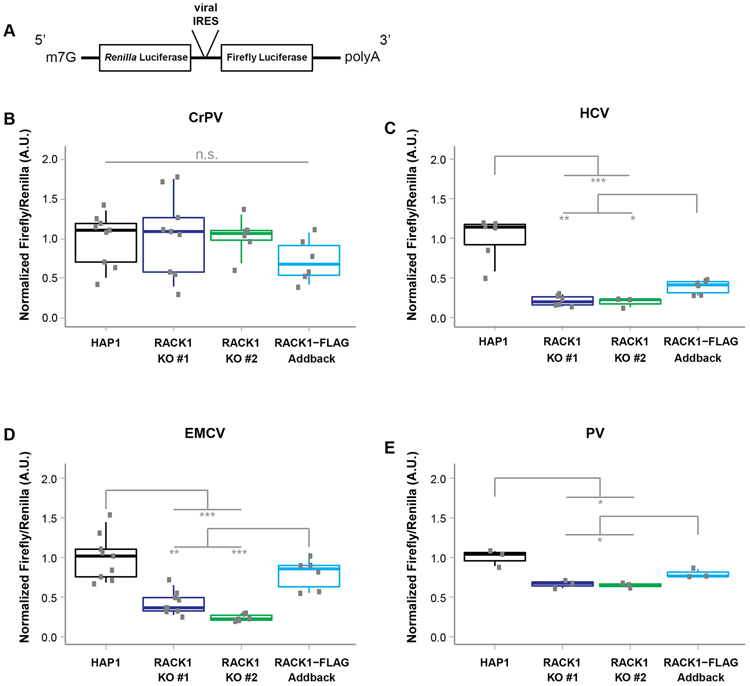
(A). Schematic overview of dual luciferase construct used in assays. The Renilla luciferase open reading frame is translated via m7G cap-dependent translation, while the viral IRES located in the intergenic region between the two coding sequences mediates translation of the Firefly luciferase open reading frame. (B-E) Translation efficiency of CrPV, HCV, EMCV and PV dual luciferase reporters in wildtype, RACK1 knockout and RACK1-FLAG addback cells. Single data points of at least three independent experiments were plotted. The box indicates the data between the interquartile range (IQR) between the 25th and 75th percentile, the median is indicated by the thick line within the box. The thin vertical bars represent the 95th percentiles ** p-value < 0.01; *** p-value < 0.001.
PV plaque diameters are decreased in cells lacking RACK1
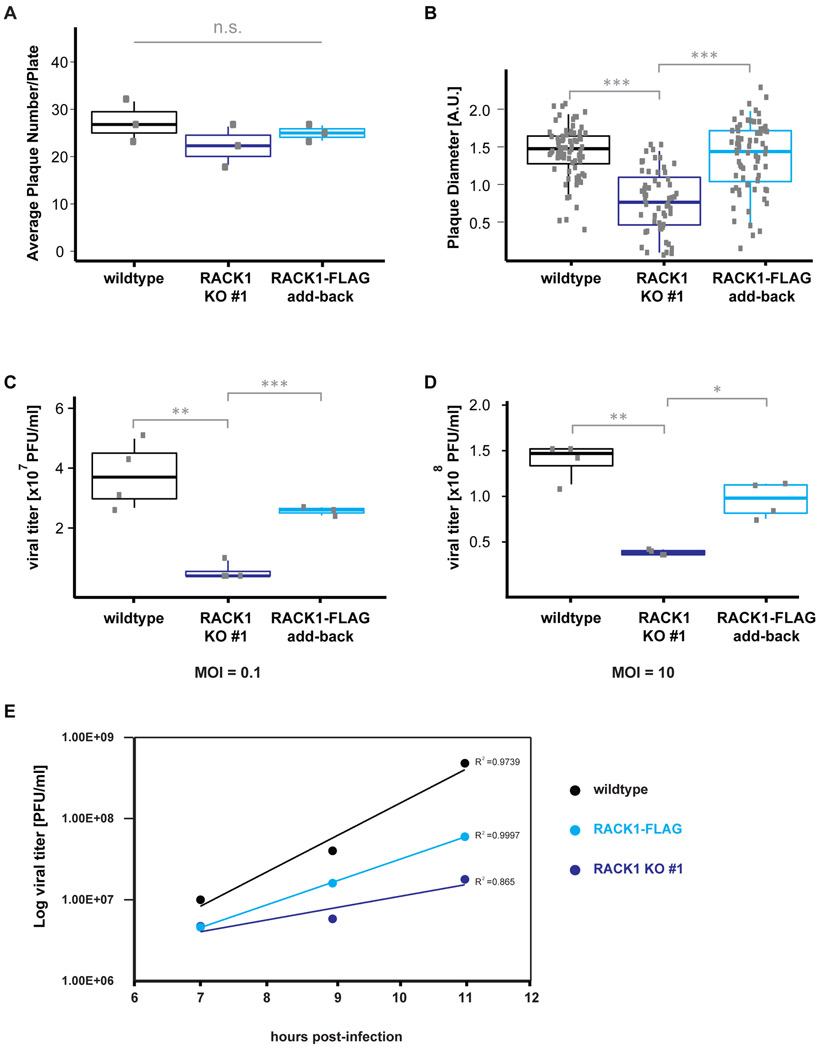
To test if the reduction of PV IRES-mediated translation impacts the virus during infection, we performed PV plaque assays in wildtype, RACK1 KO #1, and RACK1-FLAG add-back cells and measured both PV plaque numbers and plaque diameter. Following infection with the Mahoney strain of PV, we observed that the number of PV plaques was not significantly altered in any of the cell lines (figure 3A). In contrast, we observed a significant decrease in the PV plaque size in cells lacking RACK1 as compared to wildtype and RACK1-FLAG add-back cells (figure 3B). Together, these data indicate that infectious particles are similarly efficient at establishing an infection independent of cellular RACK1 levels, however, the infectious cycle and virus spread may be impaired. To further test if loss of RACK1 decreases poliovirus production, we infected cells with the Mahoney strain of poliovirus at the multiplicity of infection (MOI) of 0.1 and the MOI of 10, harvested the intracellular virus 5 hours post infection, and determined the viral titers from wildtype, RACK1 KO and RACK1-FLAG add-back cells in HeLa cells. We observed an approximately four-fold decrease in poliovirus titers in the RACK1 KO cells compared to wildtype cells, while the viral titers from RACK1-FLAG add-back cells were almost at the levels found in wildtype cells (figures 3C-D). The lower poliovirus titers correlate to the height of the virion peak detected in a sucrose gradient. The virion peak in the RACK1 KO#1 is smaller than the virion peak in wildtype and RACK1-FLAG cells, further correlating the loss of RACK1 to decreased poliovirus production (supplementary figure 2). We next performed a one-step growth curve, measuring virion release and virion titers in the media by plaque assay. We observed virion release 7 hours post infection, which agrees with the observed delayed cytopathic effect in HAP1 cells. PV virion production 11 hours post infection is highest in wildtype cells; virion production is decreased by approximately 1.5 orders of magnitude in RACK1 knockout cells and partially rescued in RACK1-FLAG addback cells (figure 3E).
