Abstract
Marfan syndrome and related disorders are a group of heritable connective tissue disorders and share many clinical features that involve cardiovascular, skeletal, craniofacial, ocular, and cutaneous abnormalities. The majority of affected individuals have aortopathies associated with early mortality and morbidity. Implementation of targeted gene panel next-generation sequencing in these individuals is a powerful tool to obtain a genetic diagnosis. Here, we report on clinical and genetic spectrum of 53 families from India with a total of 83 patients who had a clinical diagnosis suggestive of Marfan syndrome or related disorders. We obtained a molecular diagnosis in 45/53 (85%) index patients, in which 36/53 (68%) had rare variants in FBN1 (Marfan syndrome; 63 patients in total), seven (13.3%) in TGFBR1/TGFBR2 (Loeys–Dietz syndrome; nine patients in total) and two patients (3.7%) in SKI (Shprintzen–Goldberg syndrome). 21 of 41 rare variants (51.2%) were novel. We did not detect a disease-associated variant in 8 (15%) index patients, and none of them met the Ghent Marfan diagnostic criteria. We found the homozygous FBN1 variant p.(Arg954His) in a boy with typical features of Marfan syndrome. Our study is the first reporting on the spectrum of variants in FBN1, TGFBR1, TGFBR2, and SKI in Indian individuals.
Subject terms: Genotype, Genetics, Clinical genetics, Disease genetics, Cardiovascular diseases
Introduction
Heritable connective tissue disorders (HCTD) comprise a group of multisystem diseases affecting the heart, blood vessels, bone, eyes, skin, joints, and lungs. Marfan syndrome (MFS, MIM#154700), Loeys–Dietz syndrome (LDS, MIM#609192, MIM#610168, MIM#613795, MIM#614816, MIM#615582), and Shprintzen–Goldberg syndrome (SGS, MIM#182212) belong to the HCTDs and share many clinical features, such as cardiovascular, skeletal, craniofacial, ocular, and cutaneous abnormalities1. The phenotype of MFS is characterized by aortic root aneurysm or dissection, mitral valve prolapse, ectopia lentis, long bone overgrowth, joint laxity, and skin striae as the key abnormalities. Craniofacial dysmorphism includes dolichocephaly, exophthalmos, downslanted palpebral fissures, malar hypoplasia, highly arched palate, and micro- or retrognathia2. Heterozygous pathogenic variants in the FBN1 gene, encoding the extracellular matrix protein fibrillin-1, are the cause of MFS3. Pathogenic FBN1 variants are spread over the entire gene and comprise sequence-level alterations, such as missense, nonsense, frameshift, and splice variants, identified in the majority of MFS-affected cases as well as single- and multi-exon deletions in up to 5% of the affected individuals2,4. LDS has many clinical manifestations in common with MFS, however, LDS-affected patients can have characteristic craniofacial features, such as hypertelorism, abnormal uvula or cleft palate5. Typical cardiovascular features in LDS are dilatation of the aortic root at the level of the sinus of Valsalva, aneurysms affecting thoracic and abdominal aorta and arterial branches, as well as arterial tortuosity1,5. Cardiovascular manifestations tend to be more severe in LDS than in MFS6, however, a multi-center study has recently demonstrated a comparable cardiovascular outcome in individuals with MFS and LDS7. Heterozygous pathogenic variants in six genes cause LDS type 1–6: TGFBR1, TGFBR2, SMAD3, TGFB2, TGFB3, and SMAD28–13. Patients with SGS have some of the craniofacial, skeletal, skin and cardiovascular manifestations of MFS and LDS, but in addition show intellectual disability, skeletal muscle hypotonia and craniosynostosis. Mitral valve prolapse and aortic root dilatation have been reported in some cases14. In SGS-affected probands mainly de novo heterozygous pathogenic variants in the SKI gene have been identified that cluster in two regions, one encoding the R-SMAD binding domain and the other encoding the Dachshund-homology domain15–17.
Implementation of targeted gene panel next-generation sequencing (NGS) in individuals with HCTD or hereditary aortopathies in a clinical setting has been proven to be powerful in obtaining a genetic diagnosis: a pathogenic or likely pathogenic variant was identified in 3.9–35.5% of the patients tested in different centers worldwide18–26. Thus, in individuals with clinical features typical of HCTD or with a non-syndromic form of aortopathy an NGS-based molecular test is the most practical screening method to identify the disease-related sequence variant. Here, we report on our clinical and genetic findings after testing of 53 index patients from India with a clinical diagnosis suggestive of HCTD using targeted NGS and whole-exome sequencing. Although our patient cohort is small, this is the first study reporting on the spectrum of variants in FBN1, TGFBR1, TGFBR2, and SKI in Indian individuals, with about 50% novel pathogenic variants.
Results
We recruited 83 patients from 53 families with MFS, aortopathy or a related HCTD. The ages of the patients ranged from 3 months to 56 years with a median age of 14 years. The majority were males (51, 61.5%; CI 95% 51–71) and children and adolescents (53, 64% were less than 18 years of age; CI 95% 53–73). Twenty-one families (39% of 53 families; CI 95% 28–53) had more than one affected individual, including a set of monozygotic twins. Echocardiographic information was available for 77/83 individuals that included all index patients. Ophthalmological, skeletal and other information were available for 72/83 individuals.
Molecular findings in 53 unrelated Indian patients and their family members with HCTD
NGS-based genetic testing was performed in 53 unrelated Indian individuals with MFS, aortopathy or a related HCTD. Of the 53 individuals, 44 (83.0%; CI 95% 71–91) tested positive for a pathogenic variant in FBN1, TGFBR1, TGFBR2 or SKI, and 1 (1.9%; CI 95% 0.3–10) had a variant of unknown significance (VUS) in FBN1 (Table 1 and Supplementary Table S1). 36 of the 45 (80.0%; CI 95% 66–89) patients with a rare variant carried an FBN1 variant, including 21 missense (58.3%; CI 95% 42–73), five splice (13.9%; CI 95% 6–29), three small deletion/insertion (8.3%; CI 95% 3–22), two nonsense (5.6%; CI 95% 2–18), and five multi-exon deletions (13.9%; CI 95% 6–29) (Fig. 1). The known pathogenic FBN1 missense variant c.2861G > A/p.(Arg954His)27 was identified in patient 10 in a homozygous state. Both parents (first cousins) were heterozygous carriers of this FBN1 alteration. 4 (8.9%; CI 95% 4–21) patients had a TGFBR1 missense variant, 3 (6.7%; CI 95% 2–18) a TGFBR2 missense variant, and 2 (4.4%; CI 95% 1–15) a SKI missense variant. The FBN1 variants c.(1468 + 1_1469-1)_(1837 + 1_1838-1)del, c.3037G > A/p.(Gly1013Arg), and c.7828G > A/p.(Glu2610Lys) and the TGFBR1 variant c.722C > T/p.(Ser241Leu) have been identified in two non-consanguineous families each. Out of the 41 different rare variants in four genes, 20 (48.8%; CI 95% 34–63) have been previously reported in the HGMD professional and/or UMD-FBN1 database and 21 (51.2%; CI 95% 36–66) were novel (Table 1 and Supplementary Table S1). 20 of the 21 novel variants were classified as pathogenic or likely pathogenic, and the intronic FBN1 variant c.2419 + 3delinsTTTTAGATCCATATTTTAG (in family 9) was interpreted as VUS (Table 1). In 17 (37.8%; CI 95% 25–52) index patients, de novo occurrence of the pathogenic variant [ten known and seven novel variants (Table 1 and Supplementary Table S1)] was confirmed by genetic testing of the healthy parents (without confirming paternity), including 11 variants in FBN1, 3 in TGFBR1, 2 in TGFBR2, and 1 in SKI. Segregation analysis was performed in 21 families with a minimum of two affected individuals, and 29 relatives were found to carry the familial variant, including 27 individuals with an FBN1 variant. NGS of 62 genes/candidate genes related to HCTD and hereditary aortopathies (single nucleotide variant and copy number variation analysis) and MLPA analysis (all exons of FBN1, TGFBR1, TGFBR2, and 15 selected exons of COL3A1; see material and methods section for details) did not detect a disease-associated variant in 8 (15%; CI 95% 8–27) index patients.
Table 1.
