Abstract
One-carbon metabolism is a central metabolic hub that provides one-carbon units for essential biosynthetic reactions and for writing epigenetics marks. The leading role in this hub is performed by the one-carbon carrier tetrahydrofolate (THF), which accepts formaldehyde usually from serine generating one-carbon THF intermediates in a set of reactions known as the folate or one-carbon cycle. THF derivatives can feed one-carbon units into purine and thymidine synthesis, and into the methionine cycle that produces the universal methyl-donor S-adenosylmethionine (AdoMet). AdoMet delivers methyl groups for epigenetic methylations and it is metabolized to homocysteine (Hcy), which can enter the transsulfuration pathway for the production of cysteine and lastly glutathione (GSH), the main cellular antioxidant. This vital role of THF comes to an expense. THF and other folate derivatives are susceptible to oxidative breakdown releasing formaldehyde, which can damage DNA -a consequence prevented by the Fanconi Anaemia DNA repair pathway. Epigenetic demethylations catalysed by lysine-specific demethylases (LSD) and Jumonji histone demethylases can also release formaldehyde, constituting a potential threat for genome integrity. In mammals, the toxicity of formaldehyde is limited by a metabolic route centred on the enzyme alcohol dehydrogenase 5 (ADH5/GSNOR), which oxidizes formaldehyde conjugated to GSH, lastly generating formate. Remarkably, this formate can be a significant source of one-carbon units, thus defining a formaldehyde cycle that likely restricts the toxicity of one-carbon metabolism and epigenetic demethylations. This work describes recent advances in one-carbon metabolism and epigenetics, focusing on the steps that involve formaldehyde flux and that might lead to cytotoxicity affecting human health.
Keywords: Epigenetics, Formaldehyde, Fanconi anemia, One-carbon metabolism, ADH5, Glutathione
1. Introduction
Folates (Vitamin B9) are commonly found in foods in different forms, mostly conjugated to a polyglutamate chain that affects their bio-availability [1]. Diets deficient in folates underly some cases of megaloblastic anaemia and increase the risk of neural tube defects (NTDs) in newborns [2]. To prevent folate deficiency, in some countries, foods are usually supplemented with synthetic folic acid. This oxidized folate form is inactive and more stable than natural folates. In cells, the dihydrofolate reductase (DHFR) reduces folic acid to dihydrofolate (DHF) lastly generating tetrahydrofolate (THF), which is polyglutamated by the enzyme folyl-polyglutamate synthetase (FPGS) [3]. This step is essential to retain intracellular THF and to increase the activity of THF, entering the one-carbon cycle. Some enzymes such as serine hydroxymethyltransferases and the glycine cleavage system (GCS) transfer formaldehyde from serine to THF generating the key intermediate 5,10-methylene-tetrahydrofolate (5,10-CH2-THF) [4]. This intermediate, as well as DHF and THF, are intrinsically unstable and can undergo oxidative breakdown between the C-9 and N-10 bond producing three different products: a pteridine, p-aminobenzoylglutamate (pABG) and formaldehyde [[5], [6], [7]]. Therefore, the flux of one-carbon units throughout the cell also implies the movement of reactive formaldehyde posing a significant threat to cell functioning (Fig. 1). The release of reactive formaldehyde can cause genotoxicity, reactive oxygen species (ROS) production and proteotoxic stress, among others [[8], [9], [10]]. Moreover, the breakdown of THF derivatives into formaldehyde has been reported to alter the availability of one-carbon units for essential biosynthetic reactions, causing for example nucleotide imbalance that leads to replication stress and DNA damage [11,12].
Fig. 1.
Chemical formaldehyde flux and one-carbon metabolism.
Formaldehyde (red) flux throughout one-carbon metabolism is depicted indicating the metabolic pathways involved in delivering formaldehyde units throughout cellular metabolism. For simplification only the structure of tetrahydrofolate (THF) and the derivative 5,10-methylene-THF are shown (see the text and Fig. 2 for the complete set of THF derivatives). In THF, the red bond represents the carbon released as formaldehyde upon oxidative degradation. The one-carbon unit is depicted as a dotted red line in the structure of 5,10-methylene-THF. S-adenosylmethionine (AdoMet) is also shown with the reactive methyl group in red. The cartoon is a representation of histones and DNA with the methyl groups indicated in red. DMG: Dimethylglycine.
2. One-carbon cycle
One-carbon metabolism refers not only to the one-carbon cycle but also to the methionine cycle, the transsulfuration pathway, and the recently described formaldehyde cycle (Fig. 1). In the one-carbon cycle, THF is like the minute hand of a clock, delivering methyl groups to different cellular compartments and biosynthetic reactions. This cycle is compartmentalized between mitochondria, cytosol and, during S-phase, the nucleus [11,13]. The mitochondrial one-carbon cycle branch initiates with the hydroxymethylation of THF from serine catalysed by the enzyme serine-hydroxymethyltransferase 2 (SHMT2) (Fig. 2). In this organelle, glycine -through the GCS system - can also donate one carbon units to THF. Also, the metabolism of choline generates dimethylglycine (DMG) and sarcosine that can transfer formaldehyde units to THF yielding 5,10-CH2-THF, in reactions catalysed by the DMG and sarcosine dehydrogenases, respectively [14]. 5,10-CH2-THF can be used for the methylation of tRNA, which is required for mRNA translation in the mitochondria [15]. This THF derivative might also feed mitochondrial thymidylate synthase (TYMS) for de novo thymidylate synthesis [16]. Alternative, mitochondrial 5,10-CH2-THF is oxidized by the bifunctional NAD(P)-dependent enzyme methylene-THF dehydrogenase 2 (MTHFD2) or 2 like (MTHFD2L) [17], generating 10-formyl-THF (10-CHO-THF). This compound is the one-carbon unit donor in the synthesis of N-formylmethionine (fMet) for mitochondrial protein synthesis initiation [18], and it is also the substrate of methylene-THF dehydrogenase 1 like (MTHFD1L), yielding formate and THF (Fig. 2). Mitochondrial formate can translocate to the cytosol, though the identity of the transporter involved in this process remains far from clear.