Figure 3: PV plaque size is reduced in cells lacking RACK1.
(A) Analysis of PV plaque numbers. The number of visible poliovirus plaques does not differ between wildtype, RACK1 KO #1 and RACK1-FLAG cell lines. Cells were infected at identical MOI; 42h post infection, cells were stained with crystal violet and poliovirus plaques were counted. Single data points of at least three independent experiments were plotted. The box indicates the data between the interquartile range (IQR) between the 25th and 75th percentile, the median is indicated by the thick line within the box. The thin vertical bars represent the 95th percentiles. (B) PV plaques in RACK1 KO #1 cells are smaller compared to PV plaques in wildtype and RACK1-FLAG addback cells. Cells were infected at identical MOI; 42h post infection, cells were stained with crystal violet and poliovirus plaque diameters were measured. The box indicates the data between the interquartile range (IQR) between the 25th and 75th percentile, the median is indicated by the thick line within the box. The thin vertical bars represent the 95th percentiles. (C, D) Quantification of poliovirus titers after a single round of infection at MOI of 0.1 (C) and MOI of 10 (D). Poliovirus titers from RACK1 KO #1 cells are decreased to HAP1 wildtype and RACK1-FLAG addback cells. The box indicates the data between the interquartile range (IQR) between the 25th and 75th percentile, the median is indicated by the thick line within the box. The thin vertical bars represent 95th percentiles. * p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001 (E) Virion concentrations in the media of a one-step growth curve were determined by plaque assay from wildtype, RACK1 KO #1 and RACK1-FLAG addback cells.
Our results confirmed that virus production was inefficient in cells lacking RACK1 raising the question what step of the viral life cycle requires RACK1. Because of the role of RACK1 as a ribosomal protein, we investigated whether loss of RACK1 altered translation of the viral genome only prior to replication or also during the replication phase.
Loss of RACK1 impairs PV translation during the entire virus life cycle
Upon PV infection, the PV genome must be translated to give rise to the viral proteins, which include the viral proteases 2A and 3C. When levels of 2A and 3C are sufficiently high, these viral proteases cleave translation factors eIF4G and PABP, which shuts off translation in the host cell. Our observation that loss of RACK1 decreases PV IRES-mediated translation made us wonder whether loss of RACK1 may delay host-cell translation shut-off. To test whether translational shut-off was delayed in RACK1 KO cells, we performed 35S pulse-labeling experiments in mock-infected and PV-infected cells at MOI of 1. When newly synthesized proteins were metabolically labeled 185 minutes post infection and lysates resolved by SDS-PAGE we observed that wildtype and RACK1-FLAG add-back cells started to show the characteristic PV banding pattern observed in the positive control of cells infected at MOI of 10 and harvested at the same time (figure 4A) (White et al., 2011). In contrast, the protein pattern in the RACK1 KO cell lines was similar to the pattern in the mock-infected cells, in that mostly cellular proteins were visualized by metabolic labeling at the time of harvest. These data indicate that the time until PV-induced translational shut-off of the host cell was indeed delayed (figure 4A). We next determined if RACK1 KO cells are generally able to shut-off host cell translation by infecting cells at MOI of 10 and labeling newly synthesized proteins using 35S (figure 4B). We observed that all cell lines showed the characteristic PV protein products P1, 3CD and 2C 160 min post infection. Wildtype cells did no longer show the cellular protein bands marked by an asterisk, while RACK1 KO cells and RACK1-FLAG addback cell lines showed these products, indicating that translational shut-off had not proceeded as fast as in the wildtype cells (figure 4B). We further validated the metabolic labeling results by detection of viral proteins 3CD and 3D by immunoblotting. In agreement with the previous metabolic labeling results, production of PV proteins 3CD and 3D was decreased in RACK1 KO cells, while levels were higher in wildtype and RACK1-FLAG addback cell lines (figure 4C).