Number of affected family members, in silico pathogenicity predictions and ACMG classification for novel variants found in the cohort.
| Gene | Patient # | Affected family members | Nucleotide change | Affected exon(s)/intron(s) | Amino acid alteration | gnomAD MAF [%] | Pathogenicity predictions | Splice predictions | ACMG | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CADD | REVEL | M-CAP | Classification | Criteria | ||||||||
| FBN1c | 1 | 1 | c.(?_1317)_(1837 + 1_1838-1)del | Upstream of exon 1 and exons 1–15 | p.? | NA | NA | NA | NA | Not done | LP# | PVS1, PM2 |
| 3 | 1 (de novo)a | c.1130G > A | Exon 10 | p.(Cys377Tyr) | Absent | 33 | 0.927 | 0.915 | Not done | LP | PM1, PM2, PM5, PM6, PP2, PP3, PP5 | |
| 4 | 2 | c.1463G > A | Exon 12 | p.(Cys488Thr) | Absent | 32 | 0.937 | 0.892 | Not done | LP | PM1, PM2, PM5, PP2, PP3, PP5 | |
| 5b | 2 | c.(1468 + 1_1469-1)_(1837 + 1_1838-1)del | Exons 13–15 | p.? | NA | NA | NA | NA | Not done | P# | PVS1, PM2, PS4 | |
| 6b | 3 | c.(1468 + 1_1469-1)_(1837 + 1_1838-1)del | Exons 13–15 | p.? | NA | NA | NA | NA | Not done | P# | PVS1, PM2, PS4, PP1 | |
| 8 | 2 | c.1867T > A | Exon 16 | p.(Cys623Ser) | Absent | 27.6 | 0.771 | 0.493 | Not done | LP | PM1, PM2, PM5, PP2, PP3 | |
| 9 | 2 | c.2419 + 3delinsTTTTAGATCCATATTTTAG | Intron 20 | p.? | Absent | 14.96 | NA | NA | Impact on splicing | VUS# | PM2, PP3 | |
| 14 | 1 (de novo)a | c.3589 + 1G > A | Intron 29 | p.? | Absent | 34 | NA | NA | Impact on splicing | P | PVS1, PM2, PM6, PP3 | |
| 16 | 1 | c.3635G > A | Exon 30 | p.(Cys1212Tyr) | Absent | 32 | 0.965 | 0.899 | Not done | LP | PM1, PM2, PM5, PP2, PP3 | |
| 18 | 2 | c.4491C > G | Exon 37 | p.(Cys1497Trp) | Absent | 17.95 | 0.768 | 0.820 | Not done | LP | PM1, PM2, PM5, PP2, PP3 | |
| 21 | 2 | c.4817-1_4819delGATA | Intron 39/exon 40 | p.? | Absent | 37 | NA | NA | Impact on splicing | P | PVS1, PM2, PP3 | |
| 22 | 1 (de novo)a | c.5467_5474dupGAATGCAT | Exon 45 | p.(Ile1825Metfs*71) | Absent | 33 | NA | NA | Not done | P | PVS1, PM2, PM6, PP3 | |
| 25 | 3 | c.5621G > T | Exon 46 | p.(Cys1874Phe) | Absent | 33 | 0.991 | 0.981 | Not done | LP | PM1, PM2, PP1, PP2, PP3 | |
| 27 | 1 (de novo)a | c.5671 + 1G > C | Intron 46 | p.? | Absent | 33 | NA | NA | Impact on splicing | P | PVS1, PM2, PM6, PP3 | |
| 28 | 1 (de novo)a | c.5917 + 1G > T | Intron 48 | p.? | Absent | 34 | NA | NA | Impact on splicing | P | PVS1, PM2, PM6, PP3, PP5 | |
| 29 | 1 | c.5966G > C | Exon 49 | p.(Cys1989Ser) | Absent | 25.2 | 0.818 | 0.946 | Not done | LP | PM1, PM2, PM5, PP2, PP3 | |
| 30 | 3 | c.5993G > T | Exon 49 | p.(Cys1998Phe) | Absent | 33 | 0.956 | 0.928 | Not done | LP | PM1, PM2, PM5, PP1, PP2, PP3 | |
| 32 | 2 | c.(7204 + 1_7205-1)_(7819 + 1_7820-1)del | Exons 59–63 | p.? | NA | NA | NA | NA | Not done | LP# | PVS1, PM2 | |
| 34 | 2 | c.7817T > A | Exon 63 | p.(Val2606Asp) | Absent | 33 | 0.665 | 0.270 | Not done | LP | PM1, PM2, PP2, PP3 | |
| SKId | 38 | 1 (de novo)a | c.104C > G | Exon 1 | p.(Pro35Arg) | Absent | 23.6 | 0.759 | 0.964 | Not done | LP | PM1, PM2, PM5, PM6, PP3 |
| TGFBR2e | 43 | 1 | c.1453C > A | Exon 6 | p.(Arg485Ser) | Absent | 29.4 | 0.868 | 0.374 | Not done | LP | PM1, PM2, PM5, PP2, PP3 |
| 44 | 1 (de novo)a | c.1454G > C | Exon 6 | p.(Arg485Pro) | Absent | 33 | 0.937 | 0.427 | Not done | LP | PM1, PM2, PM5, PM6, PP2, PP3 | |
The functional impact of the identified variants was predicted by the Combined Annotation Dependent Depletion (CADD) tool, the Rare Exome Variant Ensemble Learner (REVEL) scoring system, and the Mendelian Clinically Applicable Pathogenicity (M-CAP) Score. CADD is a framework that integrates multiple annotations in one metric by contrasting variants that survived natural selection with simulated mutations. Reported CADD scores are phred-like rank scores based on the rank of that variant’s score among all possible single nucleotide variants of hg19, with 10 corresponding to the top 10%, 20 at the top 1%, and 30 at the top 0.1%. The larger the score the more likely the variant has deleterious effects; the score range observed here is strongly supportive of pathogenicity, with all observed variants ranking above ~ 99% of all variants in a typical genome and scoring similarly to variants reported in ClinVar as pathogenic (~ 85% of which score > 15)61. REVEL is an ensemble method predicting the pathogenicity of missense variants with a strength for distinguishing pathogenic from rare neutral variants with a score ranging from 0 to 1. The higher the score the more likely the variant is pathogenic62. M-CAP is a classifier for rare missense variants in the human genome, which combines previous pathogenicity scores (including SIFT, Polyphen-2, and CADD), amino acid conservation features and computed scores trained on mutations linked to Mendelian diseases. The recommended pathogenicity threshold is > 0.02563. Splice site prediction scores were calculated for wild-type and mutated sequences by using the programs Human Splicing Finder 3.1, NetGene2, and the Berkeley Drosophila Genome Project Database65–68. Genetic tolerance at the affected amino acid position in the protein was predicted by MetaDome64. All variants were classified according to the guidelines of the American College of Medical Genetics (ACMG) either by use of an adjusted automated interpretation by VarSome (https://varsome.com/)60or in case of whole exon deletions and Indels (#) by manual application of the guidelines.
LP likely pathogenic, NA not applicable, P pathogenic, VUS variant of unclear significance.
aPaternity not confirmed.
bApparently non-consanguineous families.
cFBN1 mRNA reference number: NM_000138.4.
dSKI mRNA reference number: NM_003036.3.
eTGFBR2 mRNA reference number: NM_001024847.2.
Figure 1.
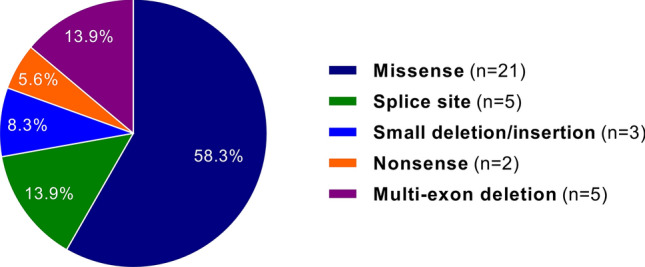
A representation of the different FBN1 variants in 36 index patients is shown.