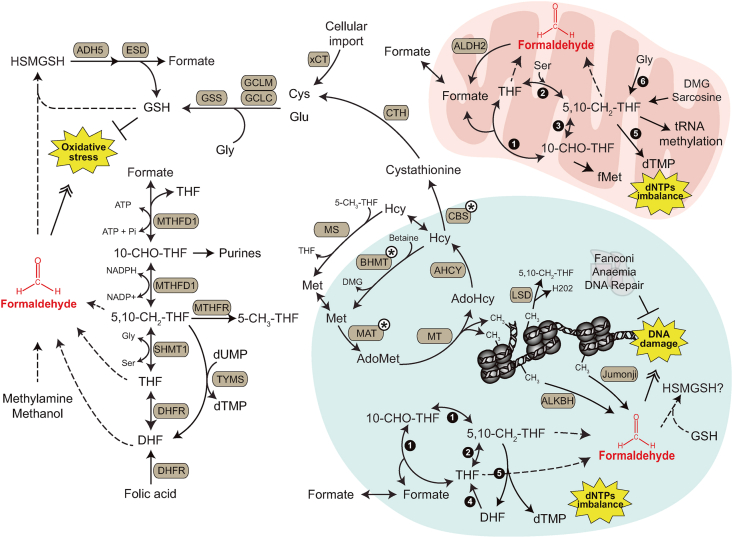
Fig. 2.
Toxic consequences of one-carbon metabolism and epigenetics.
The interconnected set of reactions that conform one-carbon metabolism is shown highlighting in yellow stars the toxic consequences that originate from one-carbon dysregulation in cytosol, mitochondria and nucleus. Dotted lines refer to spontaneous reactions. Brown-filled round boxes represent enzymes: ADH5: Alcohol dehydrogenase 5; AHCY: S-adenosylhomocysteine hydrolase; ALDH2: Aldehyde dehydrogenase 2; ALKBH: represents multiple α-ketoglutarate-dependent dioxygenases (2-OGDD) belonging to the AlkB family; BHMT: Betaine-homocysteine S-methyltransferase; CBS: Cystathionine beta-synthase; CTH: Cystathionine gamma-lyase; DHFR: Dihydrofolate reductase; ESD: Esterase D (formyl-GSH hydrolase); GCLC: Glutamate-cysteine ligase catalytic subunit; GCLM: Glutamate-cysteine ligase regulatory subunit; GCS: Glycine cleavage system; GSS: Glutathione synthase; Jumonji: JmjC-containing demethylases; LSD: Lysine-specific histone demethylase 1 or 2; MAT: Methionine adenosyltransferases I, II and III; MS: Methionine synthase; MT: DNMTs, PRMT and DOT-1L methyltransferases; MTHFD1: Methylene-THF dehydrogenase 1; MTHFR: Methyl-THF reductase; TYMS: Thymidylate synthase; SHMT1: Serine-hydroxymethyltransferase 1; xCT: Solute carrier family 7A11. Black-circles with white numbers represent nuclear (Nuc) and mitochondrial (Mit) one-carbon cycle enzymes: 1: MTHFD1 (Nuc) and MTHFD1L (Mit); 2: SHMT1 or SHMT2A (Nuc) and SHMT2 (Mit); 3: MTHFD2 or MTHFD2L (Mit); 4: DHFR (Nuc); 5: TYMS (Nuc and Mit); 6: GCS (Mit). White circles with black asterisks indicate enzymes found in both the nucleus and the cytosol. Metabolites: 10-CHO-THF: 10′-formyl-THF; 5,10-CH2-THF: 5′,10′-methylene-THF; 5-CH3-THF: 5′-methyl-THF; AdoHcy: S-adenosylhomocysteine; AdoMet: S-adenosylmethionine; DHF: Dihydrofolate; DMG: Dimethylglycine; fMet: N-formylmethionine; GSH: Glutathione; Hcy: Homocysteine; HSMGSH: S-hydroxymethyl-GSH; THF: Tetrahydrofolate.
In the cytosol, formate can re-enter the one-carbon cycle through the reversible trifunctional NADP-dependent methylene-tetrahydrofolate dehydrogenase 1 (MTHFD1), yielding 10-CHO-THF (Fig. 2) [19]. The high ratio NAPDH/NADP might favour this reductive direction from formate and THF giving first 10-CHO-THF and then 5,10-CH2-THF [20]. In cancer cells, the overflow of mitochondrial formate also favours the reductive direction of the cytosolic one-carbon cycle branch [21]. The reason for this compartmentalization has been proposed to be for uncoupling one-carbon NAD-dependent oxidations from glycolysis, which otherwise would consume cytosolic NAD, slowing glycolytic flux and affecting cellular redox homeostasis [11]. In the cytosol, 10-CHO-THF can be used for building the purine ring [22] or further oxidized to 5,10-CH2-THF. This multifunctional intermediate can reversible transfer a formaldehyde unit to glycine yielding serine and THF, a reaction catalysed by the enzyme serine-hydroxymethyltransferase 1 (SHMT1) [23]. 5,10-CH2-THF can also be directed to the enzyme thymidylate synthase (TYMS) [5,24], which catalyses the conversion of deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP), or to the methyl-THF reductase (MTHFR) yielding 5-methyl-THF (5-CH3-THF) (Fig. 2) [11]. This intermediate is the one-carbon donor for the generation of the essential amino acid residue methionine from homocysteine (Hcy), the initial step of the methionine cycle that is catalysed by the enzyme methionine synthase (MS) [25]. During S-phase, the nucleus is also a known site where one-carbon cycle takes place (Fig. 2). There, 5,10-CH2-THF plays a central role driving the synthesis of dTMP and generating DHF through the enzyme TYMS. DHF can be converted into THF by nuclear DHFR, using NADPH as a cofactor. Nuclear 5,10-CH2-THF is generated by the enzyme MTHFD1, which catalyses first the synthesis of 10-CHO-THF from THF and formate, and then the reduction of this intermediate to 5,10-CH2-THF (Fig. 2). The histone demethylase LSD1 can also produce 5,10-CH2-THF directly conjugating THF and formaldehyde (see section ‘Epigenetic demethylations and formaldehyde flux’). Moreover, THF can also generate 5,10-CH2-THF by the reversible action of nuclear enzymes SHMT1 and SHMT2A, which use serine as a formaldehyde donor [13,26]. Thus, nuclear one-carbon cycle also implies the flux of nuclear formaldehyde, a known carcinogen and a DNA-damaging molecule.
