Abstract
Cardiovascular diseases (CVDs) are the leading cause of death and a major cause of disability globally. Transcription factor EB (TFEB), as a member of the microphthalmia transcription factor (MITF) family, has been demonstrated to be a master regulator of autophagy and lysosomal biogenesis. Emerging studies suggest that TFEB regulates homeostasis in the cardiovascular system and shows beneficial effects on CVDs, including atherosclerosis, aortic aneurysm, postischemic angiogenesis, and cardiotoxicity, constituting a promising molecular target for the prevention and treatment of these diseases. Post-translational modifications regulate TFEB nuclear translocation and its transcriptional activity. Therapeutic strategies have been pursued to enhance TFEB activity and facilitate TFEB beneficial effects on CVDs. The elucidation of TFEB function and the precise underlying mechanisms will accelerate drug development and potential applications of TFEB drugs in the treatment of human diseases.
Keywords: Cardiovascular disease, Autophagy, Lysosome, Post-translational modification, Drug development
Abbreviations: TFEB, transcription factor EB; MITF, microphthalmia transcription factor; CVDs, cardiovascular diseases; TFEC, transcription factor EC; TFE3, transcription factor E3; bHLH-Zip, basic helix-loop-helix and leucine zipper domains; CLEAR, Coordinated Lysosomal Expression and Regulation; ECs, endothelial cells; VSMCs, vascular smooth muscle cells; mTOR, mammalian target of rapamycin; HPβCD, 2-Hydroxypropyl-β-cyclodextrin; PGC1α, peroxisome proliferator-activated receptor gamma, coactivator 1 alpha; PPAR, peroxisome proliferator-activated receptor; Atg5, autophagy related 5
1. Introduction
Cardiovascular diseases (CVDs) are the leading cause of death in the United States and worldwide. The mortality rate of CVD is much higher in patients with metabolic disorders, including obesity and diabetes. Transcription factor EB (TFEB) is an important transcriptional factor that enhances autophagy and lysosomal biogenesis [1,2]. Accumulating studies suggest that TFEB plays critical roles in maintaining body homeostasis, particularly in the cardiovascular, metabolic, immune, cancer, and nervous systems. In this review, we summarize the function and underlying mechanisms of TFEB in CVDs and discuss the potential of TFEB as a therapeutic target to prevent and treat these human diseases.
1.1. MITF/TFE gene family
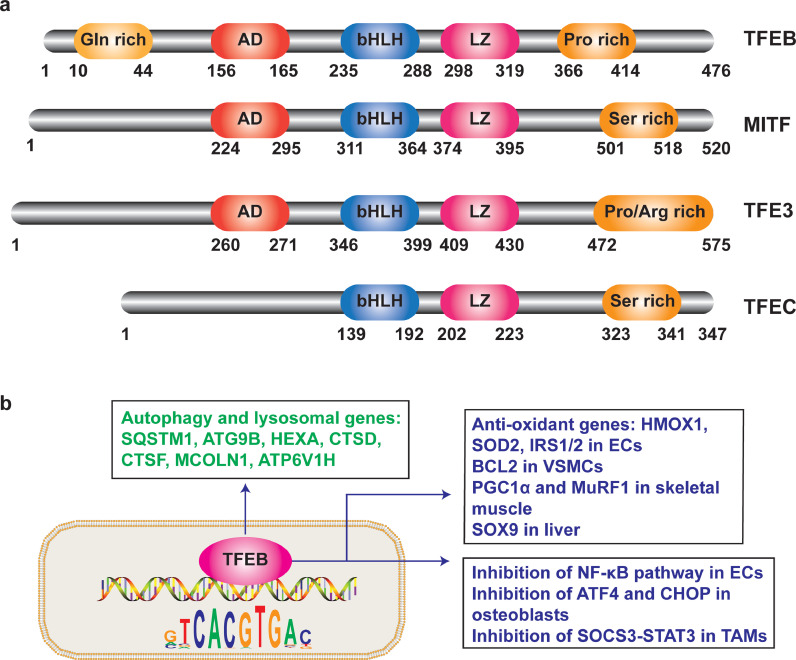
The microphthalmia transcription factor (MITF) family is comprised of TFEB, MITF, transcription factor EC (TFEC), and transcription factor E3 (TFE3). MITF family members contain similar adjacent basic helix-loop-helix and leucine zipper domains (bHLH-Zip) and regulate target genes by binding to the E-box (CANNTG) at the gene promoter (Fig. 1a). The MITF/TFE family has diverse effects on cellular processes, including organelle biogenesis, proliferation, apoptosis, nutrient sensing, metabolism, and stress adaptation [3]. Disruption of Mitf by transgene insertion leads to defects in pigmentation, eye size, bone development, mast cells, and hearing in mice [4]. Although Mitf and Tfe3 often have redundant functions in osteoclast development, Mitf/Tfe3 double knockout (KO) mice exhibited severe osteopetrosis [5].
Fig. 1.
Schematic representation of functional domains for MITF family members and TFEB target genes. (a), MITF members contain N-term transcriptional activation domain (AD), basic helix-loop-helix region (bHLH), leucine zipper (LZ), proline-rich domain (Pro-rich), or serine-rich stretch (Ser) domain. Numbers indicate amino acid location in the protein. (b), Diagram illustrating the genes and pathways regulated by TFEB. Besides autophagy and lysosomal biogenesis genes, TFEB also regulates numerous other genes and pathways. Abbreviations: ECs, endothelial cells; VSMCs, vascular smooth muscle cells; TAMs, tumor-associated macrophages.
TFEB was screened from a cDNA library in a human B-cell line by using a probe sequence in the major late promoter of the adenovirus [6]. Although the structure of MITF members is similar, TFEB has distinct effects on cell biology and multiple diseases. Tfeb-deficient mice die between 9.5–10.5 days in the uterus due to impaired placental vascularization [7]. Cell differentiation is an indispensable process in development, growth and regeneration of multicellular organisms. In the liver, TFEB induces the differentiation of murine liver stem/progenitor cells into the progenitor/cholangiocyte lineage while inhibiting hepatocyte differentiation through upregulating Sox9, a marker of precursor and biliary cells [8]. In acute promyelocytic leukemia, autophagy is critical for the differentiation of leukemic blasts. TFEB enhances autophagy and potentiates leukemic cell differentiation [9]. Osteoblasts are bone-forming cells that produce collagen type I and bone matrix proteins. TFEB enhanced osteoblast differentiation and osteoblastogenesis, resulting from the reduction of activating transcription factor 4 (ATF4) and CCAAT/enhancer-binding protein homologous protein (CHOP) [10]. These findings underscore the importance of TFEB in cell differentiation and development. The observations of severe phenotypes resulting from Mitf/Tfe member deficiency have sparked great interest in the research field. In particular, the role of TFEB in cardiovascular biology is gaining increased attention.
1.2. TFEB is a master regulator of lysosomal biogenesis and autophagy
Autophagy is a critical process for cells to clear and recycle damaged molecules and organelles. Autophagosomes fuse with lysosomes to form autolysosomes, which degrade proteins and macromolecules. TFEB regulates a variety of lysosomal genes, including HEXA, PSAP, CTSD, CTSF, MCOLN1, and ATP6V1H, by binding to the palindromic 10-base pair motif (GTCACGTGAC) in the gene promoter [1]. This motif was named Coordinated Lysosomal Expression and Regulation (CLEAR) element because of its enrichment in the promoter of lysosomal genes. TFEB also upregulates autophagy genes (e.g., UVRAG, WIPI, MAPLC3B, SQSTM1, VPS11, VPS18, and ATG9B) to promote autophagy. The enhanced autophagy and lysosomal function are essential for cells to adapt to starvation and stress conditions (Fig. 1b) [2].