Clinical findings in 74 individuals with a molecular diagnosis
Marfan syndrome
All patients with MFS and a rare FBN1 variant (including c.2419 + 3delinsTTTTAGATCCATATTTTAG interpreted as VUS) met the revised Ghent criteria. Familial occurrence was observed in 19/36 families (52%; CI 95% 37–68). Detailed phenotypic information is provided in Table 2. Aortic root dilatation (z-score > 2) was noted in 77% (CI 95% 65–86) followed by mitral valve prolapse in 67% (CI 95% 54–78) and tricuspid valve prolapse in 53% (CI 95% 41–66) of patients. We also noted atrial septal defect, pulmonary artery dilatation and cardiomegaly in three individuals each. Six patients underwent aortic or mitral valve replacement in view of valve insufficiency. We observed myopia in 60% (CI 95% 47–72) followed by lens subluxation in 49% (CI 95% 36–62), dolichostenomelia in 71% (CI 95% 58–82) and pectus abnormality in 47% (CI 95% 34–60) of individuals. In addition, there was hypotonia in seven individuals, truncal obesity in three and developmental delay or mild intellectual disability in two individuals. Developmental delay or intellectual disability was however not investigated further.
Table 2.
Clinical features in patients with Marfan or Loeys–Dietz syndrome and a clinically relevant variant.
| Features | Marfan syndrome (n = 63) | Loeys–Dietz syndrome (n = 9) |
|---|---|---|
| Ocular manifestations | ||
| Myopia | 32/53 (60%) | 1/8 (12%) |
| Ectopia lentis | 26/53 (49%) | 0/8 |
| Early cataract | 05/53 (9%) | 0/8 |
| Astigmatism | 03/53 (5%) | 2/8 (25%) |
| Microspherophakia | 02/53 (3%) | 0/8 |
| Cardiovascular manifestations | ||
| Aortic root dilatation | 45/58 (77%) | 6/8 (75%) |
| Aortic regurgitation | 14/58 (24%) | 3/8 (37%) |
| Aortic aneurysma | 04/58 (6%) | 0/8 |
| Aortic dissection | 01/58 (1.7%) | 1/8 (12%) |
| Mitral valve prolapse | 39/58 (67%) | 5/8 (62%) |
| Tricuspid valve prolapse | 31/58 (53%) | 5/8 (62%) |
| Mitral regurgitation | 33/58 (56%) | 4/8 (50%) |
| Tricuspid regurgitation | 26/58 (44%) | 3/8 (37%) |
| Bicuspid aortic valve | 01/58 (1.7%) | 1/8 (12%) |
| Skeletal findings | ||
| Pectus abnormality | 25/53 (47%) | 7/8 (87%) |
| Scoliosis | 16/53 (30%) | 3/8 (37%) |
| Thumb sign | 36/51 (70%) | 3/8 (37%) |
| Wrist sign | 35/51 (68%) | 2/8 (25%) |
| Dolichostenomelia | 38/53 (71%) | 2/8 (25%) |
| Pes planus | 27/53 (50%) | 5/8 (62%) |
| Talipes deformity | 13/53 (24%) | 3/8 (37%) |
| Genu valgum/recurvatum | 05/53 (9%) | 1/8 (12%) |
| Reduced elbow extension | 07/53 (13%) | 0/8 |
| Camptodactyly | 19/53 (35%) | 1/8 (12%) |
| Long and narrow feet | 33/53 (62%) | 5/8 (62%) |
| Metatarsus adductus | 07/53 (13%) | 1/8 (12%) |
| Craniosynostosis | 0/53 | 2/8 (25%) |
| Facial features | ||
| Long and narrow face | 36/53 (67%) | 8/8 (100%) |
| High arched palate | 34/53 (64%) | 5/8 (62%) |
| Bifid uvula/cleft palate | 0/53 | 3/8 (37%) |
| Hypertelorism | 01/53 (5%) | 8/8 (100%) |
| Exotropia | 05/53 (9%) | 0/8 |
| Dolichocephaly | 10/53 (18%) | 2/8 (25%) |
| Enophthalmos | 15/53 (28%) | 0/8 |
| Downslanted palpebral fissures | 19/53 (35%) | 8/8 (100%) |
| Malar hypoplasia | 31/53 (58%) | 4/8 (50%) |
| Micro/retrognathia | 18/53 (33%) | 8/8 (100%) |
| Low-set ears | 16/53 (30%) | 2/8 (25%) |
| Crowding of teeth | 08/53 (15%) | 2/8 (25%) |
| Other features | ||
| Skin striae | 11/53 (20%) | 2/8 (25%) |
| Skin laxity | 06/53 (11%) | 1/8 (12%) |
| Pneumothorax | 06/53 (11%) | 0/8 |
| Dural ectasia | Not tested | Not tested |
| Hernia | 07/53 (13%) | 3/8 (37%) |
| Joint laxity | 11/53 (20%) | 4/8 (50%) |
| Translucent skin | 06/53 (11%) | 0/8 |
| Decreased muscle mass | 16/53 (30%) | 4/8 (50%) |
| Joint contractures | 02/53 (3%) | 1/8 (12%) |
| Developmental delay/mild intellectual disability/motor delay | 02/53 (3%) | 4/8 (50%) |
Numerator indicates the number of individuals with a clinical feature and denominator indicates the number of individuals where information is available for the given clinical feature.
aYounger individuals with Loeys–Dietz syndrome might not manifest with aortic aneurysm.
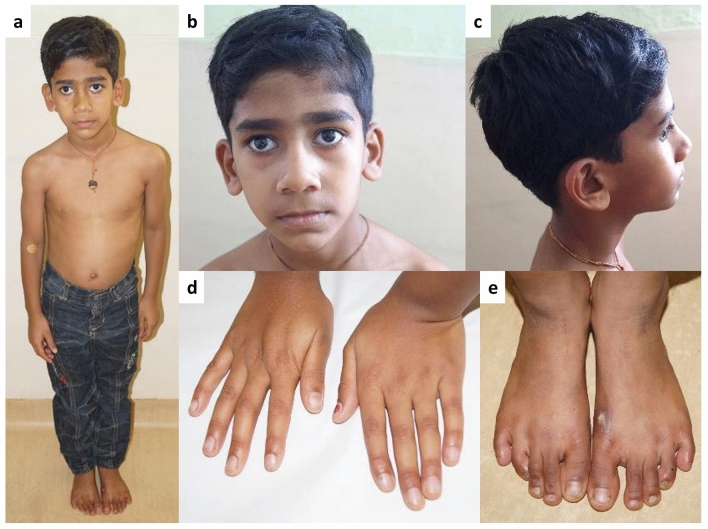
In a 5-years-8-months-old male (patient 10) we identified the homozygous pathogenic FBN1 missense variant c.2861G > A/p.(Arg954His) (Supplementary Table S1). He is the first child of a third degree consanguineous couple. His measurements were: weight of 20 kg (− 0.30 z), height of 116.5 cm (0.01 z) and head circumference of 49.5 cm (− 1.94 z). He had a long face, exotropia of the right eye, thin vermilion of the upper lip, high arched palate, bilateral lens subluxation, pes planus, mild distal joint laxity, bicuspid aortic valve, tricuspid and mitral valve prolapse and aortic sinus z-score of 2.86 (Fig. 2). Limited clinical information could be gathered via a video consultation and we specifically noted absence of breathlessness, visual problems, skin striae and chest deformity in parents. Mother however had features suggestive of Leri–Weill dyschondrosteosis (short stature, Madelung deformity, with similarly affected females and mildly affected males in the family).
Figure 2.
Photographs of patient 10 with the homozygous variant c.2861G > A, p.(Arg954His) in FBN1. He had normal stature (0.01 z) at 5 years 8 months of age with long face, exotropia of right eye, thin vermilion of upper lip (a–c), normal fingers (d) and pes planus (e).