We next asked if PV translation also remained at lower levels in RACK1 KO cells following translational shut-off. To address this question, we monitored translation of a PV-Luciferase replicon (PV-Luc) during the initial phase of translation and translation during viral replication. Instead of the viral structural proteins, the PV-Luc replicon encodes a luciferase open reading frame fused to the PV non-structural proteins. Early during infection, luciferase measurements reveal initial translation of the replicon. Once negative strand synthesis has occurred, the PV genome will start replicating, which will result in greater translation of the replicon and largely increased luciferase production. We transfected in vitro transcribed PV-Luc RNA into wildtype, RACK1 KO #1 and RACK1-FLAG cells, harvested protein lysates 3, 5, 7, and 9 hours post transfection and measured luciferase levels by luminescence. As expected, we observed robust luciferase production in wildtype cells, while luciferase levels in RACK1 KO #1 remained more than 10-fold lower (figure 4D, left panel). Consistent with our previous results, RACK1-FLAG expression in the KO cell line partially rescued luciferase expression. To help distinguish viral translation and replication stages, we immediately treated cells with guanidine hydrochloride, a drug that inhibits viral replication (Powers et al., 1969). Thus, the measured amount of luciferase produced in these experiments only represents protein production prior to viral replication. Although luciferase levels in RACK1 KO #1 and RACK1-FLAG cells were comparable at 3 hours post transfection, luciferase levels in the RACK1-FLAG cell line increased over time, reaching luciferase levels more comparable wildtype cells, while cells lacking RACK1 KO #1 failed to accumulate significant levels of luciferase (figure 4D, middle panel). When cells were treated with the translation inhibitor cycloheximide immediately after PV-Luc reporter transfection, luciferase levels measured in all cells were at background levels (figure 4D, right panel), indicating that translation of the viral genome had been completely blocked.
Taken together, these data indicate that RACK1 not only aids HCV IRES-mediated translation but also translation from the EMCV and PV IRESs as evident in dicistronic luciferase assays. Cells lacking RACK1 show reduced intracellular PV concentration and reduced plaque size, likely due to inefficient PV translation prior to as well as post host cell translational shut-off.
Discussion
The ribosomal protein RACK1 interacts with numerous cellular proteins and has been thought to function as a scaffolding protein that connects cellular signaling pathways with the ribosome and the translation machinery (Nielsen et al., 2017). In addition, it has been previously shown that RACK1 is important for HCV IRES-mediated translation (Majzoub et al., 2014), which prompted us to test whether RACK1 may be generally aiding viral IRES-mediated translation. To test the involvement of RACK1 in IRES-mediated translation, we used viral dicistronic luciferase constructs in wildtype, RACK1 KO and RACK1-FLAG cell lines (figure 2A). In agreement with previous work by Majzoub et al., translation from the CrPV IGR IRES occurred in a RACK1-independent manner (Majzoub et al., 2014), while RACK1 aids translation from the HCV EMCV and PV IRESs (figure 2B-E). Plaque assays with the Mahoney strain of PV revealed that almost all infectious particles are able to start a successful infection (figure 3A) but that PV plaques and intracellular virus concentrations are significantly smaller in cells lacking RACK1 (figure 3B-E). These findings indicate that RACK1 is dispensable for PV entry, but suggests that RACK1 enhances PV infection. Several groups have found RACK1 to regulate autophagy by directly interacting with Atg5 (Erbil et al., 2016) and by enhancing Atg14L-Beclin 1-Vps34-Vps15 complex formation (Zhao et al., 2015). However, neither Atg5 nor Beclin 1 impacted PV proliferation (Abernathy et al., 2019) making it unlikely that the observed phenotype is due to changes in the autophagy pathway. Another possibility is that RACK1 affects cap-dependent translation, which could indirectly affect PV proliferation. In contrast to others who have reported that RACK1 depletion reduces cap-dependent translation (Dobrikov et al., 2018a; Gallo et al., 2018), we do not observe major changes in cap-dependent translation in RACK1 KO cells by 35S metabolic labeling ((Jha et al., 2017), figure 4A and unpublished data). However, we observed an altered polysome profile, which indicates that translation of specific mRNAs is altered in cells lacking RACK1 (supplementary figure 1). It is further possible that the contrasting observations are due to cell line specific differences. While HAP1 cells are derived from the chronic myelogenous leukemia (CML) cell line KBM-7, all studies that found that RACK1 influenced cap-dependent and cap-independent translation were performed in HEK293, HEK293T, and HeLa cells (Dobrikov et al., 2018a, 2018b; Gallo et al., 2018). Since RACK1 stimulates global translation in a PKCβII-dependent manner (Dobrikov et al., 2018a; Grosso et al., 2008), potentially higher PKCβII mRNA expression levels found in HEK293 and HeLa cells or other cell line-specific differences might explain the opposing effect on translation ((Uhlen et al., 2015), https://www.proteinatlas.org/ENSG00000166501-PRKCB/cell). Further, Dobrikov et al. had stimulated PKCβII through 12-O-Tetradecanoyl-phorbol-13-acetate (TPA) treatment, and it is possible that RACK1 increases cap-dependent translation during PKCβII stimulation, but not in the absence of such stimulus (Dobrikov et al., 2018a).
In contrast to the moderately decreased translation of the dual luciferase construct (figure 2E), we observed more dramatic effects with significant decreases in intracellular and extracellular virus concentrations (figures 3 C-E). Such observation is not uncommon since an early translation defect as measured by the dicistronic luciferase assay is amplified by increased translation during viral replication. An early defect in translation also lengthens the time until critical quantities of poliovirus proteases 2A and 3C are produced. These two proteases cleave translation factors eIF4G and PABP, respectively, cause host cell translation shut-off and enable viral replication (Bonderoff et al., 2008; Gradi et al., 1998; Svitkin et al., 1999). Indeed, in cells lacking RACK1, viral protein production was decreased and translational shut-off of the host cell was delayed (figures 4A-C). In addition, RACK1 might also cause a defect in virion assembly or a defect in viral replication. However, the PV replicon, which does not encode the structural proteins required for virion assembly, shows a severe defect in luciferase translation in the absence of RACK1 (figure 4D), suggesting that the decrease in intracellular and extracellular virus is independent of the presence of structural proteins and makes it unlikely that RACK1 affects virion assembly. RACK1 might also affect replication, either directly due to its role in cell signaling (Adams et al., 2011; Gandin et al., 2013) or indirectly through polioviral 2A protease. Protease 2A mutants deficient in trans-, but not cis- cleavage activities are deficient in RNA replication (Yu et al., 1995), suggesting that lower levels of protease 2A might limit viral replication in RACK1 KO cells. Taken together, our data indicate that RACK1 is not essential for PV translation, but that it enhances PV translation efficiency. While we cannot exclude that RACK1 does not also function during another step of the virus life cycle, our data show that RACK1 acts as a pro-viral host protein during poliovirus infection. Following expression of RACK1-FLAG, we observed partial expression and restoration of the phenotype. The partial rescue is likely due to suboptimal lentivirus transduction and not caused by inhibition of RACK1 through the C-terminal FLAG tag since other C-terminal tags on RACK1 have been well tolerated (Gallo et al., 2018; Johnson et al., 2019), However, we cannot rule out that the positively charged FLAG-tag does not affect the RACK1-ribosome interaction, RACK1 binding to other cellular proteins, or RACK1 homodimerization or heterodimerization with eIF6 (Ceci et al., 2003; Yatime et al., 2011). Although the functional relevance of RACK1 dimerization is unclear, RACK1 homodimerization has been implicated in mediating stress signaling (Sabila et al., 2016).