3. Methionine cycle, transsulfuration and epigenetic methylations
Epigenetics refers to all chromatin, DNA and RNA changes that affect gene expression without altering their primary sequence. The methionine cycle is an utmost important part of epigenetic methylations generating the universal cellular methyl donor adenosylmethionine (AdoMet), which is used in both the nucleus and the cytosol for methylation reactions. In the DNA, the most prevalent epigenetic change is the methylation of the C5-position of cytosine (5-methylcytosine; 5 mC), predominantly at CpG dinucleotides, which is normally associated with transcriptional repression [27]. The DNA methyltransferases that catalyse the transference of a methyl group from AdoMet to a cytosine residue are known as DNMTs [28]. RNA methylations, particularly N6-methyladenosine (m6A) but also N7-methylguanosine (m7G), have emerged in the last years as another tier to control gene expression at the RNA level. The methyltransferase enzymes that participate in the methylation of RNA bases from AdoMet are known as RNA writers and have been revised elsewhere [29]. In the nucleus, histones are also altered by post-translational methylations affecting gene expression [30,31].(Fig. 2). There are at least three families of methyltransferases able to catalyse the transference of a methyl group from AdoMet to histones. These enzymes include the PRMT proteins, the proteins containing a SET domain, and those known as DOT1-like proteins (revised elsewhere [32]).
Mechanistically, after donating methyl groups in reactions catalysed by methyltransferases, AdoMet is converted to S-adenosylhomocysteine (AdoHcy), a potent inhibitor of most AdoMet -dependent methyltransferases [33]. AdoHcy is further converted to Hcy by the enzyme AdoHcy hydrolase (AHCY). Hcy can be exported out of the nucleus and recycled into methionine using 5-CH3-THF through the action of the enzyme methionine synthase (MS). Parallel to this pathway, the cytosolic and nuclear enzyme betaine-homocysteine S-methyltransferase (BHMT) can also remethylate homocysteine into methionine in either compartment, using betaine (a product of choline metabolism) as methyl donor [34,35]. Subsequently, the ATP-dependent methionine adenosyltransferase enzymes MATI, MATII or MATIII (referred as MAT in Fig. 2) will convert methionine into AdoMet. MATI and MATIII are tetramers and dimers of the isoform MATα1, respectively. All the MAT enzymes can be found in the cytosol and nucleus. Indeed, high levels of nuclear MATα1 are associated with histone 3 lysine 27 (H3K27) methylation, which in turn causes DNA methylation and gene repression in hepatocytes. MATα2 and MATβ are the catalytic and regulatory isoforms of MATII, whose expression is mostly in extrahepatic tissues [[36], [37], [38]].
In addition to being converted into methionine, Hcy can be diverted to the transsulfuration pathway for the synthesis of cysteine, a precursor of the main cellular antioxidant glutathione (GSH). Initially, cystathionine beta-synthase (CBS), which is cytosolic but can localize to the nucleus upon post-translational sumoylation, catalyses the transformation of Hcy into cystathionine (Fig. 2) [39]. Then, this metabolite is converted into cysteine through the action of cystathionine gamma-lyase (CTH) (Fig. 2) [37]. The condensation of cysteine and glutamate is the initial and rate-limiting step in the synthesis of GSH. This reaction is catalysed by the enzyme glutamate-cysteine ligase (GCL), which is composed of a catalytic (GCLC) and a regulatory unit (GCLM) (Fig. 2) [38]. Then, glycine is added by the GSH synthase (GSS) forming the reduced tripeptide GSH.
Cellular GSH synthesis controls redox homeostasis, formaldehyde toxicity and also affects epigenetics. It was reported that the upregulation of GSH synthesis can drainage cysteine from the transsulfuration pathway thus reducing the availability of AdoMet and increasing one-carbon and, consequently, formaldehyde flux. However, this effect is cell-type dependent. For example, inhibition of CBS with proparglyglycine in both rat hepatocytes and HepG2 cells lead to around a 60% decrease in intracellular levels of both free cysteine and GSH [40,41], implying that the transsulfuration pathway is a significant source of cysteine for GSH synthesis in these experimental conditions. In contrast, several cancer cells, especially those with higher expression of the cysteine glutamate transporter (xCT, also known as solute carrier family 7A11, SLC7A11), hardly use the transsulfuration pathway to produce cysteine, which instead is imported from the extracellular space as cystine and then converted into cysteine in the cytosol [43]. A recent report indicated that blocking GSH synthesis increases global 5 mC methylation in mouse hepatocytes, therefore supporting that GSH synthesis and epigenetics are connected [42]. Indeed, decades ago it was observed that folate deficiency affects the methionine cycle, reducing the formation of AdoMet with a concomitant accumulation of Hcy and AdoHcy, which can bind the catalytic region of most of the AdoMet-dependent methyltransferases blocking their activity [33]. Moreover, AdoHcy accumulation was early associated with global DNA hypomethylation in rat liver, and later in lymphocytes and liver carcinogenesis [[43], [44], [45]], whereas folate supplementation in colon cancer patients resulted in a significant decrease in global DNA hypomethylation observed in the rectal mucosa [46]. Although the experimental evidence consistently supports a connection between folate availability, AdoMet levels and DNA methylations [47], further research in this field is needed to address the impact of formaldehyde flux on epigenetics and thus on cell homeostasis.