1.3. Autophagy-independent effects of TFEB
Although many studies focused on autophagy-dependent effects of TFEB in different cells and tissues, emerging evidence indicates that besides autophagy, TFEB also regulates other diverse genes and signalling pathways. In osteoblasts, TFEB promotes cell differentiation by inhibiting ATF4 and CHOP [10]. In skeletal muscle, TFEB mediates Ang II-induced skeletal muscle wasting by transcriptional regulation of muscle-enriched E3 ubiquitin ligase muscle RING finger-1 (MuRF1) expression [11]. In tumour-associated macrophages, downregulation of TFEB promotes M2 polarization through reducing suppressor of cytokine signalling 3 (SOCS3) and signal transducer and activator of transcription 3 (STAT3) pathways [12]. In vascular endothelial cells (ECs), TFEB inhibits vascular inflammation via the upregulation of antioxidant genes, heme oxygenase-1 (HMOX1) and superoxide dismutase 2 (SOD2) [13], and inhibition of the nuclear factor kappa B (NF-κB) pathway [14]. In vascular smooth muscle cells (VSMCs), TFEB binds to the promoter of B-cell lymphoma 2 (BCL2), a potent anti-apoptotic gene [15]. In addition to autophagy and lysosomal genes, the other TFEB target genes and signalling pathways are summarized in Fig. 1b. Taken together, TFEB not only enhances autophagy and lysosome-mediated cellular clearance but also regulates diverse cellular processes largely dependent on transcriptional modulation of target genes.
2. Mechanisms mediating the regulation of TFEB
Cells can sense environmental changes, such as heat and starvation, and initiate an adaptive response to help cells maintain homeostasis. TFEB acts as a critical mediator in the context of cellular adaptation to diverse stresses. As a transcription factor, TFEB translocates to the nucleus and regulates target gene expression once upstream regulators activate it. Rapid post-translational modification and prolonged transcriptional regulation modulate TFEB activity and enable TFEB to serve as a highly dynamic effector to maintain cell homeostasis rapidly and efficiently in response to various stimuli.
2.1. Regulation of TFEB nuclear translocation
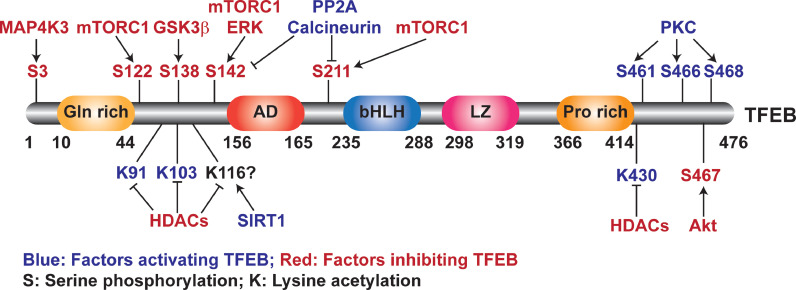
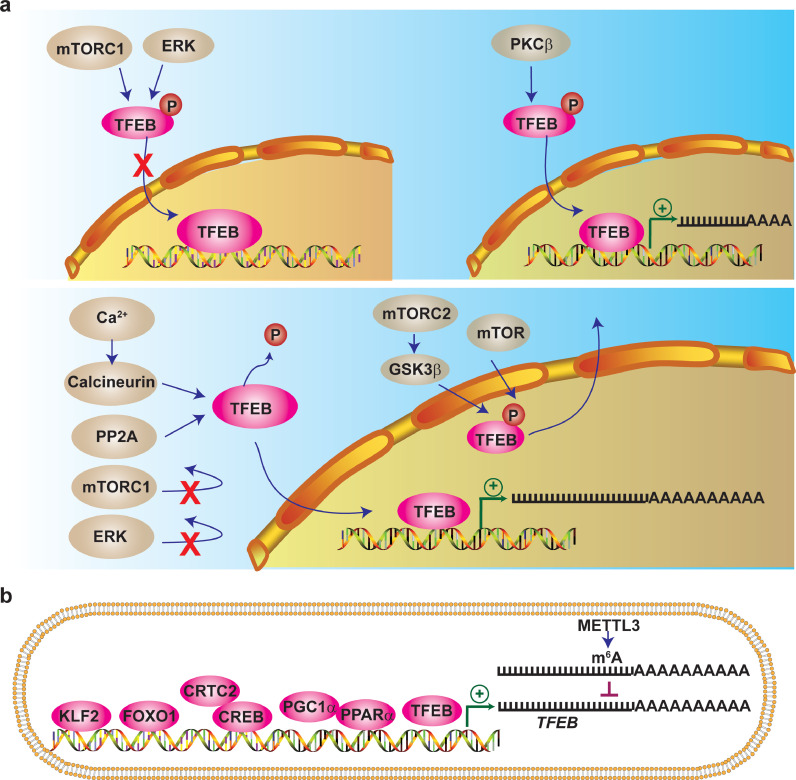
Post-translational modifications, including phosphorylation, acetylation, ubiquitination and SUMOylation, can alter protein activity, subcellular distribution, and protein interactions. Phosphorylation and acetylation sites on the TFEB protein have been identified to regulate TFEB nuclear translocation and its transcriptional activity (Fig. 2). Phosphorylation of TFEB at specific sites enables TFEB to be either retained in the cytoplasm, translocated to the nucleus, or retained in the nucleus due to impaired nuclear export. In the nucleus, TFEB binds to the promoter of target genes to regulate gene expression (Fig. 3a). TFEB phosphorylation is catalyzed by multiple kinases including the mammalian target of rapamycin complex 1 (mTORC1), [16], [17], [18] protein kinase Cβ (PKCβ) [19], extracellular signal-regulated kinase (ERK) [2], AKT serine/threonine kinase (Akt) [20], glycogen synthase kinase 3 beta (GSK3β) [21,22], and mitogen-activated protein kinase kinase kinase 3 (MAP4K3).[23] On the other hand, calcineurin (a phosphatase activated by intracellular Ca2+) [24] and protein phosphatase 2A (PP2A) [25] dephosphorylate TFEB and induce its nuclear translocation. Moreover, nuclear export is a crucial step for TFEB shuttling between the cytosol and nucleus. Chromosomal Maintenance 1 (CRM1), a major export protein that facilitates the transport of proteins across the nuclear membrane to the cytoplasm, mediates TFEB nuclear export [22,26]. mTOR-dependent TFEB phosphorylation is required for this process [27]. Another study found that TFEB nuclear export is dependent on the phosphorylation of S142 and S138 controlled by mTORC2-GSK3β [22]. Therefore, phosphorylation status can regulate TFEB nuclear translocation and transcriptional activity.
Fig. 2.
Post-translational modifications of TFEB. The activity of TFEB is strictly controlled by post-translational modifications, including phosphorylation and acetylation. Diagram illustrating the modification sites and corresponding enzymes.
Fig. 3.