Interestingly, our findings are reminiscent of ribosomal protein eS25, previously shown to mediate both viral and cellular IRES-mediated translation (Hertz et al., 2013; Landry et al., 2009). Both eS25 and RACK1 have a greater impact on HCV IRES translation, reducing IRES activity by about 75%, while PV and EMCV IRES-mediated translation is less sensitive to eS25 and RACK1 levels. Similarly, though, shRNA-mediated depletion of eS25 resulted in a modest 2-fold decrease in intracellular PV viral titers (Hertz et al., 2013), which indicates that loss of eS25 may negatively impact the viral life cycle prior and post translational shut-off, however, no PV plaque size of a direct virus plaque assay was reported.
Although both RACK1 and eS25 are ribosomal proteins that are usurped by viruses to enhance viral translation, their mechanisms appear to be distinct. In contrast to RACK1, eS25 reduction was found to inhibit all viral IRESs, including the CrPV IGR IRES (Landry et al., 2009). Landry et al. showed that the CrPV IGR IRES is unable to bind to 40S ribosomal subunits lacking eS25 (Landry et al., 2009). However, 40S ribosomal subunits lacking RACK1 directly bind the HCV IRES with an affinity similar to wildtype 40S ribosomal subunits (Johnson et al., 2019). Further, ribosomes lacking RACK1 are also able to form 80S ribosomes at high concentrations of magnesium (Johnson et al., 2019) indicating that RACK1 is neither involved in 40S nor 80S:HCV IRES complex formation. Although we cannot exclude a direct contribution of RACK1 to 40S binding or 80S complex formation with the EMCV and PV IRESs, we believe that the evidence for the HCV IRES suggests that RACK1 might employ a mechanism distinct from eS25.
Although it is unclear how RACK1 facilitates PV translation and if the way RACK1 is used by PV is identical to HCV IRES translation initiation, the similarities to eS25 roles in IRES-mediated translation are striking. These findings further raise the question which cellular RNAs are translationally regulated by RACK1 and eS25 and whether the mRNA targets are distinct. The observations that neither RACK1 nor eS25 are largely involved in canonical cap-dependent translation (Jha et al., 2017; Landry et al., 2009) suggests that the specific cellular mRNAs relying on these proteins might be translationally regulated. Hertz et al. found that cellular IRES-containing mRNAs are less efficiently translated in eS25-depleted cells (Hertz et al., 2013), indicating a role of eS25 in cap-independent translation. However, given the indirect interaction of RACK1 with the HCV IRES via eIF3 (Majzoub et al., 2014), further studies will be needed to determine if RACK1 also acts a translational enhancer of specific mRNAs (Gilbert, 2010) including mRNAs with short open reading frames as Asc1 in S. cerevisiae (Thompson et al., 2016). Overall, our results suggest that targeting RACK1 could be used as an antiviral strategy, but that more research into the cellular mRNAs that rely on RACK1 for translation is needed.
Materials and Methods
Cell culture
HAP1 cells purchased from Horizon USA (C859), HAP1-derived RACK1 knockout cell lines #1 and #2 (Jha et al., 2017), and RACK1-FLAG addback cell lines were cultured in Iscove’s modified Dulbecco’s medium (IMDM; Corning) supplemented with 5% fetal bovine serum and 2 mM L-glutamine. HEK293FT cells (Thermo Scientific) grown in DMEM (Gibco) supplemented with 5% fetal bovine serum and 2 mM L-glutamine were used to generate lentivirus particles for cellular transductions. Cultures were confirmed negative for mycoplasma using DAPI staining.
Viral-Transduction of RACK1 Add-Back Cell Line
RACK1 cDNA was cloned with primers Forward 5′ATGACTGAGCAGATGACCCTTCG3’ and Reverse 5′CTAGCGTGTGCCAARGGTCACC3’ from HeLa cells. RACK1-FLAG expression construct was a gift from Alex G Johnson and generated by PCR-amplification from cDNA with Phusion polymerase (NEB) with primers CACCATGACTGAGCAGATGACCCTTCGTG TTATCACTTATCGTCGTCATCCTTGTAATCGCGTGTGCCAATGGTCACCTGCCAC and cloned into pENTR D-TOPO vector. Using Gateway LR Clonase II (Invitrogen) RACK1-FLAG was cloned into pLenti CMV Puro DEST (w118-1) (Addgene plasmid #17452) following the manufacturer’s protocol. RACK1-FLAG was expressed in RACK1 KO #1 cell line following lentiviral transduction. For lentivirus transduction, HEK293 FT cells (ThermoFisher) were co-transfected with plasmids pCMV-dR8.2 dvpr (gag-pol; Addgene #8455), pCMV-VSV-G (envelope; Addgene #8454), pAdVantage (Promega E1711) and pLenti RACK1-FLAG using Fugene HD (Promega). The lentivirus-containing media was filtered through a 0.45 μm PES filter. Following addition of 8 μg/ml protamine sulfate the supernatant was used to transduce RACK1 KO #1 cells. Cultures transduced overnight were selected with 1 μg/ml puromycin (InvivoGen) to generate a pool of stably expressing RACK1-FLAG cells. Selection was complete when untransduced RACK1 KO #1 cells had died.