Loeys–Dietz syndrome (LDS)
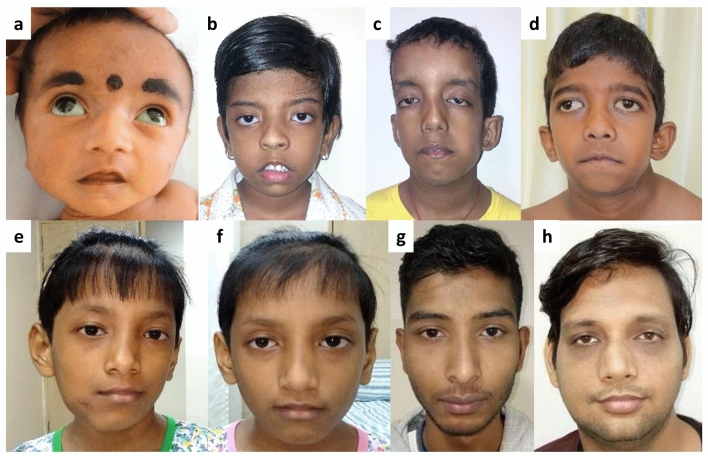
Nine patients from seven families (7/53, 13%; CI 95% 7–25) tested positive for a TGFBR1 or TGFBR2 rare variant and clinically presented with LDS. Phenotypic features are summarized in Table 2. Cardiac manifestation was observed in all individuals with aortic root dilatation in six individuals (6/8, 75%; CI 95% 41–93). One proband had a dissection of the aorta at 34 years of age. Typical facial features such as long and narrow face, hypertelorism, downslanted palpebral fissures and micro/retrognathia were seen in all eight patients (Table 2, Fig. 3). Three (37%; CI 95% 14–69) individuals had cleft palate/bifid uvula. Additionally, we noted developmental delay or motor delay (four patients), craniosysostosis (two patients), atopic dermatitis and anemia (in monozygotic twins), platybasia with basilar invagination and atlantoaxial subluxation with retroflexion (one individual) and joint dislocation (one individual) in patients with LDS.
Figure 3.
Facial photographs of patients with LDS show long faces, widely spaced eyes, downslanted palpebral fissures, thin vermilion of upper lips and micrognathia in all of them (a–h). Additionally, low set ears (b–d), ptosis (b) and wide mouth with downturned corners (d) were noted. Photographs were taken at ages: 7 months (a: Patient 44), 6 years (b: Patient 39 and c: Patient 40), 8 years (d: Patient 42), 9 years (e: Patient 45 and f: monozygotic twin of Patient 45), 16 years (g: Patient 41) and 36 years (h: Patient 43).
Shprintzen–Goldberg syndrome (SGS)
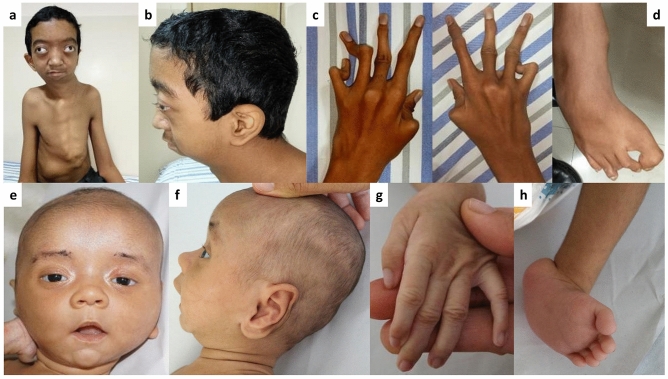
Patient 37 aged 13.5 years had height of 154 cm (− 1.09 z), weight of 35 kg (− 1.91 z) and head circumference of 53 cm (− 1.49 z). Craniosynostosis, dolichostenomelia, low-set ears, overfolded ear helix, proptosis, downslanted palpebral fissures, hypertelorism, alternative exotropia, vertical strabismus of the left eye, microcornea, depressed nasal bridge, malar flattening, thin vermilion of upper lip, micro-retrognathia, high arched palate and malocclusion of teeth were noted in him (Fig. 4a,b). He also had pectus carinatum, kyphoscoliosis, long and narrow fingers, decreased palmar creases, long and narrow feet, camptodactyly of fingers and toes, pes planus, metatarsus adductus, recurrent or incisional hernia and decreased muscle mass (Fig. 4c,d). Echocardiography revealed myxomatous prolapsing atrioventricular valves with tricuspid and mitral regurgitation and aortic root dilatation (z-score 6.7). Computed tomography of skull showed calvarial thickening in fronto-parietal bones with partially fused coronal and sagittal sutures. Anterior displacement of the atlas from the occipital condyle and atlanto-occiptal assimilation, mild levoscoliosis of the cerivothoracic vertebra and mild dextroscoliosis of the thoracic vertebra (T9) were observed on computed tomography of the spine.
Figure 4.
Clinical photographs of patients with SGS. Craniosynostosis, dolichostenomelia, low-set ears, overfolded ear helix, proptosis, downslanted palpebral fissures, hypertelorism, proptosis, alternative exotropia, vertical strabismus of left eye, microcornea, depressed nasal bridge, underdeveloped cheekbone, thin vermilion of upper lip, micro-retrognathia, pectus carinatum and kypho-scoliosis can be noted in patient 37 at age 13.5 years (a, b). He also had long and narrow fingers with camptodactyly (c), long and narrow feet with camptodactyly, pes planus and metatarsus adductus (d). The second patient (Patient 38), at 3 months of age, shows wide anterior fontanel, long ears, hairy pinnae, excess scalp skin, short and downslanted palpebral fissures, hypertelorism, depressed nasal bridge, inverted V shaped upper lip with think vermilion (e, f), long and narrow fingers (g) and bilateral talipes equinovarus (h).
The second patient (patient 38), at 3 months of age, weighed 4.3 kg (− 2.29 z), had a length of 61 cm (− 0.29 z) and head circumference of 39.5 cm (− 1.58 z). His mother had gestational diabetes mellitus. He had wide anterior fontanel, long ears, hairy pinnae, short and downslanted palpebral fissures, hypertelorism, depressed nasal bridge, inverted V-shaped upper lip with thin vermilion, high arched palate, bifid uvula, excess scalp skin, long and narrow fingers, bilateral talipes equinovarus and Mongolian spots (Fig. 4e–h). Mild pectus excavatum, skin laxity, cutis marmorata and umbilical hernia were also noted in him. Echocardiography was normal with z-score of 0.2 for the aortic root size.
Clinical findings in 8 families without a clinically relevant variant in 62 HCTD-related genes
Eight families (15%; CI 95% 8–27) did not have a clinically significant variant in 62 HCTD-related genes (disease genes and candidate genes). Clinical features of the eight index patients (46–53 and the brother of patient 52) are summarized in Table 3. None of them met the revised Ghent criteria. However, cardiac abnormalities were noted in all except one, and systemic score ≥ 7 was observed in two individuals. About half of them (4/9) fit criteria of the MASS (Mitral valve, Aorta, Skin, and Skeletal features) phenotype (MIM#604308), followed by Mitral valve prolapse syndrome (MIM%157700; #607829; %610840) (2/9), aortopathy (2/9) and an individual with Marfan-like disorder.
Table 3.
Patients with no clinically relevant variant in genes known to cause Marfan syndrome and related disorders.
| Patient # | Age at evaluation (years) | Gender | Revised Ghent criteria | Cardiac/vascular manifestations | Ocular features | Systemic score | Clinical diagnosis (pre-test) |
|---|---|---|---|---|---|---|---|
| 46 | 15 | Male | Negative | Tricuspid and mitral valve prolapse with regurgitation | Myopia | Negative | MASS phenotype |
| 47a | 15 | Female | Negative | Tricuspid and mitral valve prolapse with regurgitations | Absent | Negative | Mitral valve prolapse syndrome |
| 48 | 22 | Male | Negative | Thoracic and abdominal aortic aneurysm | Absent | Negative | Aortopathy |
| 49 | 16 | Female | Negative | Mitral valve prolapse regurgitation | Absent | Negative | MASS phenotype |
| 50 | 13 | Female | Negative | Myxomatous mitral valve with regurgitation | Absent | Negative | Mitral valve prolapse syndrome |
| 51 | 18 | Male | Negative | Myxomatous mitral valve with regurgitation | Absent | Positive | MASS phenotype |
| 52b,c | 2 | Male | Negative | Mild aortic root dilatation with ventricular septal defect | Absent | Negative | Aortopathy with facial dysmorphism |
| 52’s brotherc | 2 | Male | Negative | Absent | Absent | Negative | MASS phenotype |
| 53d | 15 | Female | Negative | Arteritis | Absent | Positive | Marfanoid disorder |
Revised Ghent criteria ‘negative’ indicates non-fulfillment. Systemic score ‘positive’ indicates systemic involvement (score ≥ 7) and ‘negative’ suggests no systemic involvement (score < 7).