Mechanisms mediating the regulation of TFEB. (a) Phosphorylation is a well-recognized post-translational modification that regulates TFEB nuclear translocation. Inhibition of mTOR or ERK, activation of PKCβ or activation of phosphatases calcineurin or PP2A induces TFEB nuclear translocation and enhances TFEB transcriptional activity. mTOR or mTORC2-GSK3β facilitates TFEB nuclear export. (b) TFEB expression is regulated at the transcriptional level. Numerous transcription factors, including Krüppel-like factor 2 (KLF2), Forkhead box O1 (FOXO1), cAMP response element-binding protein (CREB) and its co-activator, CREB-regulated transcription co-activator 2 (CRTC2), peroxisome proliferator-activated receptor alpha (PPARα), peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α) and TFEB itself, transactivate TFEB gene expression. Also, methyltransferase like 3 (METTL3) decreases TFEB mRNA expression by N6-methyladenosine (m6A) modification.
Besides phosphorylation, acetylation of lysine residues (K91, K103, K116, and K430) has been reported to increase TFEB activity in HCT116 cells, a human colorectal carcinoma cell line [28]. However, another study reported that TFEB K116 deacetylation by SIRT1 promotes its transcriptional activity in microglia [29]. The discrepancy may be due to the nature of stimuli and cell types used. SUMOylation is a reversible post-translational modification in which small ubiquitin-like modifier (SUMO) proteins are covalently conjugated to lysine residues in specific target proteins. SUMOylation of TFEB was reported in COS-7 cells. However, the mechanisms and functional changes related to SUMOylation remain to be explored .[30]. Collectively, post-translational modifications of the TFEB protein provide a molecular basis for the modulation of TFEB activity in cells (Fig. 3a).
2.2. Regulation of TFEB expression and mRNA stability
Multiple transcription factors that regulate TFEB at the transcriptional level have been identified in specific cells under different conditions (Fig. 3b). Krüppel-like factor 2 (KLF2) increases TFEB transcription in human umbilical vein endothelial cells (HUVECs) under laminar shear stress [14]. In the liver, starvation induces TFEB expression via a positive autoregulatory loop in mice [31], and cAMP response element-binding protein (CREB) and its co-activator, CREB-regulated transcription co-activator 2 (CRTC2), cooperatively increase TFEB transcription [32]. In the brain, TFEB mRNA expression is compromised in the mouse Huntington's disease model. PGC-1α can bind to the TFEB promoter to rescue its transcription and attenuate neurodegeneration in Huntington's disease [33]. In mouse astrocytes, the PPARα-RXRα-PGC1α complex increases TFEB transcription by binding to the PPAR response element in the TFEB promoter [34]. In adipocytes, Forkhead box O1 (FOXO1) directly increases TFEB transcription [35]. In addition to gene transcription, the regulation of mRNA stability is one of the critical control steps in dynamic gene expression. Methyltransferase like 3 (METTL3) destabilizes TFEB RNA through N6-methyladenosine (m6A) mRNA modification following ischemia/reperfusion (I/R) in mouse hearts [36]. Taken together, TFEB is a druggable target based on the feasibility of post-translational modifications and alterations in expression.
3. TFEB and CVDs
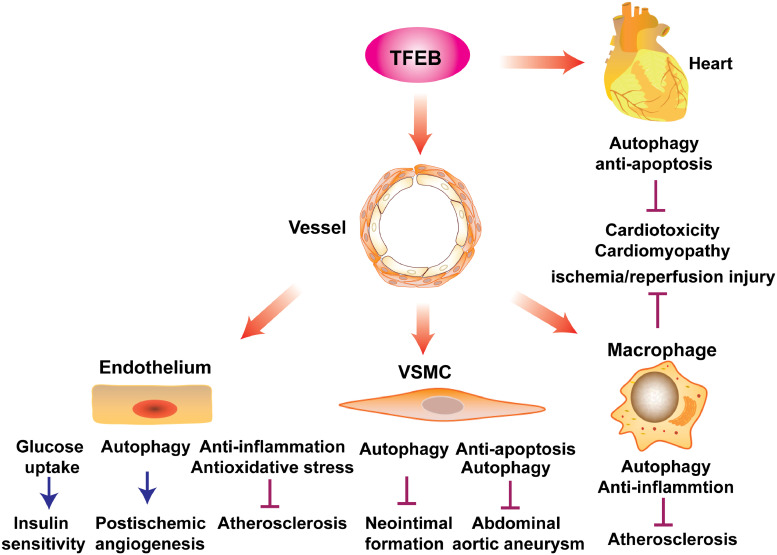
Although CVDs are heterogeneous in nature, they share some common pathological features, such as atherosclerosis and inflammation [37,38]. Recent studies have revealed that TFEB is critical to maintaining vascular and heart homeostasis (Fig. 4). TFEB possesses cell type-specific effects and shares some common pathways (e.g., promotes autophagy and cell survival) in different cells. Cell-specific knockout and transgenic animal models combined with various disease models have provided useful information for understanding the roles of TFEB in CVDs and developing new therapeutic approaches.
Fig. 4.
The role of TFEB in cardiovascular diseases (CVDs) and metabolic diseases. TFEB regulates the function of endothelial cells (ECs), vascular smooth muscle cells (VSMCs), macrophages, and cardiomyocytes in various CVDs, underscoring a critical role of TFEB in regulating cardiovascular homeostasis.
3.1. TFEB in endothelial cells (ECs)
ECs serve as a barrier between blood flow and the vascular wall. Endothelial dysfunction is an early event that initiates atherosclerotic development. TFEB is upregulated by atheroprotective laminar shear stress in vascular ECs both in vitro and in vivo [13,14]. In ECs, TFEB displays anti-inflammatory, anti-atherosclerotic, and pro-angiogenic effects. Using an EC-specific TFEB transgenic mouse model (mouse Tie2 promoter-driven TFEB transgene), we demonstrated that TFEB inhibits EC inflammation and attenuates atherosclerosis in apolipoprotein E (ApoE) knockout (KO) mice [13]. Similar results have shown that TFEB overexpression diminishes vascular inflammation in diabetic db/db mice [14]. Recently, we demonstrated that TFEB in ECs improves systemic glucose tolerance in vivo using EC-Tfeb transgenic and EC-Tfeb KO (VE-cadherin Cre X floxed Tfeb) mice. TFEB increases glucose uptake in ECs and promotes insulin transport across ECs. Mechanistically, TFEB upregulates insulin receptor substrate-1 (IRS1) and IRS2, and activates the Akt signaling pathway in ECs [39]. Angiogenesis plays a critical role in embryonic development, wound healing, and other physiological and pathological responses. Using EC-Tfeb transgenic and EC-Tfeb KO mouse models, we found that endothelial TFEB promotes postischemic angiogenesis via activating AMP-activated protein kinase α (AMPKα) signalling and enhancing autophagy [40]. EC-specific Tfeb KO mice also showed impaired endothelial proliferation and vasculature defects [41]. Therefore, TFEB is critical to regulating vascular EC biology and potentially reversing endothelial dysfunction-related vascular diseases.
3.2. TFEB in vascular smooth muscle cells (VSMCs)
VSMCs are the major cell type in the tunica media of vessels. During atherosclerotic development, VSMCs contribute to many atherosclerotic plaque cells, including foam cells, macrophage-like cells, and mesenchymal stem cell-like cells. Moreover, VSMC plasticity and apoptosis are involved in the pathogenesis of aortic aneurysms [42] and neointimal formation [43]. TFEB has been demonstrated to be essential to regulate VSMC function. Trehalose (α-D-glucopyranosyl α-D-glucopyranoside), a TFEB activator, enhances TFEB-mediated autophagy in cultured VSMCs and attenuates VSMC proliferation and migration. A particular mouse study suggests that trehalose inhibits VSMC migration and proliferation, and suppresses neointimal formation in partially ligated carotid arteries [44]. Another study found that TFEB upregulates cathepsin S and is required for nicotine-induced VSMC migration. The discrepancy of TFEB in VSMC migration might result from different stimuli and characteristics of VSMCs [45]. Recently, we found that TFEB is significantly reduced in both thoracic aortic aneurysm (TAA) and abdominal aortic aneurysm (AAA) lesions. TFEB potently inhibits VSMC apoptosis by upregulating B-cell lymphoma 2 (BCL2) directly. VSMC-selective Tfeb KO promotes AAA development in multiple mouse models, including the proprotein convertase subtilisin/kexin type 9 (PCSK9)/angiotensin II (AngII) model and β-aminopropionitrile (BAPN) model [15]. Although it has been found that TFEB regulates VSMC migration, proliferation and apoptosis, the roles of TFEB in VSMC biology and vascular diseases remain to be fully explored.