4. Epigenetic demethylations and formaldehyde flux
So far, the formaldehyde flux from the one-carbon cycle to epigenetic methylations has been discussed. Next, attention will be drawn to the epigenetic demethylation reactions that generate formaldehyde. Histone and DNA methylations were originally thought to be irreversible and only lost by passive mechanisms during DNA replication. This view changed with the identification of demethylating enzymes [48]. There are three main families of demethylases that can generate formaldehyde as part of their catalytic mechanism: (i) the Jumonji (JmjC) family of histone demethylases; (ii) the AlkB family; and (iii) the lysine-specific (LSD) family of demethylases. Most histone demethylations are under the control of the Jumonji family, which belongs to the superfamily of Fe(II)-, and 2-oxoglutarate-dependent dioxygenases (2-OGDDs) [30]. The catalytic mechanism of 2-OGDD depends on oxygen, which is reduced to superoxide, oxidizing a catalytic Fe(II) ion. The superoxide attacks 2-oxoglutarate (2-OG), generating an intermediate that decarboxylates into succinate while oxidizing the methyl carbon to formaldehyde [49]. Cell metabolism has been reported to influence the activity of 2-OGDD enzymes. Accumulation of succinate can inhibit 2-OGDD enzymes, while fluctuations in 2-OG can also alter their activity [50]. On the other hand, active nucleic acid demethylation was originally reported in Escherichia coli with the identification of the DNA repair protein AlkB. This protein is able to oxidize alkyl groups on damaged DNA bases, regenerating the undamaged base and releasing the methyl group as formaldehyde [51]. The AlkB family is conserved in humans and also belongs to the 2-OGDD superfamily of demethylases [52]. The third group of demethylases that can generate formaldehyde is the LSD family of histone demethylases. There are two identified proteins belonging to this family, LSD1 and LSD2, which are amine oxidases containing a flavin-adenine dinucleotide (FAD) cofactor as an electron acceptor. They catalyse the demethylation of mono- and demethylated lysine residues generating formaldehyde and hydrogen peroxide (H2O2) [53]. Remarkably, in the nucleus, LSD1 was found to contain a THF molecule in the active site close to the FAD cofactor, which suggests that THF accepts formaldehyde originated from the catalytic demethylation likely generating 5,10-CH2-THF(Fig. 2) [54]. It is interesting to remark that 5 mC in the DNA is demethylated through successive oxidations of 5 mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5 fC) and 5-carboxylcytosine (5caC) catalysed by the Ten to Eleven Translocation (TET) proteins, lastly releasing the methyl unit as carbon dioxide. Although TET proteins also belong to the 2-OGDD family, they carry out the oxidative demethylation on the DNA with no release of free formaldehyde [48,55].
5. Oxidative THF breakdown
In addition to the canonical formaldehyde flux from the one-carbon cycle to epigenetic methylations, the molecular backbones of THF and of some THF derivatives can undergo spontaneous oxidative degradation releasing formaldehyde [5,7]. THF backbone consists of a pterine unit and a pABG moiety linked by a methylene group, which is the source of free formaldehyde upon THF decomposition (solid red line in THF structure, Fig. 1) [5]. It is still unclear to what extent cellular THF experiences this oxidative degradation. However, a significant fraction of cellular THF will likely decompose. Insights have been provided recently describing that pABG from THF and DHF accumulates in absence of the mitochondrial one-carbon branch or upon inhibition of DHFR by exposing cancer cells to the antifolate chemotherapeutic drug methotrexate [56]. The degradation of THF can, at least in part, be prevented by the enzyme quinoid dihydropteridine reductase (QDPR), which participates in tetrahydrobiopterin metabolism, and has been recognized as a metabolite repairing enzyme [57]. In vitro, the decomposition of THF, DHF and 5,10-CH2-THF is accelerated in presence of oxidant agents such as H2O2 or by increasing the temperature [7]. Cellular H2O2 can be generated mainly in the peroxisomes [58]. It can also originate from oxidative protein refolding in the endoplasmic reticulum -a reaction catalysed by protein disulphide isomerases (PDI) that are re-oxidized by ERO1-alpha, generating H2O2 [59], from FAD-dependent demethylations [60], and can also be produced from ROS generated in the mitochondrial electron chain (ETC) [61]. Therefore, cells need to sustain cellular redox homeostasis not only to prevent general oxidative damage but also to limit the oxidative degradation of THF derivatives, which would otherwise impair vital biosynthetic pathways and lead to the accumulation of toxic formaldehyde.
6. The formaldehyde cycle and human health
The surplus generation of formaldehyde from THF degradation can pose a significant threat to cells. Formaldehyde is a well-established genotoxin classified by the World Health Organization (WHO) as a human carcinogen present in the environment and obtained from methanol metabolism, from methylamine and from multiple cellular demethylations (Fig. 1), reaching blood concentrations close to 50 μM [[62], [63], [64]]. This aldehyde was shown to cause a plethora of DNA lesions such as base damage, DNA-protein, DNA-interstrand and DNA-intrastrand crosslinks [65]. Formaldehyde was also reported to damage proteins causing proteotoxic stress and triggering the activation of Heat Shock Transcription Factor 1 (HSF1) [10]. The strong electrophilicity of formaldehyde makes this molecule very reactive against electron-rich moieties, especially those containing thiol groups such as GSH and free cysteine, and also likely against thiol-rich proteins such as thioredoxins [66]. Indeed, formaldehyde rapidly reacts with GSH, yielding S-hydroxymethyl-GSH (HSMGSH) [9]. This spontaneous reaction blocks the redox-active thiol group of GSH impairing its antioxidant function and causing accumulation of ROS [9]. Cells evolved the alcohol dehydrogenase 5 enzyme (ADH5/GSNOR) to metabolize HSMGSH, recovering redox-active GSH and lastly generating formate (Fig. 2) [62,67]. This formate -originated through ADH5- can be incorporated into purines and thymine, thus becoming another meaningful source of one-carbon units, and defining a formaldehyde cycle that converts a toxin into a one-carbon source for anabolic reactions [7].