MASS phenotype Mitral valve, Aorta, Skin, and Skeletal features.
aPatient 47 had poor scholastic performance.
bPatient 52 had developmental delay.
cThe difference in the phenotypes of patient 52 and his brother could suggest variable expression.
dPatient 53 had microtia and perauricular tag.
Discussion
We describe the clinical spectrum and genetic findings in 83 individuals from 53 Indian families with MFS, LDS and SGS. This is so far the largest cohort of Indian patients with a definitive molecular diagnosis for an aortopathy in a total of 45 index patients. The identification of clinically significant variants in MFS and related disorders reduces the uncertainty in diagnosis in individuals with a suspected diagnosis and guides appropriate management of their cardiovascular and ocular complications. Our study also provides the mutation spectrum in Indian patients with these three types of HCTDs and adds 21 novel rare variants.
We could not find any published reports on Indian patients with LDS or SGS with a molecular diagnosis. Our report now adds seven patients with LDS, two novel TGFBR2 variants, two patients with SGS and a novel disease-causing variant in the R-SMAD binding domain of SKI to the literature. We did not note any unusual clinical features in our small cohort of individuals with LDS and SGS.
Previously, only two publications have reported pathogenic variants in FBN1 in individuals from India28,29. One reported a fetus with arthrogryposis, multiple joint dislocations, scoliosis and facial dysmorphism who carried the variant p.(Pro2002Ser). Incidentally the fetus had a variant in FBN2 too (NM_001999.3:c.2945G > T; p.(Cys982Phe)). However, segregation of the variants in the family was not performed28. The second family comprised 27 individuals with ectopia lentis in whom the FBN1 missense variant p.(Arg240Cys) segregated29. Absence of other cardinal manifestations of MFS suggests occurrence of autosomal dominant “isolated” ectopia lentis 1 in the family (MIM#129600).
Several large cohorts on MFS and related disorders have been published previously with patients originating from European countries or China23,30–36. They report a definitive molecular diagnosis in 40–95% of individuals, depending on the inclusion criteria and the testing strategy. We obtained a molecular diagnosis in 45/53 (85%; CI 95% 73–92) families by NGS, including targeted multiple gene panel and whole-exome sequencing, and multiplex ligation-dependent probe amplification.
In our cohort, 36 of the 45 (80.0%; CI 95% 66–89) patients carried a rare variant in FBN1, and the majority of them were missense (21, 58.3%; CI 95% 42–73) in concordance with the literature34,37, and 13 substituted or introduced a cysteine residue. Also, we observed splicing variants and multi-exon deletions (five each or 13.9% each; CI 95% 6–29 each), small deletion/insertion (three, 8.3%; CI 95% 3–22) and nonsense (two, 5.6%; CI 95% 2–18) variants in FBN1. In the literature only ~ 5% of probands with an FBN1 pathogenic variant have been reported to carry a deletion or duplication2,33, which is 2.8-fold lower in our relatively small cohort of Indian patients. We noted missense variants in four (8.9%; CI 95% 4–21) patients in TGFBR1, three (6.7%; CI 95% 2–18) in TGFBR2, and two (4.4%; CI 95% 1–15) in SKI. Overall, there were 41 rare variants in four genes, with four variants identified in more than one family indicating the private nature of the remaining variants37. 51.2% (CI 95% 36–66) of the rare variants were novel, which is similar to the percentages reported in other studies (46.6–67.5%)19,23,26,31,32,34.
Disease-causing variants in exon 24–32 have been associated with early-onset and rapidly progressive MFS2,30,38. Seven (7/36) index patients of our cohort have pathogenic variants in this region of the FBN1 gene. Their age at diagnosis ranged from 4 months to 7 years except one (23-years-old). Four of them had de novo variants whereas three were familial. We also report on a proband (patient 11) and his paternal half-sister with the FBN1 nonsense variant c.3012C > A/p.(Tyr1004*). The father did not carry this variant in leukocyte-derived DNA indicating germline mosaicism in him. Five of the seven (71%; CI 95% 36–92) had ocular, cardiac and skeletal manifestations. We also had a 4-months-old infant with early onset MFS in our cohort. She had atrioventricular valve prolapse with severe mitral regurgitation, ostium secundum type of atrial septal defect measuring 11 mm, dilated chambers of heart with dilated aortic root, scoliosis, skin laxity and long and narrow fingers. She succumbed to cardiac failure at 6 months of age.
Bi-allelic FBN1 variants have been reported in 16 families (eight with homozygous and eight with compound heterozygous variants) with MFS39–45. We also document a patient (patient 10) with the homozygous FBN1 missense variant c.2861G > A/p.(Arg954His). Similar to the present individual, all the families reported in the literature with homozygous variants were consanguineous, except one (7/8, 87.5%; CI 95% 53–98). The initially described seven patients with bi-allelic variants had a severe clinical course with early age of onset ranging from day 7 to 22 years39–42,44,45. However, Arnaud et al. reported nine families with bi-allelic variants in FBN1 with classical and mild clinical features with age at diagnosis ranging from 8 to 53 years43. In the 16 reported families with bi-allelic FBN1 variants 17 missense variants, two frameshift and one nonsense variant have been identified43. Together with the p.(Arg954His) variant detected in patient 10 reported here, the vast majority of bi-allelic variants represent amino acid substitutions (18/21; 85.7%; CI 95% 65–95). Patient 10 at 5-years-8-months presented with typical facial features, bilateral ectopia lentis, bicuspid aortic valve with z-score of ≥ 2 and atrioventricular valve prolapse, the classical form of MFS. Although we were unable to perform a detailed clinical examination of patient 10’s parents who are heterozygous carriers of the p.(Arg954His) variant, the same heterozygous variant has been previously reported in a 58-year-old female with skeletal features, ectopia lentis but no cardiovascular abnormalities27. In the gnomAD browser, the variant was listed in 1 out of 251,154 alleles. Heterozygous carriers of the p.(Arg2726Trp) variant, who have a second pathogenic FBN1 variant on the other allele in three families, only had isolated skeletal features typical of MFS and/or high stature43. In addition, incomplete penetrance has been reported for individuals carrying the p.(Arg2726Trp) variant in the heterozygous state46, which is in line with a worldwide minor allele frequency (MAF) of 0.067% for this FBN1 variant (gnomAD browser). Interestingly, a worldwide MAF of 0.02% and 0.12% (gnomAD browser) for the FBN1 alterations p.(Pro1424Ala) and p.(Ala986Thr)43, respectively, also suggests incomplete penetrance in individuals carrying either of the variants in the heterozygous state and full penetrance in individuals with one of the two aforementioned FBN1 variants in trans with a second pathogenic variant. Although further studies are needed to study the effect of recessive FBN1 missense variants on fibrillin-1 function, several of the 18 missense variants identified in a homozygous or compound heterozygous state may act as hypomorphic alleles.
Eight families (15%; CI 95% 8–27) did not have a clinically significant variant in genes known to cause MFS or associated with HCTD (62 genes on NGS panel), and similar observations were reported in the literature33,34. Targeted panel NGS testing has considerable limitations in the detection of single- and multi-exon deletions/duplications and structural variants as well as non-coding and regulatory variants. Thus, clinically relevant variants might have been missed in one or several of the eight index patients. None of the eight patients met the revised Ghent criteria. The majority of the negative patients have atrio-ventricular valve prolapse with regurgitation. We observed poor scholastic performance (P47)/developmental delay (P52), ectopic and horseshoe kidney with polycystic ovaries (P50) and microtia and pre-auricular tag (P53) in some of them. Whole-exome or whole-genome sequencing will be performed in the eight families to identify the genetic cause underlying the disease in the index patients.
In conclusion, we describe the first and largest cohort of patients with MFS or related disorders from India and provide a base for further genetic testing in this large population. About half of them harbored a novel variant, which has expanded the mutation spectrum of these disorders. Biallelic FBN1 missense variants can be present in individuals with classic MFS and may point to hypomorphic FBN1 alleles manifesting only when present in the homozygous or compound heterozygous state. Identification of clinically significant variants reduces uncertainty in diagnosis in suspected individuals and guides appropriate management of their cardiovascular and ocular complications. Yet genetically unsolved patients with MFS-like conditions in this cohort suggests further genetic heterogeneity and the presence of phenocopies.