3.3. TFEB in macrophages
Macrophages have been well recognized as important immune effector cells in inflammation, injury, regeneration, and remodelling in the vasculature and heart under pathological conditions [46]. Macrophages link certain risk factors, including obesity, diabetes, and immune dysfunction, to CVDs by sensing microenvironmental changes and triggering intracellular signalling cascades. Similar to its roles in ECs, TFEB exhibits anti-inflammatory and anti-atherosclerotic effects in macrophages. In the ApoE KO mouse atherosclerosis model, the accumulation of excessive intracellular cholesterol impairs normal lysosomal function in macrophages. Macrophage-specific TFEB overexpression promotes autophagy and lysosomal biogenesis in atherosclerotic plaques and reduces lesion size in an autophagy-related 5 (Atg5) and ubiquitin-binding protein p62 (p62)-dependent manner in ApoE KO mice [47]. TFEB overexpression in macrophages restores normal lysosomal function and reduces inflammasome activation and interleukin-1β (IL1β) production [48]. Intriguingly, the inhibition of IL1β is autophagy-independent as Atg5 KO did not abolish this effect [48]. Macrophage TFEB also attenuates ventricular dysfunction after cardiac IR injury in a partially lysosomal acid lipase-dependent and Atg5-independent manner [49]. In the setting of atherosclerosis and myocardial infarction, TFEB has diverse effects on macrophages in vivo. However, the impact of TFEB is unlikely to be fully addressed by a single mechanism. Taken together, macrophage TFEB plays critical roles in the regulation of vascular and cardiac homeostasis. Further, it is noteworthy that macrophage TFEB is involved in not only CVDs but also in immune defence and cancer development. Macrophage-specific TFEB overexpression suppresses breast tumour growth in mice. TFEB knockdown facilitates macrophage M2 polarization through inhibition of suppressor of cytokine signalling 3 (SOCS3) and activation of signal transducer and activator of transcription 3 (STAT3) in the tumour microenvironment [12].
3.4. TFEB in cardiomyocytes
Cardiomyocytes are responsible for generating contractile force in the heart. Autophagy is critical to maintaining heart homeostasis and is involved in a broad spectrum of cardiac diseases. TFEB activity is dynamically altered in the heart in response to microenvironmental changes. Fasting induces TFEB nuclear translocation and refeeding leads to a rapid decline in nuclear TFEB content in the mouse myocardium in vivo. Next, TFEB is essential for repetitive starvation-dependent attenuation of cell death induced by hypoxia-reoxygenation in neonatal rat cardiomyocytes in vitro [50]. Lipopolysaccharides (LPS) induce autophagy and TFEB nuclear translocation in the heart of young mice but not aged mice, which may result in increased susceptibility to LPS-induced myocardial injury in aged mice [51].
In the heart, growing evidence shows that TFEB enhances the survival of cardiomyocytes under pathological conditions. TFEB restores mitochondrial biogenesis via induction of peroxisome proliferator-activated receptor-gamma, co-activator 1 alpha (PGC1α), and suppresses BCL-2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3)-induced cardiomyocyte death in vitro [50]. The R120G mutant of αB-crystallin (CryAB R120G) in cardiomyocytes impairs autophagic flux and causes desmin-related proteotoxicity and cardiomyopathy. Forced TFEB overexpression increases autophagic flux and remarkably attenuates CryAB R120G overexpression-induced accumulation of protein aggregates and cell death in neonatal rat ventricular cardiomyocytes [52]. A recent study found that adeno-associated virus (AAV)-mediated TFEB overexpression can also attenuate cardiac dysfunction and myocardial hypertrophy in those mice overexpressing CryAB R120G in cardiomyocytes [53]. Monoamine oxidase-A (MAO-A) activation promotes ROS production, inhibits TFEB nuclear translocation, and blocks autophagic flux in cardiomyocytes. AAV-mediated TFEB overexpression attenuates autophagic blockade, cardiomyocyte death, and heart failure in MAO-A transgenic mice [54]. Thus, TFEB protects cardiomyocytes against proteotoxicity and cell death through enhancing autophagy and lysosomal function.
Emerging evidence suggests that impaired TFEB activity contributes to cardiotoxicity and cardiomyopathy under pathological conditions. Insulin resistance-induced hyperglycemia and fatty acid overutilization lead to glucolipotoxicity in the heart. Glucolipotoxicity downregulates TFEB and leads to diminished lysosomal function in cardiomyocytes [55]. Reduced nuclear TFEB content and elevated lipid (diacylglycerol and triacylglycerol) accumulation concomitantly occurred in the heart from high-fat high-sucrose diet-fed mice. Cardiomyocyte-specific Tfeb KO disturbs not only autophagy and lysosomal function but also metabolic pathways, rendering the heart susceptible to nutrient overload-induced injury in mice [55].
Cancer chemotherapy drugs can also induce cardiotoxicity and further lead to heart failure. Doxorubicin (DOX) is a chemotherapy drug that blocks the enzyme, topoisomerase 2. DOX represses TFEB expression, and restoration of TFEB prevents DOX-induced ROS production, caspase activation, and cardiomyocyte death [56]. Patients treated with proteasome inhibition chemotherapies readily develop cardiotoxicity. The ubiquitin-proteasome system and autophagy-lysosomal pathway cooperatively regulate proteostasis. A recent study demonstrated that Mcoln1-calcineurin-TFEB-p62 mediates the activation of the autophagy-lysosomal pathway during proteasome malfunction, and p62 deficiency exacerbates proteasome inhibition-induced ventricle malfunction in mice [57]. Taken together, TFEB is critical in the pathogenesis of cardiomyopathy and is becoming a potential target for the treatment of heart diseases.
4. Pharmacological modulation of TFEB and clinical implications
Defective autophagy and lysosomal dysfunction have been implicated in many diseases, including CVDs (e.g., atherosclerosis and cardiomyopathy), metabolic disorders (e.g., hepatic steatosis, obesity, diabetes), and neurodegenerative diseases. The restoration of autophagy and lysosomal function becomes a promising strategy to treat these diseases. Herein, TFEB, as a master regulator of autophagy and lysosomal biogenesis, is a potential molecular target to enhance or restore the capacity of cellular clearance and to protect cells against pathological conditions. Either elevating the expression or inducing nuclear translocation is a strategy to increase TFEB activity. TFEB drugs/compounds have been identified in many cell types and tissues with distinct mechanisms. Based on the rapidly accumulated TFEB studies, we summarize the synthetic and natural TFEB activators that have effects both in vitro and in vivo (Table 1).
Table 1.