The formaldehyde cycle not only provides one-carbon units for anabolic reactions but also prevents a rise in the endogenous formaldehyde level. Indeed, mice lacking ADH5 were shown to accumulate N2-hydroxymethyl-deoxyguanine [8], a product of the reaction between formaldehyde and deoxyguanine on the DNA [68]. In these mice, DNA crosslink repair becomes essential, and the simultaneous inactivation of the Fanconi Anaemia DNA Repair pathway and ADH5 precipitates bone marrow failure (BMF), liver and kidney dysfunction, and leukaemia [8]. Furthermore, formaldehyde has been proposed to underly lethality, progeria and hepatocellular carcinoma in patients with Ruijs-Aalfs syndrome, a genetic disease caused by mutations in the gene coding for the DNA dependent protease Spartan (DVC1) [62,69]. Formaldehyde can also reduce the half-life of the tumour suppressor BRCA2, a significant threat for BRCA2-mutation carriers, who might present an increased rate of mutation and cancer development in case of exposure to environmental formaldehyde [70]. In humans, bi-allelic mutations in the gene coding for the formaldehyde metabolizing enzyme ADH5 were associated with an inherited bone marrow failure syndrome observed within individuals carrying a negative dominant mutation in the gene ALDH2 (ALDH2*2 polymorphism) [71]. This mutation, called the flush mutation because it causes redness of the face upon alcohol consumption, reduced the capacity of the mitochondrial enzyme ALDH2 to metabolize aldehydes [72,73]. It remains to be explored whether ALDH2 has any role in preventing the toxicity of mitochondrial formaldehyde from one-carbon metabolism.
7. THF toxicity through TYMS
The one-carbon flux implies the movement of reactive formaldehyde, which can affect several cellular structures. Interestingly, it was described that free formaldehyde can spontaneously condensate with THF, producing 5,10-CH2-THF [74,75]. This reaction might alter the endogenous level of 5,10-CH2-THF, affecting multiple biochemical reactions that depend on this THF derivative. Remarkably, cells lacking the main formaldehyde metabolizing enzyme ADH5 are very sensitive to formaldehyde and to exogenous THF [12], which might seem counterintuitive to the initial observation about spontaneous condensation between formaldehyde and THF. This reaction would reduce free formaldehyde; thus, it could be expected ADH5 to be at least dispensable for THF tolerance, which is in stark contrast with the experimental observations. Moreover, for this reaction to be meaningful in vivo, THF has to outcompete GSH, which is a cellular nucleophile present at millimolar levels [76]. It is still possible that added THF can react with endogenous formaldehyde, transiently increasing 5,10-CH2-THF and thus affecting the activity of TYMS. In support of this hypothesis, Rosado and col. showed that the toxicity of THF in ΔADH5 cells can be partially suppressed by adding dUMT or by inhibiting TYMS with the antimetabolite 5-fluor-uracyl (5-FU) [12]. Furthermore, the authors showed that inhibiting TYMS can also alleviate the THF-dependent phosphorylation of the DNA damage marker H2AX, supporting that TYMS reaction is involved in DNA damage caused by THF. It would be interesting to resolve whether an increase in endogenous formaldehyde affect the stoichiometry of 5,10-CH2-THF, leading to TYMS-dependent nucleotide imbalance and genome instability.
8. Conclusions and perspectives
The folate derivative THF plays a central role in one-carbon metabolism by accepting a formaldehyde molecule from several donors and distributing it to vital biosynthetic reactions such as purine and pyrimidine synthesis, and to AdoMet for methylations and writing of epigenetic marks. A dysregulation of THF metabolism can cause significant cellular impairment by altering those synthetic reactions, something well documented for CpG methylations in conditions of dietary folate deficiency. Epigenetic demethylation reactions through LSD, Jumonji and Alk demethylases generate formaldehyde, posing a significant threat to the genome. A two-tier control mechanism functions to prevent formaldehyde-caused damage. In one tier, endogenous formaldehyde is metabolized into formate, reducing its free concentration. The second tier consists of the Fanconi Anaemia DNA repair pathway that repairs DNA damage inflicted by formaldehyde (Fig. 2). In addition, THF, DHF and 5,10-CH2-THF were shown to be toxic for cancer and haematopoietic cells lacking the formaldehyde catabolic enzyme ADH5 [8,12]. Two hypotheses have been proposed to explain THF-induced cytotoxicity. On one hand, the oxidative breakdown of THF can release formaldehyde, which has been shown to poison ADH5-deficient cells. The underlying cause of this toxicity might be a combination of elevated ROS through GSH redox homeostasis imbalance and DNA damage, which will trigger cell death (Fig. 2). The second hypothesis to explain THF toxicity proposes that THF condensates with endogenous formaldehyde increasing the level of 5,10-CH2-THF, which might lead to a hyperactivation of TYMS, nucleotide imbalance and DNA replication stress (Fig. 2) [12]. Independently of the underlying mechanism, the increased toxicity of THF in ADH5-lacking cells might be used as a therapeutic intervention in cancer by combining THF and the ADH5 inhibitor N6022 [77], particularly in those cancer cells deficient in DNA crosslink repair that have been reported to be sensitive to THF [7]. Remarkably, cells deficient in one-carbon cycle enzymes can still survive consuming formate generated from endogenous formaldehyde through ADH5 [7]. This survival mechanism might be important in cancer cells that become resistant to anti-folates such as methotrexate, which is a chemotherapeutic drug that blocks the one-carbon cycle by inhibiting DHFR [78]. Thus, the combination of anti-folates and N6022 might help to overcome chemotherapy resistance in cancer cells.
Acknowledgements
The authors would like to acknowledge Guillermo Burgos-Barragan, Julia Qüesta and Carolina Perez Castro for their valuable suggestions on the manuscript. AEM and CU are CONICET fellows. LBP is funded by CONICET (PUE22920160100010CO), FOCEM MERCOSUR (COF 03/11) and ANPyCT (PICT-PRH 2017–4668), and receives support from the MPI for Metabolism Research (Cologne, Germany) and MPI for Biophysical Chemistry (Göttingen, Germany).