Methods
Study approval
The study was approved by the Institutional Ethics Committee, Kasturba Medical College and Hospital, Manipal (IEC No: 118/2016) and Narayana Health Academic Ethics Committee, Narayana Health Hospitals, Bangalore (NH/AEC-CL-2017-191). Informed consent for clinical data, samples and publication of photographs was obtained from parents/legal guardians of patients or the patients themselves. All experiments were performed in accordance with relevant guidelines and regulations.
Patient cohort and data collection
We recruited pediatric, adolescent and adult patients referred for genetics counseling at Kasturba Hospital, Manipal, India and Narayana Hrudayalaya Hospitals, Bangalore, India with features suggestive of MFS, aortopathy or related HCTDs over a period of 5 years. Clinical data and samples for all individuals were obtained with informed consent of patients’ parents/legal guardians or the patients themselves, including written consent to use photographs in this report. Clinical data that included a three-generation pedigree and family history of similarly affected individuals (specifically for the presence of tall stature, ocular abnormalities or visual defects and cardiac surgeries) were noted. We performed physical examination and recorded anthropometry for all patients. We collected echocardiographic information and calculated z-score for the aortic root measurements. Ophthalmological evaluation comprised a slit-lamp examination. We performed radiographic assessment and other imaging whenever necessary. Revised Ghent criteria was used for the diagnosis of MFS47,48. We collected two millilitres of blood samples from patients and their available family members including parents and siblings for genomic DNA isolation.
The lower and upper limits of the 95% confidence interval (CI 95%) for a proportion were calculated with the VassarStats tool (http://vassarstats.net/index.html) according to the method previously described49.
Molecular genetic analysis
Genomic DNA was isolated from leukocytes by standard procedures. For targeted NGS of the DNA sample of patients 1, 2, 5–8, 10–15, 17–24, 26, 28–37, 39–41, 43, and 45–53, we initially selected the coding region and adjacent intronic sequences of 18 genes (ACTA2 (NM_001613.2), BGN (NM_001711.5), CBS (NM_000071.2, NM_001321072.1), COL3A1 (NM_000090.3), FBN1 (NM_000138.4), FBN2 (NM_001999.3), LOX (NM_002317.6), MFAP5 (NM_003480.3), MYH11 (NM_001040113.1), MYLK (NM_053025.3), NOTCH1 (NM_017617.4), PRKG1 (NM_017617.4, NM_001098512.2), SKI (NM_003036.3), SMAD3 (NM_005902.3), TGFB2 (NM_001135599.2), TGFB3 (NM_003239.3), TGFBR1 (NM_004612.3), and TGFBR2 (NM_001024847.2)) related to syndromic and non-syndromic forms of aortopathies and connective tissue disorders. Enrichment of the regions of interest (ROI) was performed with the Illumina Rapid Capture Custom Enrichment kit or the Illumina Nextera Flex for Enrichment kit according to the manufacturer’s instructions. Briefly, following fragmentation of genomic DNA, fragmented DNA was amplified and patient-specific (index) adapters were added by PCR. Samples from 12 patients were combined into one single hybridization mix containing target-specific capture probes. The DNA-probe hybrids were then captured with streptavidin beads, and non-targeted DNA fragments as well as unspecific binding were removed by heated washes. Next, the captured DNA library was eluted from the beads, purified and amplified by PCR. For generation of clusters and subsequent sequencing of the targeted DNA samples on a flow cell, a sequencing reagent kit from Illumina was used. High-throughput NGS data were generated on an Illumina sequencing platform26.
SALSA MLPA kits P065-C1 and P066-C1 Marfan Syndrome, P148-B3 TGFBR1-TGFBR2, and P155-D2 COL3A1 (MRC-Holland) were used according to the manufacturer’s instructions to detect single and multiple exon deletions/duplications in FBN1 (all 66 exons), TGFBR1 (all nine exons), TGFBR2 (all eight exons) and COL3A1 (exons 1, 2, 4, 5, 9, 11, 14, 17, 20, 23, 28, 36, 43, 47, and 51). PCR products were separated on an automated capillary DNA sequencer (ABI 3500; Applied Biosystems). MLPA data were analysed with the Sequence Pilot module MLPA software (JSI Medical Systems)26.
For patients 46–53 without a pathogenic variant in one of the aforementioned 18 genes, the analysis was extended to the 44 additional genes on the customized NGS panel [ADAMTS10 (NM_030957.3), ADAMTS2 (NM_014244.4), B3GALT6 (NM_080605.3), B4GALT7 (NM_007255.2), CDKL1 (NM_004196.4), CHST14 (NM_130468.3), COL1A1 (NM_000088.3), COL1A2 (NM_000089.3), COL2A1 (NM_001844.4), COL4A1 (NM_001845.5), COL4A5 (NM_000495.4), COL5A1 (NM_000093.4), COL5A2 (NM_000393.3), DCHS1 (NM_003737.3), DIDO1 (NM_033081.2), DUOX2 (NM_014080.4), EFEMP2 (NM_016938.4), ELN (NM_001278939.1), EMILIN1 (NM_007046.3), FBLN5 (NM_006329.3), FKBP14 (NM_017946.3), FLNA (NM_001110556.1), FLNC (NM_001458.4), FOXE3 (NM_012186.2), FOXS1 (NM_004118.3), GATA5 (NM_080473.4), KDR (NM_002253.2), LRP1 (NM_002332.2), LTBP2 (NM_000428.2), LTBP4 (NM_003573.2), MAT2A (NM_005911.5), PEAR1 (NM_001080471.1), PLK1 (NM_005030.5), PLOD1 (NM_000302.3), PLOD3 (NM_001084.4), PRDM5 (NM_018699.3), SLC2A10 (NM_030777.3), SLC39A13 (NM_152264.4), SMAD2 (NM_005901.5), SMAD4 (NM_005359.5), SOX18 (NM_018419.2), TNXB (NM_019105.6), ULK4 (NM_017886.3), ZNF469 (NM_001127464.2)]. ROI sequences were aligned to the human reference genome (hg19) and visualized and evaluated by the Sequence Pilot module SeqNext software (JSI Medical Systems). NGS data of patients 46–53 were analysed for single nucleotide variants and copy number variations in all 62 panel genes.
Enrichment of the regions of interest for patients 3, 9, 25, 27, 38, 42 and 44 was performed with a custom Haloplex enrichment kit according to the manufacturer’s protocol (Agilent Technologies) as described previously19. Compared to the original kit described in Proost et al. (2015) the custom Haloplex enrichment kit contained additional probes for PRKG1 (ENST00000401604), TGFB3 (ENST00000238682), MAT2A (ENST00000306434) and MFAP5 (ENST00000359478) for patients 3, 9, 25, 27, 38, 42 and 44, FOXE3 (ENST00000335071) for patients 9, 25, 27, 38 and 44 and ELN (ENST00000358929), FBN2 (ENST00000262464) and SMAD2 (ENST00000402690) for patients 27 and 38. The concentration of each library was measured by Qubit fluorometric quantification (Life Technologies). For generation of clusters and subsequent sequencing of the targeted DNA samples on a flow cell, a sequencing reagent kit from Illumina was used. High-throughput NGS data were generated on an Illumina sequencing platform. ROI sequences were aligned to the human reference genome (hg19) and visualized and evaluated by the Sequence Pilot module SeqNext software (JSI Medical Systems)19.
Whole-exome sequencing (WES) in patients 4 and 16 was performed either with Nextera Rapid Capture Exomes (Illumina) or Agilent SureSelect V6 (Agilent Technologies) kit. Massively parallel sequencing was done on an Illumina NextSeq Platform. There was an average coverage depth of 110×, with ~ 94% of bases covered at > 20× and the data was analysed using an in-house pipeline based on Burrows-Wheeler Aligner (v0.7.15)50 and Genome Analysis Toolkit Best Practices pipeline (v3.6)51. We used ANNOVAR to annotate the variant call format (vcf) files52,53. We integrated annotated data with phenotypes catalogued in Online Mendelian Inheritance in Man, human phenotype ontology (HPO) terms, and allele frequency details from in-house variant database of 870 exomes of Indians. Rare variants were retrieved with minor allele frequency of < 1% in population databases [Exome Aggregation Consortium (ExAC) and gnomAD54,55] and our in-house data. Variants were prioritized for the phenotypes56.