Molecules targeting TFEB and their respective mechanisms and function.
| Synthetic molecules targeting TFEB | ||||
|---|---|---|---|---|
| Name | Mechanism | In vitro study | In vivo study | Ref |
| 3,4-dimethoxychalcone (3,4-DC) | Inhibits mTOR Promotes TFEB nuclear translocation |
HepG2 and U2OS cells: Enhances autophagy |
Attenuates myocardial infarction and improves the efficacy of chemotherapy drugs | [59] |
| Alexidine, and ikarugamycin | Alexidine and Ikarugamycin: Ca2+-CaMKKβ/AMPK/mTORC1 Promote TFEB nuclear translocation |
HeLa cells: Enhance autophagy |
Attenuate metabolic disorders in HFD-fed mice and extend lifespan in C. elegans | [62] |
| Ezetimibe | Activates AMPK Promotes TFEB nuclear translocation |
Mouse hepatocytes and human hepatoma cells: Increases autophagy and ameliorates lipid accumulation and apoptosis Macrophages: Inhibits the NLRP3 inflammasome-IL1β pathway |
Attenuates lipid accumulation, inflammation, and fibrosis in liver-specific Atg7 wild-type and haploinsufficient mice | [61] |
| Formononetin | Activates AMPK Promotes TFEB nuclear translocation |
HepG2 and mouse hepatocytes: Facilitates lysosome biogenesis and lipophagy |
Inhibits HFD-induced hepatic steatosis and lipid disorders in mice | [65] |
| Gemfibrozil | Activates PPARα-RXRα-PGC1α Increases TFEB expression |
Astrocytes: Enhances lysosomal biogenesis |
Increases lysosomal biogenesis in the cortex in mice | [34] |
| MSL | Promotes TFEB nuclear translocation | HeLa cells: Enhances autophagy and decreases intracellular lipid accumulation |
Improves metabolic disorders in ob/ob mice | [63] |
| Tubastatin A | Increases TFEB acetylation Promotes TFEB nuclear translocation |
NRK-52E cells: Attenuates cell death | Protective in rat experimental kidney disease | [66] |
| Natural molecules targeting TFEB | ||||
| Aspirin | Activates PPARα Increases TFEB transcription |
Mouse primary astrocytes: Increases lysosomal biogenesis |
Reduces the amyloid burden in the hippocampus of 5XFAD mice | [67] |
| Carbon monoxide | Activates PERK-calcineurin Promotes TFEB nuclear translocation |
Hepatocytes: Increases mitophagy |
Attenuates inflammatory liver injury induced by LPS/D-GalN in mice | [64] |
| Curcumin analog-C1 | Binding to N-terminal of TFEB protein Promotes TFEB nuclear translocation |
N2a cells and HeLa cells: Promotes autophagy flux and lysosomal degradation/biogenesis |
Activates TFEB and enhances autophagy in rat brains | [68] |
| Digoxin | Digoxin: Ca2+-Calcineurin Promotes TFEB nuclear translocation |
HeLa cells: Enhances autophagy |
Attenuates metabolic disorders in HFD-fed mice and extend lifespan in C. elegans | [62] |
| Gypenoside XVII | Promotes TFEB nuclear translocation by releasing TFEB from TFEB/14-3-3 complexed | PC12: Increases autophagy and eliminates AβPP, Aβ40, and Aβ42 protein | Prevents the formation of Aβ plaques in the hippocampus and cortex of APP/PS1 mice | [69] |
| HEP14 and HEP15 | Activate PKC-GSK3β Promote TFEB nuclear translocation |
HeLa cells: Enhance lysosome biogenesis |
Attenuate the formation of amyloid β (Aβ) plaques in APP/PS1 mouse brains | [21] |
| HPβCD | Depletes intracellular cholesterol and inhibits mTORC1 Promotes TFEB nuclear translocation |
VSMCs: Enhances autophagy and inhibits apoptosis HeLa, LINCL fibroblasts, and macrophages: Enhances autophagy and suppresses macrophage M2 polarization. |
Inhibits abdominal aortic aneurysm formation and progression in mice Inhibits breast tumour growth in mice |
[12,15,60] |
| Procyanidin B2 | Possible direct binding Promotes TFEB nuclear translocation |
HepG2: Enhances lysosomal function |
Attenuates HFD-induced hepatic steatosis in mice | [70] |
| Trehalose | Activates calcium-dependent phosphatase PPP3/calcineurin or inhibits Akt Promotes TFEB nuclear translocation |
NSC34 cells, macrophages, and HeLa cells: Promotes clearance of neurotoxic misfolded proteins Enhances autophagic flux |
Prolongs the lifespan in the Batten disease mouse model. Attenuates atherosclerosis and cardiac remodelling in mice. Suppresses vascular neointimal formation |
[47,58,71,72] |
Abbreviation: HFD: high-fat diet; HPβCD: 2-Hydroxypropyl-β-cyclodextrin; HEP14: 5β-O-angelate-20-deoxyingenol and HEP15: 3β-O-angelate-20-deoxyingenol; Trehalose: α-D-glucopyranosyl α-D-glucopyranoside; MSL: 4-(4-fluorophenyl) sulfonyl-5-methylthio-2-phenyloxazole; PC12: rat pheochromocytoma cell line; PERK, protein kinase RNA-like endoplasmic reticulum kinase; CAMKKβ, calcium/calmodulin-dependent protein kinase kinase 2; PPARα, peroxisome proliferator-activated receptor-alpha; RXRα, retinoid X receptor-alpha; AMPK, 5′ AMP-activated protein kinase; GSK3β, glycogen synthase kinase 3 beta; PKC, protein kinase C.
TFEB activators have been demonstrated to benefit CVDs. Trehalose reduces atherosclerosis in ApoE KO mice [47] and improves cardiac remodelling after myocardial infarction [58]. 3,4-dimethoxychalcone (3,4-DC) induces autophagy in mouse hearts and ameliorates myocardial infarction after ischemic injury [59]. 2-Hydroxypropyl-β-cyclodextrin (HPβCD), an FDA-approved cyclodextrin derivative currently used to increase the solubility of lipophilic drugs, was revealed as a TFEB activator and enhanced the autophagic clearance of intracellular proteolipid aggregates.[60] Recently, using VSMC-Tfeb knockout mice, we demonstrated that HPβCD inhibits abdominal aortic aneurysm formation in a VSMC TFEB-dependent manner. HPβCD-dependent activation of TFEB inhibits VSMC apoptosis via upregulation of BCL2 [15].
TFEB activation also has shown its beneficial effects on metabolic diseases and neurodegenerative diseases. Ezetimibe, a prescribed cholesterol-lowering drug, activates TFEB and further attenuates hepatic lipid accumulation and inflammation in methionine- and choline-deficient (MCD) diet-fed mice [61]. A nanotechnology-enabled high-throughput screen identified three novel compounds (digoxin, alexidine, and ikarugamycin) as TFEB activators. An oral supplement of digoxin or intravenous injection of alexidine or ikarugamycin attenuates hepatic steatosis, obesity, and hyperglycemia in HFD-fed mice [62]. Another study emphasized the inhibitory effect of TFEB activation on metabolic disorders. [4-(4-fluorophenyl) sulfonyl-5-methylthio-2-phenyloxazole] (MSL) was found to activate TFEB and to improve obesity, hyperglycemia, hepatic steatosis, and adipose inflammation in ob/ob diabetic mice [63]. TFEB activation also improves neurodegenerative diseases in mice. Trehalose promotes the clearance of proteolipid aggregates and prolongs the lifespan in the mouse model of Batten disease [20]. 5β-O-angelate-20-deoxyingenol (HEP14) and 3β-O-angelate-20-deoxyingenol (HEP15) are natural products isolated and purified from the aerial parts of Euphorbia peplus L. Administration of HEP14 activates TFEB and ameliorates amyloid-beta (Aβ) plaque formation in APP/PS1 mice, an Alzheimer's disease model [21]. Not limited to the diseases mentioned above, emerging studies demonstrated that TFEB activation affects cancer growth and progression. Also, HPβCD suppresses macrophage M2 polarization and inhibits breast tumour growth in mice [12].