References
- 1.Visentin M., Diop-Bove N., Zhao R., Goldman I.D. The intestinal absorption of folates. Annu. Rev. Physiol. 2014;76:251–274. doi: 10.1146/annurev-physiol-020911-153251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barua S., Kuizon S., Junaid M.A. Folic acid supplementation in pregnancy and implications in health and disease. J. Biomed. Sci. 2014;21:1–9. doi: 10.1186/s12929-014-0077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McBurney M.W., Whitmore G.F. Isolation and biochemical characterization of folate deficient mutants of Chinese hamster cells. Cell. 1974;2:173–182. doi: 10.1016/0092-8674(74)90091-9. [DOI] [PubMed] [Google Scholar]
- 4.Garrow T.A., Brenner A.A., Whitehead V.M., Chen X.N., Duncan R.G., Korenberg J.R., Shane B. Cloning of human cDNAs encoding mitochondrial and cytosolic serine hydroxymethyltransferases and chromosomal localization. J. Biol. Chem. 1993;268:11910–11916. [PubMed] [Google Scholar]
- 5.Chippel D., Scrimgeour K.G. Oxidative degradation of dihydrofolate and tetrahydrofolate. Can. J. Biochem. 1970;48:999–1009. doi: 10.1139/o70-156. [DOI] [PubMed] [Google Scholar]
- 6.Fox J.T., Stover P.J. Chapter 1 folate-mediated one-carbon metabolism. Vitam. Horm. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- 7.Burgos-barragan G., Wit N., Meiser J., Dingler F.A., Pietzke M., Mulderrig L., Pontel L.B., Rosado I.V., Brewer T.F., Cordell R.L., Monks P.S., Chang C.J., Vazquez A., Patel K.J. Mammals divert endogenous genotoxic formaldehyde into one-carbon metabolism. Nature. 2017;548:549–554. doi: 10.1038/nature23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pontel L.B., Rosado I.V., Burgos-Barragan G., Garaycoechea J.I., Yu R., Arends M.J., Chandrasekaran G., Broecker V., Wei W., Liu L., Swenberg J.A., Crossan G.P., Patel K.J. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol. Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umansky C., Morellato A., Scheidegger M., Rieckher M., Martinefski M.R., Fernandez G.A., Kolesnikova K., Vesting A.J., Karakasilioti I., Reingruber H., Wei Y., He R., Bollini M., Monge M.E., Schumacher B., Pontel L.B. 2020. Endogenous Formaldehyde Scavenges Cellular Glutathione Resulting in Cytotoxic Redox Disruption; p. 2020. BioRxiv. 05.14.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortega-Atienza S., Rubis B., McCarthy C., Zhitkovich A. Formaldehyde is a potent proteotoxic stressor causing rapid Heat Shock transcription factor 1 activation and lys48-linked polyubiquitination of proteins. Am. J. Pathol. 2016;186:2857–2868. doi: 10.1016/j.ajpath.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ducker G.S., Rabinowitz J.D. One-carbon metabolism in health and disease. Cell Metabol. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.José C.B.G., Isabel A.B., José T.M., Piruat P.M.J.I., Caballero-velázquez T., Pérez-simón J.A., V Rosado I. 2018. Genotoxicity of Tetrahydrofolic Acid to Hematopoietic Stem and Progenitor Cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Field M.S., Kamynina E., Chon J., Stover P.J. Nuclear folate metabolism. Annu. Rev. Nutr. 2014;38:219–243. doi: 10.1146/annurev-nutr-071714-034441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter D.H., Cook R.J., Wagner C. Enzymatic properties of dimethylglycine dehydrogenase and sarcosine dehydrogenase from rat liver. Arch. Biochem. Biophys. 1985;243:396–407. doi: 10.1016/0003-9861(85)90516-8. [DOI] [PubMed] [Google Scholar]
- 15.Morscher R.J., Ducker G.S., Li S.H.J., Mayer J.A., Gitai Z., Sperl W., Rabinowitz J.D. Mitochondrial translation requires folate-dependent tRNA methylation. Nature. 2018;554:128–132. doi: 10.1038/nature25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersona D.D., Quintero C.M., Stovera P.J. Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15163–15168. doi: 10.1073/pnas.1103623108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin M., Momb J., Appling D.R. Human mitochondrial MTHFD2 is a dual redox cofactor-specific methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase. Canc. Metabol. 2017;5 doi: 10.1186/s40170-017-0173-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker E.J., Hershman S.G., Köhrer C., Belcher-Timme C.A., Patel J., Goldberger O.A., Christodoulou J., Silberstein J.M., McKenzie M., Ryan M.T., Compton A.G., Jaffe J.D., Carr S.A., Calvo S.E., RajBhandary U.L., Thorburn D.R., Mootha V.K. Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metabol. 2011;14:428–434. doi: 10.1016/j.cmet.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tibbetts A.S., Appling D.R. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 20.Ducker G.S., Chen L., Morscher R.J., Teng X., Kang Y., Rabinowitz J.D., Ghergurovich J.M., Esposito M. Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway article reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metabol. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietzke M., Meiser J., Vazquez A. Formate metabolism in health and disease. Mol. Metab. 2020;33:23–37. doi: 10.1016/j.molmet.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hum D.W., Bellli A.W., Rozenii R., Mackenzies R.E. 1988. Primary Structure of a Human Trifunctional Enzyme: Isolation of a cDNA Encoding Methylenetetrahydrofolate Dehydrogenase-Methenyltetrahydrofolate Cyclohydrolase-Formyltetrahydrofolate Synthetase*. [PubMed] [Google Scholar]
- 23.Herbig K., Chiang E.P., Lee L.R., Hills J., Shane B., Stover P.J. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J. Biol. Chem. 2002;277:38381–38389. doi: 10.1074/jbc.M205000200. [DOI] [PubMed] [Google Scholar]
- 24.An S., Kumar R., Sheets E.D., Benkovic S.J. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;80:320. doi: 10.1126/science.1152241. 103–106. [DOI] [PubMed] [Google Scholar]