Polysome Profile Analysis
Polysome profile analyses were performed on a 10 cm dish of approximately 80%-90% confluency in 10-50% sucrose gradients containing either in 500 mM KCl, 15 mM Tris-HCl pH 7.5, 15 mM MgCl2, 1 mg/ml heparin (Sigma) and 100 μg/ml cycloheximide (Sigma) or 500 mM KCl; 15 mM Tris-HCl, pH 7.5; 2 mM MgCl2; 1 mg/ml heparin (Sigma), 2 mM puromycin as previously described by Fuchs et al. (Fuchs et al., 2011).
Immunoblotting
Total protein lysate was harvested in RIPA buffer (Fuchs et al., 2011), and proteins separated by 12% SDS-PAGE were transferred to a nitrocellulose membrane (GE Healthcare) for 70 Vh at 4°C. Following transfer, membranes were blocked in 1% milk in PBS for 30 minutes at room temperature, washed three times in phosphate buffered saline with 0.1% (w/v) Tween 20 (PBS/T) for 10 minutes each and placed in primary antibody overnight at 4°C. Primary antibodies used were rabbit RACK1 (1:2000 dilution, Cell Signaling #4716S), FLAG-HRP (1:1000, Sigma-Aldrich, #F2555), L13a (1:1000, Cell Signaling #2765S), actin (1:1000, Sigma-Aldrich #A2066), RPS25 (1:1000, abcam, #ab102940), and PV 3CD (kind gift from Karla Kirkegaard). Following overnight incubation, membranes were washed three times in PBS/T for 10 minutes each. For visualization of the HRP-conjugated FLAG antibody, membranes were directly imaged on a BioRad ChemiDoc XRS+. For all other antibodies, membranes were incubated in a 1:10,000 dilution of goat anti-rabbit secondary (Jackson) in 5% milk and PBS/T. Membranes were washed a final 3 times in PBS/T for 10 minutes each prior to being imaged on the BioRad ChemiDoc XRS+.
Dual-Luciferase Assays
Dicistronic DNA constructs with Renilla and Firefly luciferase sequences flanked a viral IRES sequence. Viral IRESs evaluated were hepatitis C virus (HCV), cricket paralysis virus (CrPV), poliovirus (PV), and encephalomyocarditis virus (EMCV) (all gifts from Peter Sarnow, Stanford, USA). Approximately 20,000 cells of each cell line were seeded in the wells of a 96-well plate. For each construct, 100 ng DNA was transfected using lipofectamine 3000 reagent in accordance with the manufacturer’s instructions (ThermoFisher). After 24 hours, cells were lysed in 50 μl 1X passive lysis buffer (Promega) and 20 μl sample was read for 10s in a Glomax 20/20 luminometer (Promega) using the dual-luciferase assay reagent (Promega #E1910). Averages of the Firefly over Renilla ratio of at least three independent replicates and the 95th percentiles were calculated and plotted.
Poliovirus Plaque Assays
Approximately 2 million cells were seeded into 60 mm dishes the day prior to infection. Cells were washed in PBS+ (PBS supplemented with 10 mg/ml MgCl2 and 10 mg/ml CaCl2) and 100 μl diluted poliovirus was used to infect cells for 30 minutes at 37°C, 5% CO2. Cells were covered with 1% bactoagar-media mixture (DMEM supplemented with 5% fetal bovine serum and 2 mM L-glutamine). After 40 hours at 37°C, 5% CO2, the agar layer and cells were fixed and stained for 30 minutes at RT with a crystal violet solution containing 3.7% formaldehyde, 0.1% crystal violet and 20% ethanol. PV plaque sizes were determined by measuring the plaque diameter in pixels using ImageJ (NIH). Poliovirus plaques of three independent replicates were counted. The mean, interquartile range and 95th percentiles were plotted.
Poliovirus Infectivity Assays
HAP1 wildtype, RACK1 KO and RACK1-FLAG add-back cells were infected with the Mahoney strain of poliovirus at a multiplicity of infection (MOI) of 0.1 or 10 in PBS+. Infections were carried out for 30 min at 37°C and 5% CO2. After removal of the virus, complete medium was added, and cells were incubated at 37°C and 5% CO2 for 6 h. Cells were harvested into PBS+, and intracellular poliovirus was released by three freeze-thaw cycles. For the one-step growth curve, virus-containing media was harvested and used for subsequent plaque assays. To determine the virus titers,10-fold serial dilutions of the virus stocks were created and 100 μl of the dilution was used to infect HeLa cells for 30 minutes at 37°C, 5% CO2. Following infection, the cells were covered with 1% bactoagar-media mixture (DMEM supplemented with 5% fetal bovine serum and 2 mM L-glutamine) and incubated for 40 hours at 37°C, 5% CO2. The agar layer was then removed, and cells were fixed and stained for 5 minutes at RT with a 0.1% crystal violet solution in 20% ethanol. PV plaques from three independent replicates were counted. The mean, interquartile range and 95th percentiles were plotted.
35S metabolic labeling
Wildtype, RACK1 KO #1 and #2, and RACK1-FLAG expressing cells were either mock-infected or infected with PV Mahoney at MOI of 1 or MOI of 10 for 30 minutes at 37°C, 5% CO2. Cells were incubated for the indicated times at 37°C, 5% CO2, then media was exchanged for DMEM lacking cysteine and methionine (Corning®). After starvation for 30 min, cells were metabolically labeled for 10 min with 10 μCi EasyTag™ EXPRESS35 Protein Labeling Mix (Perkin Elmer). Total cell lysate was harvested in RIPA buffer (Fuchs et al., 2011) and separated by 10% SDS-PAGE. The dried gel was exposed to a phosphor-screen (GE) and scanned using a Typhoon scanner (GE).