5. Outstanding questions
Genetic or pharmacological activation of TFEB inhibits CVDs (e.g., atherosclerosis, ischemic vascular injury, aortic aneurysm, and cardiotoxicity), metabolic disorders, and neurodegenerative diseases, indicating that TFEB is becoming a promising potential molecular target for the treatment of these diseases. TFEB has both autophagy-dependent and independent effects, shedding light on the comprehensive impact of TFEB in different cells and tissues under physiological and pathological conditions.
5.1. Tissue-specific effects of TFEB
The tissue distribution of TFEB can accordingly affect target gene expression. RNA-sequencing or ChIP-sequencing could be applied to explore the target genes of TFEB in different cell types. In mammalian cells, the initiation of transcription requires not only the recruitment of basic RNA polymerase II machinery but also transcription factors and co-activators. The cell-specific expression pattern of co-factors could also determine the accessibility and efficiency of TFEB in the regulation of target genes. The exploration of co-activators for TFEB may provide an approach to manipulate TFEB activity in a cell-specific manner.
5.2. Potential TFEB-independent effects of TFEB targeting drugs
Some drugs/compounds and multiple signalling pathways (mTOR, Akt, PKC, Ca2+) have been found to increase either TFEB expression or nuclear translocation. Of note, we must acknowledge that these drugs/chemical compounds may activate other specific molecular targets and pathways, not only TFEB. In-depth studies are required to avoid the unwanted TFEB-independent effects in the application of these TFEB activators.
5.3. Selectively boosting TFEB
TFEB exerts diverse effects via distinct mechanisms in different cells and tissues. TFEB has been implicated in the pathogenesis of a variety of tumours such as renal cell carcinoma [73], breast cancer [74] and Birt–Hogg–Dubé syndrome (a hereditary diseases characterized by increased risk of multiple tumours) [75]. The fusion of the TFEB gene and the anonymous non-protein-encoding Alpha gene by chromosome translocation t(6;11) (p21.1; q12) has been reported in the renal tumour [73]. The role and underlying mechanisms of TFEB in cancer biology remains to be fully understood. It is critical to take full advantage of the beneficial impacts of TFEB but avoid possible detrimental effects under pathological conditions. The strategy of boosting TFEB activity should be applied selectively in a cell/tissue-specific manner.
Conclusion
Emerging studies have expanded our knowledge of TFEB in cell biology and many diseases. TFEB plays critical roles in both the heart and vasculature in autophagy-dependent and independent manners. However, the function and underlying mechanisms of TFEB in cardiovascular biology remain to be fully understood. Further investigations need to be performed to establish TFEB as a potential therapeutic target for the prevention and treatment of CVDs. Mechanistic studies revealed that TFEB activity could be modulated by post-translational modifications and alterations in expression. TFEB drugs and chemical compounds are being pursued with translational feasibility for the treatment of many diseases, including CVDs.
Search strategy and selection criteria
Data for this review were collected through PubMed. The following search terms were used: TFEB, endothelial cells, macrophages, smooth muscle cells, heart, atherosclerosis, angiogenesis, neurodegenerative disease, adipose tissue, liver metabolism, diabetes, drug development. Only articles published in English were included.
Contributors
Y. Fan conceived the idea of the review article. H. Lu, Y. Fan, and J. Sun performed the literature search. H. Lu and Y. Fan wrote the manuscript. Y. Fan and H. Lu drew the figures. M. Hamblin, Y.E. Chen, and Y. Fan revised the manuscript. All authors approved the final version of the manuscript.
Declaration of Competing Interests
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by the National Institutes of Health R01HL138094 and R01HL145176 (to Y. Fan), R01HL068878, R01HL137214, and R01HL134569 (to Y. E. Chen), and American Heart Association 17PRE33400179 (to H. Lu). The funders had no roles in paper design, data collection, data analysis, interpretation, or paper writing. We also would like to acknowledge all the other studies in this field that were not cited or discussed solely due to space limitations.
References
- 1.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 2.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slade L, Pulinilkunnil T. The MiTF/TFE family of transcription factors: master regulators of organelle signaling, metabolism, and stress adaptation. Mol Cancer Res. 2017;15(12):1637–1643. doi: 10.1158/1541-7786.MCR-17-0320. [DOI] [PubMed] [Google Scholar]
- 4.Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74(2):395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 5.Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci USA. 2002;99(7):4477–4482. doi: 10.1073/pnas.072071099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr CS, Sharp PA. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol Cell Biol. 1990;10(8):4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development. 1998;125(23):4607–4616. doi: 10.1242/dev.125.23.4607. [DOI] [PubMed] [Google Scholar]
- 8.Pastore N, Huynh T, Herz NJ, Calcagni A, Klisch TJ, Brunetti L. TFEB regulates murine liver cell fate during development and regeneration. Nat Commun. 2020;11(1):2461. doi: 10.1038/s41467-020-16300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orfali N, O'Donovan TR, Cahill MR, Benjamin D, Nanus DM, McKenna SL. All-trans retinoic acid (ATRA)-induced TFEB expression is required for myeloid differentiation in acute promyelocytic leukemia (APL) Eur J Haematol. 2020;104(3):236–250. doi: 10.1111/ejh.13367. [DOI] [PubMed] [Google Scholar]
- 10.Yoneshima E, Okamoto K, Sakai E, Nishishita K, Yoshida N, Tsukuba T. The transcription factor EB (TFEB) regulates osteoblast differentiation through ATF4/CHOP-dependent pathway. J Cell Physiol. 2016;231(6):1321–1333. doi: 10.1002/jcp.25235. [DOI] [PubMed] [Google Scholar]
- 11.Du Bois P, Pablo Tortola C, Lodka D, Kny M, Schmidt F, Song K. Angiotensin II induces skeletal muscle atrophy by activating TFEB-mediated MuRF1 expression. Circ Res. 2015;117(5):424–436. doi: 10.1161/CIRCRESAHA.114.305393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang L, Hodge J, Saaoud F, Wang J, Iwanowycz S, Wang Y. Transcriptional factor EB regulates macrophage polarization in the tumor microenvironment. Oncoimmunology. 2017;6(5) doi: 10.1080/2162402X.2017.1312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu H, Fan Y, Qiao C, Liang W, Hu W, Zhu T. TFEB inhibits endothelial cell inflammation and reduces atherosclerosis. Sci Signal. 2017;10(464):eaah4214. doi: 10.1126/scisignal.aah4214. [DOI] [PubMed] [Google Scholar]
- 14.Song W, Zhang CL, Gou L, He L, Gong YY, Qu D. Endothelial TFEB (Transcription Factor EB) restrains IKK (IkappaB Kinase)-p65 pathway to attenuate vascular inflammation in diabetic db/db mice. Arterioscler Thromb Vasc Biol. 2019;39(4):719–730. doi: 10.1161/ATVBAHA.119.312316. [DOI] [PubMed] [Google Scholar]