Identified sequence variants have been searched in the following databases: HGMD Professional versions 2017.1-2019.2 (https://portal.biobase-international.com/hgmd/pro/start.php)57,58, UMD-FBN1 (http://www.umd.be/FBN1/)37, and gnomAD v2.1.1. (https://gnomad.broadinstitute.org/)54. Classification of novel variants as pathogenic variants, likely pathogenic variants and variants of unknown significance (VUS) was performed according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology standards and guidelines59, either with the help of VarSome (https://varsome.com/)60 or by manual application of the guidelines. The functional impact of novel variants was assessed by the pathogenicity prediction programs CADD (http://cadd.gs.washington.edu/score)61, REVEL (https://sites.google.com/site/revelgenomics/downloads)62, and M-CAP (http://bejerano.stanford.edu/MCAP/)63. Genetic tolerance at the affected amino acid position in the protein was predicted by MetaDome (https://stuart.radboudumc.nl/metadome/)64. Splice site prediction scores for novel intronic variants were calculated for wild-type and mutated sequences by using the in silico tools Human Splicing Finder 3.1 (http://umd.be/HSF3/HSF.shtml), NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/), and the Berkeley Drosophila Genome Project Database (https://www.fruitfly.org/seq_tools/splice.html)65–68.
Sanger sequencing was performed for validation of pathogenic, likely pathogenic sequence variants and VUS identified by NGS and for regions of interest covered by less than 20 reads. Segregation analysis of pathogenic and likely pathogenic variants in affected and/or healthy family members of the index patient was performed by Sanger sequencing using an automated capillary DNA sequencer (ABI 3500; Applied Biosystems). Sequence electropherograms were analysed using the Sequence Pilot module SeqPatient software (JSI Medical Systems).
All novel variants were deposited in the LOVD Database, where they are available under the DB-ID numbers 0000667876 to 0000667897, 0000708485 and 0000708486.
Supplementary Information
Acknowledgements
We thank all the patients and families for their kind participation in the study. The project entitled “Improving the clinical care of children and young adults with Marfan syndrome and related disorders by molecular genetic testing through next generation sequencing” was jointly funded by the Indian Council of Medical Research (File No. 5/7/1508/2016 to K.M.G.) and the Federal Ministry of Education and Research (01DQ17003 to K.K.). This research was also supported by funding from the University of Antwerp (GOA, Methusalem-OEC Grant “Genomed” FFB190208), the Fund for Scientific Research, Flanders (FWO, Belgium, G.0356.17), The Dutch Heart Foundation (2013T093), and the Marfan Foundation. B.L. is senior clinical investigator of the Fund for Scientific Research, Flanders, holds a consolidator Grant from the European Research Council (Genomia—ERC-COG-2017-771945) and is a member of European Reference Network on rare vascular disorders (VASCERN). L.V.H. and J.M. are supported by the Fund for Scientific Research Flanders as PhD and postdoctoral researchers, respectively.
Author contributions
K.M.G. and K.K. designed and supervised the study. K.M.G., S.J.P., K.M.A., P.V.S., V.S.K., S.S., S.M., S.K.V. and A.S. collected family details, performed clinical and/or echocardiographic evaluation and recruited patients for this study. S.S.N. and K.M.G. collected and summarized clinical data. L.V.D.H., J.M., L.V.L., B.L., P.E.S., F.K., I.R., A.W.F. and K.K. evaluated targeted gene panel next-generation sequencing, Sanger sequencing and MLPA data and interpreted the variants. N.K. performed whole-exome sequencing analysis. P.E.S. prepared Fig. 1, Table 1 and Supplementary Table S1. S.S.N. and K.M.G. prepared Figs. 2, 3, 4, Tables 2 and 3. S.S.N., P.E.S., K.M.G. and K.K. wrote the manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
All data generated or analysed during this study are included in this published article (and its “Supplementary Information File”).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shalini S. Nayak and Pauline E. Schneeberger.
Contributor Information
Katta M. Girisha, Email: girish.katta@manipal.edu
Kerstin Kutsche, Email: kkutsche@uke.de.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80755-7.
References
- 1.Meester JAN, et al. Differences in manifestations of Marfan syndrome, Ehlers–Danlos syndrome, and Loeys–Dietz syndrome. Ann. Cardiothorac. Surg. 2017;6:582–594. doi: 10.21037/acs.2017.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dietz, H. C. Marfan Syndrome. In (eds Adam M. P. et al.) GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2020. 2001 Apr 18 [updated 2017 Oct 12].
- 3.Dietz HC, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 4.Cannaerts E, van de Beek G, Verstraeten A, Van Laer L, Loeys B. TGF-beta signalopathies as a paradigm for translational medicine. Eur. J. Med. Genet. 2015;58:695–703. doi: 10.1016/j.ejmg.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Loeys, B. L. & Dietz, H. C. Loeys–Dietz Syndrome. In (eds M. P. Adam et al.) GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2020. 2008 Feb 28 [updated 2018 Mar 1].
- 6.Attias D, et al. Comparison of clinical presentations and outcomes between patients with TGFBR2 and FBN1 mutations in Marfan syndrome and related disorders. Circulation. 2009;120:2541–2549. doi: 10.1161/CIRCULATIONAHA.109.887042. [DOI] [PubMed] [Google Scholar]
- 7.Mühlstädt K, et al. Case-matched comparison of cardiovascular outcome in Loeys–Dietz Syndrome versus Marfan Syndrome. J. Clin. Med. 2019 doi: 10.3390/jcm8122079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Micha D, et al. SMAD2 mutations are associated with arterial aneurysms and dissections. Hum. Mutat. 2015;36:1145–1149. doi: 10.1002/humu.22854. [DOI] [PubMed] [Google Scholar]
- 9.Bertoli-Avella AM, et al. Mutations in a TGF-beta ligand, TGFB3, cause syndromic aortic aneurysms and dissections. J. Am. Coll. Cardiol. 2015;65:1324–1336. doi: 10.1016/j.jacc.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boileau C, et al. TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of Marfan syndrome. Nat. Genet. 2012;44:916–921. doi: 10.1038/ng.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindsay ME, et al. Loss-of-function mutations in TGFB2 cause a syndromic presentation of thoracic aortic aneurysm. Nat. Genet. 2012;44:922–927. doi: 10.1038/ng.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Laar IM, et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat. Genet. 2011;43:121–126. doi: 10.1038/ng.744. [DOI] [PubMed] [Google Scholar]
- 13.Loeys BL, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat. Genet. 2005;37:275–281. doi: 10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- 14.Greally, M. T. Shprintzen–Goldberg Syndrome. In (eds M. P. Adam et al.). GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2020. 2006 Jan 13 [updated 2020 Apr 9]. [PubMed]
- 15.Schepers D, et al. The SMAD-binding domain of SKI: A hotspot for de novo mutations causing Shprintzen–Goldberg syndrome. Eur. J. Hum. Genet. 2015;23:224–228. doi: 10.1038/ejhg.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carmignac V, et al. In-frame mutations in exon 1 of SKI cause dominant Shprintzen–Goldberg syndrome. Am. J. Hum. Genet. 2012;91:950–957. doi: 10.1016/j.ajhg.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyle AJ, et al. Mutations in the TGF-beta repressor SKI cause Shprintzen–Goldberg syndrome with aortic aneurysm. Nat. Genet. 2012;44:1249–1254. doi: 10.1038/ng.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poninska JK, et al. Next-generation sequencing for diagnosis of thoracic aortic aneurysms and dissections: Diagnostic yield, novel mutations and genotype phenotype correlations. J. Transl. Med. 2016;14:115. doi: 10.1186/s12967-016-0870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Proost D, et al. Performant mutation identification using targeted next-generation sequencing of 14 thoracic aortic aneurysm genes. Hum. Mutat. 2015;36:808–814. doi: 10.1002/humu.22802. [DOI] [PubMed] [Google Scholar]