Poliovirus Reporter Translation Assay
The luciferase-expressing, poliovirus-derived replicon plasmid prib(+)Luc-Wt was linearized with Mlu I (Vogt and Andino, 2010) and in vitro transcribed with HiScribe T7 Quick High Yield RNA Synthesis Kit (NEB) as previously described (Avanzino et al., 2017). Wildtype, RACK1 KO #1 and #2, and RACK1-FLAG expressing cells were trypsinized, resuspended and gently pelleted. For each transfection reaction, approximately 3x106 cells were resuspended in 500 μl of media and reverse transfected in suspension with Lipofectamine 3000 following the manufacturer’s transfection protocol for a 6-well plate. Immediately after transfection, three aliquots of 250 μl of transfected cells were mixed with 500 μl of IMDM media each. To inhibit translation, one aliquot of transfected cells was treated with 25 μg/ml cyloheximide (Sigma). In a second aliquot, PV replicon replication was inhibited with 2 mM guanidine hydrochloride. Aliquots of 150 μl were removed 3, 5, 7 and 9 h post transfection and luminescence was measured with luciferase assay reagent (Promega) using a Glomax luminometer (Promega).
Supplementary Material
Supplementary Figure 1: Polysome gradient analysis of wildtype, RACK1 KO #1 and RACK1-FLAG cell lines appear similar. Polysome gradients from wildtype, RACK1 KO #1 and RACK1-FLAG cell lines. The RACK1 KO #1 polysome profile shows a shift of the heaviest polysomes to the left and a slight decrease in polysome levels when compared to wildtype and RACK1-FLAG polysome profiles.
Supplementary Figure 2: Poliovirus virion peak height compared between RACK1 cell lines. Polysome-associated virion peak is higher in wildtype and RACK1-FLAG polysome profiles as compared to RACK1 KO #1 polysome profile. Data from three biological replicates is shown.
Acknowledgements
We thank Peter Sarnow (Stanford University) and Karla Kirkegaard (Stanford University) for their respective kind gifts of viral IRES plasmids and 3CD antibody. Alex G Johnson (Stanford University) was so kind to share the RACK1-FLAG lentivirus plasmid. The prib(+)Luc-Wt plasmid was a kind gift from Raul Andino (UCSF). We further thank Sangeetha Selvam, Cara Pager, and John Cleary for discussions and for critically reading the manuscript. This work was supported by a National Institutes of Health Grant R03 AI144839 to GF, start-up funds from the University at Albany, State University of New York, and the University at Albany Faculty Research Awards Program (FRAP) (to GF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare that they have no conflict of interest with the contents of this article. The content is solely the responsibility of the authors.
References
- Abernathy E, Mateo R, Majzoub K, van Buuren N, Bird SW, Carette JE, Kirkegaard K, 2019. Differential and convergent utilization of autophagy components by positive-strand RNA viruses. PLOS Biol. 17, e2006926 10.1371/journal.pbio.2006926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DR, Ron D, Kiely PA, 2011. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun. Signal. 10.1186/1478-811X-9-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger AM, Armache J-P, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R, 2013. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–5. 10.1038/nature12104 [DOI] [PubMed] [Google Scholar]
- Avanzino BC, Fuchs G, Fraser CS, 2017. Cellular cap-binding protein, eIF4E, promotes picornavirus genome restructuring and translation. Proc. Natl. Acad. Sci. U. S. A 114 10.1073/pnas.1704390114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balvay L, Rifo RS, Ricci EP, Decimo D, Ohlmann T, 2009. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta - Gene Regul. Mech 1789, 542–557. 10.1016/j.bbagrm.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Black TL, Safer B, Hovanessian A, Katze MG, 1989. The cellular 68,000-Mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: implications for translational regulation. J. Virol 63, 2244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Sabatini D, 1971. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. U. S. A 68, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonderoff JM, Larey JL, Lloyd RE, 2008. Cleavage of poly(A)-binding protein by poliovirus 3C proteinase inhibits viral internal ribosome entry site-mediated translation. J. Virol 82, 9389–99. 10.1128/JVI.00006-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR, 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–3. 10.1038/nature10348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhäuser N, Carlo Marchisio P, Biffo S, 2003. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426, 579–584. 10.1038/nature02160 [DOI] [PubMed] [Google Scholar]
- Chang BY, Chiang M, Cartwright CA, 2001. The interaction of Src and RACK1 is enhanced by activation of protein kinase C and tyrosine phosphorylation of RACK1. J. Biol. Chem 276, 20346–56. 10.1074/jbc.M101375200 [DOI] [PubMed] [Google Scholar]
- Clarke PA, Schwemmle M, Schickinger J, Hilse K, Clemens MJ, 1991. Binding of Epstein-Barr virus small RNA EBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucleic Acids Res. 19, 243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle SM, Gilbert WV, Doudna JA, 2009. Direct Link between RACK1 Function and Localization at the Ribosome In Vivo. Mol. Cell. Biol 29, 1626–1634. 10.1128/MCB.01718-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S, Bonderoff JM, Chumakov KM, Lloyd RE, Hellen CUT, 2008. Cleavage of eukaryotic initiation factor eIF5B by enterovirus 3C proteases. Virology 378, 118–122. 10.1016/j.virol.2008.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikov MI, Dobrikova EY, Gromeier M, 2018a. Ribosomal RACK1:Protein Kinase C βII Phosphorylates Eukaryotic Initiation Factor 4G1 at S1093 To Modulate Cap-Dependent and -Independent Translation Initiation. Mol. Cell. Biol 38 10.1128/MCB.00304-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikov MI, Dobrikova EY, Gromeier M, 2018b. Ribosomal RACK1:Protein Kinase C βII Modulates Intramolecular Interactions between Unstructured Regions of Eukaryotic Initiation Factor 4G (eIF4G) That Control eIF4E and eIF3 Binding. Mol. Cell. Biol 38 10.1128/MCB.00306-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbil S, Oral O, Mitou G, Kig C, Durmaz-Timucin E, Guven-Maiorov E, Gulacti F, Gokce G, Dengjel J, Sezerman OU, Gozuacik D, 2016. RACK1 Is an Interaction Partner of ATG5 and a Novel Regulator of Autophagy. J. Biol. Chem 291, 16753–65. 