- 15.Lu H, Sun J, Liang W, Chang Z, Rom O, Zhao Y. Cyclodextrin prevents abdominal aortic aneurysm via activation of vascular smooth muscle cell transcription factor EB. Circulation. 2020;142(5):483–498. doi: 10.1161/CIRCULATIONAHA.119.044803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5(228):ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8(6):903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferron M, Settembre C, Shimazu J, Lacombe J, Kato S, Rawlings DJ. A RANKL-PKCbeta-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27(8):955–969. doi: 10.1101/gad.213827.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmieri M, Pal R, Nelvagal HR, Lotfi P, Stinnett GR, Seymour ML. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017;8(1):14338. doi: 10.1038/ncomms14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Xu M, Ding X, Yan C, Song Z, Chen L. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nature cell biology. 2016;18(10):1065–1077. doi: 10.1038/ncb3407. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Friedrichsen HJ, Andrews S, Picaud S, Volpon L, Ngeow K. A TFEB nuclear export signal integrates amino acid supply and glucose availability. Nat Commun. 2018;9(1):2685. doi: 10.1038/s41467-018-04849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu CL, Lee EX, Gordon KL, Paz EA, Shen WC, Ohnishi K. MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB. Nat Commun. 2018;9(1):942. doi: 10.1038/s41467-018-03340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nature Cell Biol. 2015;17(3):288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martina JA, Puertollano R. Protein phosphatase 2A stimulates activation of TFEB and TFE3 transcription factors in response to oxidative stress. J Biol Chem. 2018;293(32):12525–12534. doi: 10.1074/jbc.RA118.003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silvestrini MJ, Johnson JR, Kumar AV, Thakurta TG, Blais K, Neill ZA. Nuclear export inhibition enhances HLH-30/TFEB activity, autophagy, and lifespan. Cell Rep. 2018;23(7):1915–1921. doi: 10.1016/j.celrep.2018.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napolitano G, Esposito A, Choi H, Matarese M, Benedetti V, Di Malta C. mTOR-dependent phosphorylation controls TFEB nuclear export. Nat Commun. 2018;9(1):3312. doi: 10.1038/s41467-018-05862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Wang J, Zhou Z, Park JE, Wang L, Wu S. Importance of TFEB acetylation in control of its transcriptional activity and lysosomal function in response to histone deacetylase inhibitors. Autophagy. 2018;14(6):1043–1059. doi: 10.1080/15548627.2018.1447290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao J, Zheng L, Zhang Q, Li X, Zhang X, Li Z. Deacetylation of TFEB promotes fibrillar Abeta degradation by upregulating lysosomal biogenesis in microglia. Protein Cell. 2016;7(6):417–433. doi: 10.1007/s13238-016-0269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE. Sumoylation of MITF and its related family members TFE3 and TFEB. J Biol Chem. 2005;280(1):146–155. doi: 10.1074/jbc.M411757200. [DOI] [PubMed] [Google Scholar]
- 31.Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nature cell biology. 2013;15(6):647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516(7529):108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA. PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4(142) doi: 10.1126/scitranslmed.3003799. 142ra97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh A, Jana M, Modi K, Gonzalez FJ, Sims KB, Berry-Kravis E. Activation of peroxisome proliferator-activated receptor alpha induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders. J Biol Chem. 2015;290(16):10309–10324. doi: 10.1074/jbc.M114.610659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Tao Z, Zheng LD, Brooke JP, Smith CM, Liu D. FoxO1 interacts with transcription factor EB and differentially regulates mitochondrial uncoupling proteins via autophagy in adipocytes. Cell Death Discov. 2016;2:16066. doi: 10.1038/cddiscovery.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15(8):1419–1437. doi: 10.1080/15548627.2019.1586246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orekhov AN, Ivanova EA, Markin AM, Nikiforov NG, Sobenin IA. Genetics of Arterial-Wall-Specific Mechanisms in Atherosclerosis: Focus on Mitochondrial Mutations. Curr Atheroscler Rep. 2020;22(10):54. doi: 10.1007/s11883-020-00873-5. [DOI] [PubMed] [Google Scholar]
- 38.Markin AM, Sobenin IA, Grechko AV, Zhang D, Orekhov AN. Cellular mechanisms of human atherogenesis: focus on chronification of inflammation and mitochondrial mutations. Front Pharmacol. 2020;11:642. doi: 10.3389/fphar.2020.00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun J, Lu H, Liang W, Zhao G, Ren L, Hu D, et al. Endothelial TFEB (transcription factor EB) improves glucose tolerance via upregulation of IRS (insulin receptor substrate) 1 and IRS2. Arteriosclerosis, Thrombosis,Vasc Biol. 0(0):ATVBAHA.120.315310. [DOI] [PMC free article] [PubMed]
- 40.Fan Y, Lu H, Liang W, Garcia-Barrio MT, Guo Y, Zhang J. Endothelial TFEB (Transcription Factor EB) positively regulates postischemic angiogenesis. Circ Res. 2018;122(7):945–957. doi: 10.1161/CIRCRESAHA.118.312672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doronzo G, Astanina E, Cora D, Chiabotto G, Comunanza V, Noghero A. TFEB controls vascular development by regulating the proliferation of endothelial cells. EMBO J. 2019;38(3):e98250. doi: 10.15252/embj.201798250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clement M, Chappell J, Raffort J, Lareyre F, Vandestienne M, Taylor AL. Vascular smooth muscle cell plasticity and autophagy in dissecting aortic aneurysms. Arterioscler Thromb Vasc Biol. 2019;39(6):1149–1159. doi: 10.1161/ATVBAHA.118.311727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lacolley P, Regnault V, Nicoletti A, Li Z, Michel JB. The vascular smooth muscle cell in arterial pathology: a cell that can take on multiple roles. Cardiovasc Res. 2012;95(2):194–204. doi: 10.1093/cvr/cvs135. [DOI] [PubMed] [Google Scholar]
- 44.Wang YT, Li X, Chen J, McConnell BK, Chen L, Li PL. Activation of TFEB ameliorates dedifferentiation of arterial smooth muscle cells and neointima formation in mice with high-fat diet. Cell Death Dis. 2019;10(9):676. doi: 10.1038/s41419-019-1931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ni H, Xu S, Chen H, Dai Q. Nicotine modulates CTSS (Cathepsin S) synthesis and secretion through regulating the autophagy-lysosomal machinery in atherosclerosis. Arterioscler Thromb Vasc Biol. 2020;40(9):2054–2069. doi: 10.1161/ATVBAHA.120.314053. [DOI] [PubMed] [Google Scholar]