- 25.Sanderson S.M., Gao X., Dai Z., Locasale J.W. Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat. Rev. Canc. 2019;19:625–637. doi: 10.1038/s41568-019-0187-8. [DOI] [PubMed] [Google Scholar]
- 26.Anderson D.D., Eom J.Y., Stover P.J. Competition between sumoylation and ubiquitination of serine hydroxymethyltransferase 1 determines its nuclear localization and its accumulation in the nucleus. J. Biol. Chem. 2012;287:4790–4799. doi: 10.1074/jbc.M111.302174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulis M., Esteller M. DNA methylation and cancer. Adv. Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 28.Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 30.Kooistra S., Helin K. Post-translational modifications: molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 31.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Greer E.L., Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffman D., Marion D., Cornatzer W., Duerre J. S-Adenosylmethionine and S-adenosylhomocystein metabolism in isolated rat liver. Effects of L-methionine, L-homocystein, and adenosine. J. Biol. Chem. 1980;255:10822–10827. https://www.jbc.org/content/255/22/10822 accessed June 21, 2020. [PubMed] [Google Scholar]
- 34.Obeid R. The metabolic burden of methyl donor deficiency with focus on the betaine homocysteine methyltransferase pathway. Nutrients. 2013;5:3481–3495. doi: 10.3390/nu5093481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Miguelsanz J., Vallecillo N., Garrido F., Reytor E., Pérez-Sala D., Pajares M.A. Betaine homocysteine S-methyltransferase emerges as a new player of the nuclear methionine cycle. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1165–1182. doi: 10.1016/j.bbamcr.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee R.V., Matthews R.G. Cobalamin‐dependent methionine synthase. Faseb. J. 1990;4:1450–1459. doi: 10.1096/fasebj.4.5.2407589. [DOI] [PubMed] [Google Scholar]
- 37.Markham G.D., Pajares M.A. Structure-function relationships in methionine adenosyltransferases. Cell. Mol. Life Sci. 2009;66:636–648. doi: 10.1007/s00018-008-8516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu S.C., Mato J.M. S-adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabil O., Zhou Y., Banerjee R. Human cystathionine beta-synthase is a target for sumoylation. Biochemistry. 2006;45:13528–13536. doi: 10.1021/bi0615644. [DOI] [PubMed] [Google Scholar]
- 40.Mosharov E., Cranford M.R., Banerjee R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry. 2000;39:13005–13011. doi: 10.1021/bi001088w. [DOI] [PubMed] [Google Scholar]
- 41.Beatty P.W., Reed D.J. Involvement of the cystathionine pathway in the biosynthesis of glutathione by isolated rat hepatocytes. Arch. Biochem. Biophys. 1980;204:80–87. doi: 10.1016/0003-9861(80)90009-0. [DOI] [PubMed] [Google Scholar]
- 42.Parsanathan R., Jain S.K. Glutathione deficiency induces epigenetic alterations of vitamin D metabolism genes in the livers of high-fat diet-fed obese mice. Sci. Rep. 2019;9:14784. doi: 10.1038/s41598-019-51377-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi P., Melnyk S., Pogribna M., Pogribny I.P., Hine R.J., James S.J. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J. Biol. Chem. 2000;275:29318–29323. doi: 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]
- 44.Cox R., Prescott C., Irving C.C. The effect of S-adenosylhomocysteine on DNA methylation in isolated rat liver nuclei. BBA Sect. Nucleic Acids Protein Synth. 1977;474:493–499. doi: 10.1016/0005-2787(77)90070-3. [DOI] [PubMed] [Google Scholar]
- 45.Mirbahai L., Southam A.D., Sommer U., Williams T.D., Bignell J.P., Lyons B.P., Viant M.R., Chipman J.K. Disruption of DNA methylation via S-adenosylhomocysteine is a key process in high incidence liver carcinogenesis in fish. J. Proteome Res. 2013;12:2895–2904. doi: 10.1021/pr400195u. [DOI] [PubMed] [Google Scholar]
- 46.Cravo M.L., Pinto A.G., Chaves P., Cruz J.A., Lage P., Nobre Leitão C., Costa Mira F. Effect of folate supplementation on DNA methylation of rectal mucosa in patients with colonic adenomas: correlation with nutrient intake. Clin. Nutr. 1998;17:45–49. doi: 10.1016/s0261-5614(98)80304-x. [DOI] [PubMed] [Google Scholar]
- 47.Serefidou M., Venkatasubramani A.V., Imhof A. The impact of one carbon metabolism on histone methylation. Front. Genet. 2019;10:764. doi: 10.3389/fgene.2019.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamadema N., Burr S., Brewer A.C. Dynamic regulation of epigenetic demethylation by oxygen availability and cellular redox. Free Radic. Biol. Med. 2019;131:282–298. doi: 10.1016/j.freeradbiomed.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 50.Xiao M., Yang H., Xu W., Ma S., Lin H., Zhu H., Liu L., Liu Y., Yang C., Xu Y., Zhao S., Ye D., Xiong Y., Guan K.-L. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aravind L., Koonin E.V. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fedeles B.I., Singh V., Delaney J.C., Li D., Essigmann J.M. The AlkB family of Fe(II)/α-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 2015;290:20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 54.Luka Z., Moss F., Loukachevitch L.V., Bornhop D.J., Wagner C. Histone demethylase LSD1 is a folate-binding protein. Biochemistry. 2011;50:4750–4756. doi: 10.1021/bi200247b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasmussen K.D., Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng Y., Lin T.Y., Lee G., Paddock M.N., Momb J., Cheng Z., Li Q., Fei D.L., Stein B.D., Ramsamooj S., Zhang G., Blenis J., Cantley L.C. Mitochondrial one-carbon pathway supports cytosolic folate integrity in cancer cells. Cell. 2018;175:1546–1560. doi: 10.1016/j.cell.2018.09.041. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pollock R.J., Kaufman S. Dihydrofolate reductase is present in brain. J. Neurochem. 1978;30:253–256. doi: 10.1111/j.1471-4159.1978.tb07059.x. [DOI] [PubMed] [Google Scholar]