10.1074/jbc.M115.708081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez PM, Sessions OM, Wagner EJ, Gromeier M, Garcia-Blanco MA, 2005. The Polypyrimidine Tract Binding Protein Is Required for Efficient Picornavirus Gene Expression and Propagation. J. Virol 79, 6172–6179. 10.1128/JVI.79.10.6172-6179.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Diges C, Kohlstaedt LA, Wehner KA, Sarnow P, 2011. Proteomic Analysis of Ribosomes: Translational Control of mRNA Populations by Glycogen Synthase GYS1. J. Mol. Biol 410, 118–130. 10.1016/j.jmb.2011.04.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo S, Ricciardi S, Manfrini N, Pesce E, Oliveto S, Calamita P, Mancino M, Maffioli E, Moro M, Crosti M, Berno V, Bombaci M, Tedeschi G, Biffo S, 2018. RACK1 Specifically Regulates Translation through Its Binding to Ribosomes. Mol. Cell. Biol 38 10.1128/MCB.00230-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Senft D, Topisirovic I, Ronai ZA, 2013. RACK1 Function in Cell Motility and Protein Synthesis. Genes Cancer 4, 369–377. 10.1177/1947601913486348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbasi VR, Weaver CM, Hill S, Friedman DB, Link AJ, 2004. Yeast Asc1p and Mammalian RACK1 Are Functionally Orthologous Core 40S Ribosomal Proteins That Repress Gene Expression. Mol. Cell. Biol 24, 8276–8287. 10.1128/MCB.24.18.8276-8287.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadge GD, Swaminathan S, Katze MG, Thimmapaya B, 1991. Binding of the adenovirus VAI RNA to the interferon-induced 68-kDa protein kinase correlates with function. Proc. Natl. Acad. Sci. U. S. A 88, 7140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV, 2010. Alternative Ways to Think about Cellular Internal Ribosome Entry. J. Biol. Chem 285, 29033–29038. 10.1074/jbc.R110.150532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A, Svitkin YV, Imataka H, Sonenberg N, 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. U. S. A 95, 11089–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso S, Volta V, Sala LA, Vietri M, Marchisio PC, Ron D, Biffo S, 2008. PKCβII modulates translation independently from mTOR and through RACK1. Biochem. J 415, 77–85. 10.1042/BJ20080463 [DOI] [PubMed] [Google Scholar]
- Guo J, Chen J-G, 2008. RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant Biol. 8, 108 10.1186/1471-2229-8-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang S, Valerius O, Hall H, Zeng Q, Li J-F, Weston DJ, Ellis BE, Chen J-G, 2011. Involvement of Arabidopsis RACK1 in Protein Translation and Its Regulation by Abscisic Acid. PLANT Physiol. 155, 370–383. 10.1104/pp.110.160663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebner CM, Wilson R, Rader J, Bidder M, Laimins LA, 2006. Human papillomaviruses target the double-stranded RNA protein kinase pathway. J. Gen. Virol 87, 3183–3193. 10.1099/vir.0.82098-0 [DOI] [PubMed] [Google Scholar]
- Hertz MI, Landry DM, Willis AE, Luo G, Thompson SR, 2013. Ribosomal protein s25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol. Cell. Biol 33, 1016–26. 10.1128/MCB.00879-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz MI, Thompson SR, 2011. Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology 411, 355 10.1016/J.VIROL.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannot G, Bajan S, Giguère NJ, Bouasker S, Banville IH, Piquet S, Hutvagner G, Simard MJ, 2011. The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 12, 581–586. 10.1038/embor.2011.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Rollins MG, Fuchs G, Procter DJ, Hall EA, Cozzolino K, Sarnow P, Savas JN, Walsh D, 2017. Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature 546 10.1038/nature22814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AG, Lapointe CP, Wang J, Corsepius NC, Choi J, Fuchs G, Puglisi JD, 2019. RACK1 on and off the ribosome 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas JL, Smith MA, Pronovost SM, Beckerle MC, 2007. Characterization of RACK1 function inDrosophila development. Dev. Dyn 236, 2207–2215. 10.1002/dvdy.21217 [DOI] [PubMed] [Google Scholar]
- Kershner L, Welshhans K, 2017. RACK1 is necessary for the formation of point contacts and regulates axon growth. Dev. Neurobiol 77, 1038–1056. 10.1002/dneu.22491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, 2008. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci 33, 274–283. https://doi.org/S0968-0004(08)00091-1 [pii] 10.1016/j.tibs.2008.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Joachims M, Lloyd RE, 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol 76, 2062–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE, 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol 24, 1779–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR, 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23, 2753–2764. https://doi.org/23/23/2753 [pii] 10.1101/gad.1832209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Pettiford S, Jiang B, Jacobs BL, 1994. Products of the porcine group C rotavirus NSP3 gene bind specifically to double-stranded RNA and inhibit activation of the interferon-induced protein kinase PKR. J. Virol 68, 3821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS-Y, Burdeinick-Kerr R, Whelan SPJ, 2013. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc. Natl. Acad. Sci. U. S. A 110, 324–9. 10.1073/pnas.1216454109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majzoub K, Hafirassou ML, Meignin C, Goto A, Marzi S, Fedorova A, Verdier Y, Vinh J, Hoffmann JA, Martin F, Baumert TF, Schuster C, Imler J-L, 2014. RACK1 Controls IRES-Mediated Translation of Viruses. Cell 159, 1086–1095. 10.1016/j.cell.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda R, Ikeuchi K, Nomura S, Inada T, 2014. Protein quality control systems associated with no-go and nonstop mRNA surveillance in yeast. Genes to Cells 19, 1–12. 10.1111/gtc.12106 [DOI] [PubMed] [Google Scholar]
- Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, Spahn CM, 2011. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. https://doi.org/gkr114 [pii] 10.1093/nar/gkr114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MH, Flygaard RK, Jenner LB, 2017. Structural analysis of ribosomal RACK1 and its role in translational control. Cell. Signal. 35, 272–281. 10.1016/j.cellsig.2017.01.026 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N, 2007. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 35, 1514–1521. 10.1093/nar/gkl1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM, 2016. The integrated stress response. EMBO Rep. 17, 1374–1395. 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N, 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325. [DOI] [PubMed] [Google Scholar]
- Powers CD, Miller BA, Kurtz H, Ackermann WW, 1969. Specific effect of guanidine in the programming of poliovirus inhibition of deoxyribonucleic acid synthesis. J. Virol 3, 337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano N, Veronese M, Manfrini N, Zolla L, Ceci M, 2019. Ribosomal RACK1 promotes proliferation of neuroblastoma cells independently of global translation upregulation. Cell. Signal 53, 102–110. 10.1016/j.cellsig.2018.09.020 [DOI] [PubMed] [Google Scholar]
- Sabila M, Kundu N, Smalls D, Ullah H, 2016. Tyrosine Phosphorylation Based Homo-dimerization of Arabidopsis RACK1A Proteins Regulates Oxidative Stress Signaling Pathways in Yeast. Front. Plant Sci 7, 176 10.3389/fpls.2016.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CMT, Nissen P, Frank J, 2004. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol 11, 957–962. 10.1038/nsmb822 [DOI] [PubMed] [Google Scholar]
- Sharp TV, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud CG, Hilse K, Clemens MJ, 1993. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 21, 4483–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J, 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science (80-. ). 291, 1959–1962. [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ, 2017. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol. Cell 65, 751–760.e4. 10.1016/j.molcel.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Gradi A, Imataka H, Morino S, Sonenberg N, 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol 73, 3467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MK, Rojas-Duran MF, Gangaramani P, Gilbert WV , 2016. The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. Elife 5 10.7554/eLife.11154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F, 2015. Tissue-based map of the human proteome. Science (80-. ). 347, 1260419–1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Ventoso I, MacMillan SE, Hershey JW, Carrasco L, 1998. Poliovirus 2A proteinase cleaves directly the eIF-4G subunit of eIF-4F complex. FEBS Lett. 435, 79–83. [DOI] [PubMed] [Google Scholar]
- Vogt DA, Andino R, 2010. An RNA element at the 5’-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS Pathog. 6, e1000936 10.1371/journal.ppat.1000936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volta V, Beugnet A, Gallo S, Magri L, Brina D, Pesce E, Calamita P, Sanvito F, Biffo S, 2013. RACK1 depletion in a mouse model causes lethality, pigmentation deficits and reduction in protein synthesis efficiency. Cell. Mol. Life Sci 70, 1439–1450. 10.1007/s00018-012-1215-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner P, Shnitsar I, Urlaub H, Borchers A, 2011. RACK1 is a novel interaction partner of PTK7 that is required for neural tube closure. Development 138, 1321–1327. 10.1242/dev.056291 [DOI] [PubMed] [Google Scholar]
- Wek RC, 2018. Role of eIF2α Kinases in Translational Control and Adaptation to Cellular Stress. Cold Spring Harb. Perspect. Biol 10, a032870 10.1101/cshperspect.a032870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang H-Y, Anthony TG, 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans 34, 7–11. 10.1042/BST20060007 [DOI] [PubMed] [Google Scholar]
- White JP, Reineke LC, Lloyd RE, 2011. Poliovirus Switches to an eIF2-Independent Mode of Translation during Infection. J. Virol 85, 8884–8893. 10.1128/JVI.00792-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Collier M, Loerke J, Ismer J, Schmidt A, Hilal T, Sprink T, Yamamoto K, Mielke T, Bürger J, Shaikh TR, Dabrowski M, Hildebrand PW, Scheerer P, Spahn CMT, 2015. Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA. EMBO J. 34, 3042–58. 10.15252/embj.201592469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatime L, Hein KL, Nilsson J, Nissen P, 2011. Structure of the RACK1 dimer from saccharomyces cerevisiae. J. Mol. Biol 411, 486–498. 10.1016/j.jmb.2011.06.017 [DOI] [PubMed] [Google Scholar]
- Yu SF, Benton P, Bovee M, Sessions J, Lloyd RE, 1995. Defective RNA replication by poliovirus mutants deficient in 2A protease cleavage activity. J. Virol 69, 247–252. 10.1128/jvi.69.1.247-252.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang Q, Qiu G, Zhou S, Jing Z, Wang J, Wang W, Cao J, Han K, Cheng Q, Shen B, Chen Y, Zhang WJ, Ma Y, Zhang J, 2015. RACK1 Promotes Autophagy by Enhancing the Atg14L-Beclin 1-Vps34-Vps15 Complex Formation upon Phosphorylation by AMPK. Cell Rep. 13, 1407–1417. 10.1016/j.celrep.2015.10.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Polysome gradient analysis of wildtype, RACK1 KO #1 and RACK1-FLAG cell lines appear similar. Polysome gradients from wildtype, RACK1 KO #1 and RACK1-FLAG cell lines. The RACK1 KO #1 polysome profile shows a shift of the heaviest polysomes to the left and a slight decrease in polysome levels when compared to wildtype and RACK1-FLAG polysome profiles.
Supplementary Figure 2: Poliovirus virion peak height compared between RACK1 cell lines. Polysome-associated virion peak is higher in wildtype and RACK1-FLAG polysome profiles as compared to RACK1 KO #1 polysome profile. Data from three biological replicates is shown.