- 20.Wooderchak-Donahue W, et al. Clinical utility of a next generation sequencing panel assay for Marfan and Marfan-like syndromes featuring aortopathy. Am. J. Med. Genet. 2015;167A:1747–1757. doi: 10.1002/ajmg.a.37085. [DOI] [PubMed] [Google Scholar]
- 21.Ziganshin BA, et al. Routine genetic testing for thoracic aortic aneurysm and dissection in a clinical setting. Ann. Thorac. Surg. 2015;100:1604–1611. doi: 10.1016/j.athoracsur.2015.04.106. [DOI] [PubMed] [Google Scholar]
- 22.Weerakkody R, et al. Targeted genetic analysis in a large cohort of familial and sporadic cases of aneurysm or dissection of the thoracic aorta. Genet. Med. 2018;20:1414–1422. doi: 10.1038/gim.2018.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang M, et al. Identification of novel clinically relevant variants in 70 Southern Chinese patients with thoracic aortic aneurysm and dissection by next-generation sequencing. Sci. Rep. 2017;7:10035. doi: 10.1038/s41598-017-09785-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campens L, et al. Gene panel sequencing in heritable thoracic aortic disorders and related entities—Results of comprehensive testing in a cohort of 264 patients. Orphanet. J. Rare Dis. 2015;10:9. doi: 10.1186/s13023-014-0221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overwater E, et al. Results of next-generation sequencing gene panel diagnostics including copy-number variation analysis in 810 patients suspected of heritable thoracic aortic disorders. Hum. Mutat. 2018;39:1173–1192. doi: 10.1002/humu.23565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renner S, et al. Next-generation sequencing of 32 genes associated with hereditary aortopathies and related disorders of connective tissue in a cohort of 199 patients. Genet. Med. 2019;21:1832–1841. doi: 10.1038/s41436-019-0435-z. [DOI] [PubMed] [Google Scholar]
- 27.Söylen B, et al. Prevalence of dural ectasia in 63 gene-mutation-positive patients with features of Marfan syndrome type 1 and Loeys–Dietz syndrome and report of 22 novel FBN1 mutations. Clin. Genet. 2009;75:265–270. doi: 10.1111/j.1399-0004.2008.01126.x. [DOI] [PubMed] [Google Scholar]
- 28.Aggarwal S, Das Bhowmik A, Tandon A, Dalal A. Exome sequencing reveals blended phenotype of double heterozygous FBN1 and FBN2 variants in a fetus. Eur. J. Med. Genet. 2018;61:399–402. doi: 10.1016/j.ejmg.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Vanita V, et al. A recurrent FBN1 mutation in an autosomal dominant ectopia lentis family of Indian origin. Mol. Vis. 2007;13:2035–2040. [PubMed] [Google Scholar]
- 30.Faivre L, et al. Effect of mutation type and location on clinical outcome in 1013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am. J. Hum. Genet. 2007;81:454–466. doi: 10.1086/520125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Attanasio M, et al. FBN1 mutation screening of patients with Marfan syndrome and related disorders: Detection of 46 novel FBN1 mutations. Clin. Genet. 2008;74:39–46. doi: 10.1111/j.1399-0004.2008.01007.x. [DOI] [PubMed] [Google Scholar]
- 32.Hung CC, et al. Mutation spectrum of the fibrillin-1 (FBN1) gene in Taiwanese patients with Marfan syndrome. Ann. Hum. Genet. 2009;73:559–567. doi: 10.1111/j.1469-1809.2009.00545.x. [DOI] [PubMed] [Google Scholar]
- 33.Baetens M, et al. Applying massive parallel sequencing to molecular diagnosis of Marfan and Loeys–Dietz syndromes. Hum. Mutat. 2011;32:1053–1062. doi: 10.1002/humu.21525. [DOI] [PubMed] [Google Scholar]
- 34.Mannucci L, et al. Mutation analysis of the FBN1 gene in a cohort of patients with Marfan Syndrome: A 10-year single center experience. Clin. Chim. Acta. 2020;501:154–164. doi: 10.1016/j.cca.2019.10.037. [DOI] [PubMed] [Google Scholar]
- 35.Vanem TT, et al. Marfan syndrome: Evolving organ manifestations-A 10-year follow-up study. Am. J. Med. Genet. 2020;182:397–408. doi: 10.1002/ajmg.a.61441. [DOI] [PubMed] [Google Scholar]
- 36.Bombardieri E, et al. Marfan syndrome and related connective tissue disorders in the current era in Switzerland in 103 patients: Medical and surgical management and impact of genetic testing. Swiss Med. Wkly. 2020;150:w20189. doi: 10.4414/smw.2020.20189. [DOI] [PubMed] [Google Scholar]
- 37.Collod-Beroud G, et al. Update of the UMD-FBN1 mutation database and creation of an FBN1 polymorphism database. Hum. Mutat. 2003;22:199–208. doi: 10.1002/humu.10249. [DOI] [PubMed] [Google Scholar]
- 38.Carande EJ, Bilton SJ, Adwani S. A case of neonatal Marfan syndrome: A management conundrum and the role of a multidisciplinary team. Case Rep. Pediatr. 2017;2017:8952428. doi: 10.1155/2017/8952428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karttunen L, Raghunath M, Lönnqvist L, Peltonen L. A compound-heterozygous Marfan patient: Two defective fibrillin alleles result in a lethal phenotype. Am. J. Hum. Genet. 1994;55:1083–1091. [PMC free article] [PubMed] [Google Scholar]
- 40.de Vries BB, Pals G, Odink R, Hamel BC. Homozygosity for a FBN1 missense mutation: Clinical and molecular evidence for recessive Marfan syndrome. Eur. J. Hum. Genet. 2007;15:930–935. doi: 10.1038/sj.ejhg.5201865. [DOI] [PubMed] [Google Scholar]
- 41.Hilhorst-Hofstee Y, et al. The clinical spectrum of missense mutations of the first aspartic acid of cbEGF-like domains in fibrillin-1 including a recessive family. Hum. Mutat. 2010;31:E1915–1927. doi: 10.1002/humu.21372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Dijk FS, et al. Compound-heterozygous Marfan syndrome. Eur. J. Med. Genet. 2009;52:1–5. doi: 10.1016/j.ejmg.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Arnaud P, et al. Homozygous and compound heterozygous mutations in the FBN1 gene: Unexpected findings in molecular diagnosis of Marfan syndrome. J. Med. Genet. 2017;54:100–103. doi: 10.1136/jmedgenet-2016-103996. [DOI] [PubMed] [Google Scholar]
- 44.Hogue J, et al. Homozygosity for a FBN1 missense mutation causes a severe Marfan syndrome phenotype. Clin. Genet. 2013;84:392–393. doi: 10.1111/cge.12073. [DOI] [PubMed] [Google Scholar]
- 45.Khan AO, Bolz HJ, Bergmann C. Results of fibrillin-1 gene analysis in children from inbred families with lens subluxation. J. Aapos. 2014;18:134–139. doi: 10.1016/j.jaapos.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 46.Buoni S, et al. The FBN1 (R2726W) mutation is not fully penetrant. Ann. Hum. Genet. 2004;68:633–638. doi: 10.1046/j.1529-8817.2004.00113.x. [DOI] [PubMed] [Google Scholar]
- 47.Loeys BL, et al. The revised Ghent nosology for the Marfan syndrome. J. Med. Genet. 2010;47:476–485. doi: 10.1136/jmg.2009.072785. [DOI] [PubMed] [Google Scholar]
- 48.von Kodolitsch Y, et al. Perspectives on the revised Ghent criteria for the diagnosis of Marfan syndrome. Appl. Clin. Genet. 2015;8:137–155. doi: 10.2147/TACG.S60472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newcombe RG. Two-sided confidence intervals for the single proportion: Comparison of seven methods. Stat. Med. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Girisha KM, et al. Biallelic variants p.Arg1133Cys and p.Arg1379Cys in COL2A1: Further delineation of phenotypic spectrum of recessive Type 2 collagenopathies. Am. J. Med. Genet. A. 2020;182:338–347. doi: 10.1002/ajmg.a.61414. [DOI] [PubMed] [Google Scholar]
- 57.Stenson PD, et al. The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stenson PD, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 59.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kopanos C, et al. VarSome: The human genomic variant search engine. Bioinformatics. 2019;35:1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ioannidis NM, et al. REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jagadeesh KA, et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 64.Wiel L, et al. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum. Mutat. 2019;40:1030–1038. doi: 10.1002/humu.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-O. [DOI] [PubMed] [Google Scholar]
- 66.Desmet FO, et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hebsgaard SM, et al. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its “Supplementary Information File”).