- 46.Poznyak AV, Wu WK, Melnichenko AA, Wetzker R, Sukhorukov V, Markin AM. Signaling pathways and key genes involved in regulation of foam cell formation in atherosclerosis. Cells. 2020;9(3):584. doi: 10.3390/cells9030584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sergin I, Evans TD, Zhang X, Bhattacharya S, Stokes CJ, Song E. Exploiting macrophage autophagy-lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun. 2017;8:15750. doi: 10.1038/ncomms15750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emanuel R, Sergin I, Bhattacharya S, Turner J, Epelman S, Settembre C. Induction of lysosomal biogenesis in atherosclerotic macrophages can rescue lipid-induced lysosomal dysfunction and downstream sequelae. Arterioscler Thromb Vasc Biol. 2014;34(9):1942–1952. doi: 10.1161/ATVBAHA.114.303342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Javaheri A, Bajpai G, Picataggi A, Mani S, Foroughi L, Evie H. TFEB activation in macrophages attenuates postmyocardial infarction ventricular dysfunction independently of ATG5-mediated autophagy. JCI Insight. 2019;4(21) doi: 10.1172/jci.insight.127312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma X, Liu H, Murphy JT, Foyil SR, Godar RJ, Abuirqeba H. Regulation of the transcription factor EB-PGC1alpha axis by beclin-1 controls mitochondrial quality and cardiomyocyte death under stress. Mol Cell Biol. 2015;35(6):956–976. doi: 10.1128/MCB.01091-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li F, Lang F, Zhang H, Xu L, Wang Y, Hao E. Role of TFEB mediated autophagy, oxidative stress, inflammation, and cell death in endotoxin induced myocardial toxicity of young and aged mice. Oxid Med Cell Longev. 2016;2016 doi: 10.1155/2016/5380319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan B, Zhang H, Cui T, Wang X. TFEB activation protects against cardiac proteotoxicity via increasing autophagic flux. J Mole Cell Cardiol. 2017;113:51–62. doi: 10.1016/j.yjmcc.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma X, Mani K, Liu H, Kovacs A, Murphy JT, Foroughi L. Transcription factor EB activation rescues advanced alphaB-crystallin mutation-induced cardiomyopathy by normalizing desmin localization. J Am Heart Assoc. 2019;8(4) doi: 10.1161/JAHA.118.010866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santin Y, Sicard P, Vigneron F, Guilbeau-Frugier C, Dutaur M, Lairez O. Oxidative stress by monoamine oxidase-a impairs transcription factor EB activation and autophagosome clearance, leading to cardiomyocyte necrosis and heart failure. Antioxidants Redox Signal. 2016;25(1):10–27. doi: 10.1089/ars.2015.6522. [DOI] [PubMed] [Google Scholar]
- 55.Trivedi PC, Bartlett JJ, Mercer A, Slade L, Surette M, Ballabio A. Loss of function of transcription factor EB remodels lipid metabolism and cell death pathways in the cardiomyocyte. Biochim Biophys Acta Mol Basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165832. [DOI] [PubMed] [Google Scholar]
- 56.Bartlett JJ, Trivedi PC, Yeung P, Kienesberger PC, Pulinilkunnil T. Doxorubicin impairs cardiomyocyte viability by suppressing transcription factor EB expression and disrupting autophagy. The Biochem J. 2016;473(21):3769–3789. doi: 10.1042/BCJ20160385. [DOI] [PubMed] [Google Scholar]
- 57.Pan B, Li J, Parajuli N, Tian Z, Wu P, Lewno MT. The Calcineurin-TFEB-p62 Pathway Mediates the Activation of Cardiac Macroautophagy by Proteasomal Malfunction. Circ Res. 2020;127(4):502–518. doi: 10.1161/CIRCRESAHA.119.316007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sciarretta S, Yee D, Nagarajan N, Bianchi F, Saito T, Valenti V. Trehalose-induced activation of autophagy improves cardiac remodeling after myocardial infarction. J Am Coll Cardiol. 2018;71(18):1999–2010. doi: 10.1016/j.jacc.2018.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen G, Xie W, Nah J, Sauvat A, Liu P, Pietrocola F. 3,4-Dimethoxychalcone induces autophagy through activation of the transcription factors TFE3 and TFEB. EMBO Mol Med. 2019;11(11):e10469. doi: 10.15252/emmm.201910469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song W, Wang F, Lotfi P, Sardiello M, Segatori L. 2-Hydroxypropyl-beta-cyclodextrin promotes transcription factor EB-mediated activation of autophagy: implications for therapy. J Biol Chem. 2014;289(14):10211–10222. doi: 10.1074/jbc.M113.506246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim SH, Kim G, Han DH, Lee M, Kim I, Kim B. Ezetimibe ameliorates steatohepatitis via AMP activated protein kinase-TFEB-mediated activation of autophagy and NLRP3 inflammasome inhibition. Autophagy. 2017;13(10):1767–1781. doi: 10.1080/15548627.2017.1356977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang C, Niederstrasser H, Douglas PM, Lin R, Jaramillo J, Li Y. Small-molecule TFEB pathway agonists that ameliorate metabolic syndrome in mice and extend C. elegans lifespan. Nat Commun. 2017;8(1):2270. doi: 10.1038/s41467-017-02332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lim H, Lim YM, Kim KH, Jeon YE, Park K, Kim J. A novel autophagy enhancer as a therapeutic agent against metabolic syndrome and diabetes. Nat Commun. 2018;9(1):1438. doi: 10.1038/s41467-018-03939-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim HJ, Joe Y, Rah SY, Kim SK, Park SU, Park J. Carbon monoxide-induced TFEB nuclear translocation enhances mitophagy/mitochondrial biogenesis in hepatocytes and ameliorates inflammatory liver injury. Cell Death Dis. 2018;9(11):1060. doi: 10.1038/s41419-018-1112-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Zhao H, Li X, Wang Q, Yan M, Zhang H. Formononetin alleviates hepatic steatosis by facilitating TFEB-mediated lysosome biogenesis and lipophagy. J Nutr Biochem. 2019;73 doi: 10.1016/j.jnutbio.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 66.Brijmohan AS, Batchu SN, Majumder S, Alghamdi TA, Thieme K, McGaugh S. HDAC6 inhibition promotes transcription factor EB activation and is protective in experimental kidney disease. Front Pharmacol. 2018;9:34. doi: 10.3389/fphar.2018.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chandra S, Jana M, Pahan K. Aspirin induces lysosomal biogenesis and attenuates amyloid plaque pathology in a mouse model of alzheimer's disease via PPARalpha. J Neurosci. 2018;38(30):6682–6699. doi: 10.1523/JNEUROSCI.0054-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song JX, Sun YR, Peluso I, Zeng Y, Yu X, Lu JH. A novel curcumin analog binds to and activates TFEB in vitro and in vivo independent of MTOR inhibition. Autophagy. 2016;12(8):1372–1389. doi: 10.1080/15548627.2016.1179404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meng X, Luo Y, Liang T, Wang M, Zhao J, Sun G. Gypenoside XVII enhances lysosome biogenesis and autophagy flux and accelerates autophagic clearance of amyloid-beta through TFEB activation. J Alzheimer's Dis. 2016;52(3):1135–1150. doi: 10.3233/JAD-160096. [DOI] [PubMed] [Google Scholar]
- 70.Su H, Li Y, Hu D, Xie L, Ke H, Zheng X. Procyanidin B2 ameliorates free fatty acids-induced hepatic steatosis through regulating TFEB-mediated lysosomal pathway and redox state. Free Radic Biol Med. 2018;126:269–286. doi: 10.1016/j.freeradbiomed.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 71.Rusmini P, Cortese K, Crippa V, Cristofani R, Cicardi ME, Ferrari V. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy. 2019;15(4):631–651. doi: 10.1080/15548627.2018.1535292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y-T, Li X, Chen J, McConnell BK, Chen L, Li P-L. Activation of TFEB ameliorates dedifferentiation of arterial smooth muscle cells and neointima formation in mice with high-fat diet. Cell Death & Disease. 2019;10(9):676. doi: 10.1038/s41419-019-1931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci USA. 2003;100(10):6051–6056. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei H, Wang C, Croce CM, Guan JL. p62/SQSTM1 synergizes with autophagy for tumor growth in vivo. Genes Dev. 2014;28(11):1204–1216. doi: 10.1101/gad.237354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Napolitano G, Di Malta C, Esposito A, de Araujo MEG, Pece S, Bertalot G. A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dube syndrome. Nature. 2020;585(7826):597–602. doi: 10.1038/s41586-020-2444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]