- 58.Fransen M., Nordgren M., Wang B., Apanasets O. Role of peroxisomes in ROS/RNS-metabolism: implications for human disease. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2012;1822:1363–1373. doi: 10.1016/j.bbadis.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 59.Ramming T., Okumura M., Kanemura S., Baday S., Birk J., Moes S., Spiess M., Jenö P., Bernèche S., Inaba K., Appenzeller-Herzog C. A PDI-catalyzed thiol-disulfide switch regulates the production of hydrogen peroxide by human Ero1. Free Radic. Biol. Med. 2015;83:361–372. doi: 10.1016/j.freeradbiomed.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 60.Forneris F., Binda C., Vanoni M.A., Mattevi A., Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579:2203–2207. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 61.Boveris A., Oshino N., Chance B. The cellular production of hydrogen peroxide. Biochem. J. 1972;128:617–630. doi: 10.1042/bj1280617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reingruber H., Pontel L.B. Formaldehyde metabolism and its impact on human health. Curr. Opin. Toxicol. 2018;9:28–34. doi: 10.1016/J.COTOX.2018.07.001. [DOI] [Google Scholar]
- 63.Luo W., Li H., Zhang Y., Ang C.Y. Determination of formaldehyde in blood plasma by high-performance liquid chromatography with fluorescence detection. J. Chromatogr. B Biomed. Sci. Appl. 2001;753:253–257. doi: 10.1016/s0378-4347(00)00552-1. [DOI] [PubMed] [Google Scholar]
- 64.IARC working group on the evaluation of carcinogenic risks to humans., formaldehyde, 2-butoxyethanol and 1-tert-Butoxypropan-2-ol. IARC Work. Gr. Eval. Carcinog. Risks to Humans. 2006;88:1–478. [PMC free article] [PubMed] [Google Scholar]
- 65.Kawanishi M., Matsuda T., Yagi T. Genotoxicity of formaldehyde: molecular basis of DNA damage and mutation. Front. Environ. Sci. . 2014;2:36. https://www.frontiersin.org/article/10.3389/fenvs.2014.00036 [Google Scholar]
- 66.Pietzke M., Burgos-Barragan G., Wit N., Tait-Mulder J., Sumpton D., Mackay G.M., Patel K.J., Vazquez A. Amino acid dependent formaldehyde metabolism in mammals. Commun. Chem. 2020;3:78. doi: 10.1038/s42004-020-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosado I.V., Langevin F., Crossan G.P., Takata M., Patel K.J. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat. Struct. Mol. Biol. 2011;18:1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- 68.Lu K., Collins L.B., Ru H., Bermudez E., Swenberg J.A. Distribution of DNA adducts caused by inhaled formaldehyde is consistent with induction of nasal carcinoma but not leukemia. Toxicol. Sci. 2010;116:441–451. doi: 10.1093/toxsci/kfq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stingele J., Bellelli R., Alte F., Hewitt G., Sarek G., Maslen S.L., Tsutakawa S.E., Borg A., Kjær S., Tainer J.A., Skehel J.M., Groll M., Boulton S.J. Mechanism and regulation of DNA-protein crosslink repair by the DNA-dependent metalloprotease SPRTN. Mol. Cell. 2016;64:688–703. doi: 10.1016/j.molcel.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan S.L.W., Chadha S., Liu Y., Gabasova E., Perera D., Ahmed K., Constantinou S., Renaudin X., Lee M.Y., Aebersold R., Venkitaraman A.R. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell. 2017;169:1105–1118. doi: 10.1016/j.cell.2017.05.010. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dingler F.A., Wang M., Mu A., Millington C.L., Oberbeck N., Watcham S., Pontel L.B., Kamimae-Lanning A.N., Langevin F., Nadler C., Cordell R.L., Monks P.S., Yu R., Wilson N.K., Hira A., Yoshida K., Mori M., Okamoto Y., Okuno Y., Muramatsu H., Shiraishi Y., Kobayashi M., Moriguchi T., Osumi T., Kato M., Miyano S., Ito E., Kojima S., Yabe H., Yabe M., Matsuo K., Ogawa S., Göttgens B., Hodskinson M.R.G., Takata M., Patel K.J. Two aldehyde clearance systems are essential to prevent lethal formaldehyde accumulation in mice and humans. Mol. Cell. 2020;80(6):996–1012. doi: 10.1016/j.molcel.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harada S., Agarwal D.P., Goedde H.W. Aldehyde ehydrogenase deficiency as cause of facial flushing reaction to alcohol in Japanese. Lancet. 1981;318:982. doi: 10.1016/S0140-6736(81)91172-7. [DOI] [PubMed] [Google Scholar]
- 73.Wang R.-S., Nakajima T., Kawamoto T., Honma T. Effects of aldehyde dehydrogenase-2 genetic polymorphisms on metabolism of structurally different aldehydes in human liver. Drug Metab. Dispos. 2002;30:69. doi: 10.1124/dmd.30.1.69. LP – 73. [DOI] [PubMed] [Google Scholar]
- 74.Kallen R.G., Jencks W.P. The mechanism of the condensation of formaldehyde with tetrahydrofolic acid. J. Biol. Chem. 1966;241:5851–5863. [PubMed] [Google Scholar]
- 75.He H., Noor E., Ramos-Parra P.A., García-Valencia L.E., Patterson J.A., Díaz de la Garza R.I., Hanson A.D., Bar-Even A. Vivo rate of formaldehyde condensation with tetrahydrofolate. Metabolites. 2020;10 doi: 10.3390/metabo10020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morgan B., Ezeriņa D., Amoako T.N.E., Riemer J., Seedorf M., Dick T.P. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat. Chem. Biol. 2013;9:119–125. doi: 10.1038/nchembio.1142. [DOI] [PubMed] [Google Scholar]
- 77.Green L.S., Chun L.E., Patton A.K., Sun X., Rosenthal G.J., Richards J.P. Mechanism of inhibition for N6022, a first-in-class drug targeting S-nitrosoglutathione reductase. Biochemistry. 2012;51:2157–2168. doi: 10.1021/bi201785u. [DOI] [PubMed] [Google Scholar]
- 78.Osborn M.J., Freeman M., Huennekens F.M. Inhibition of dihydrofolic reductase by aminopterin and amethopterin. Proc. Soc. Exp. Biol. Med. 1958;97:429–431. doi: 10.3181/00379727-97-23764. [DOI] [PubMed] [Google Scholar]