Abstract
Background
Bacterial infections of the lung, skin, bloodstream and other tissues are common in patients with systemic lupus erythematosus (lupus) and are often more severe and invasive than similar infections in control populations. A variety of studies have explored the changes in bacterial abundance in lupus patients, the rates of infection and the influence of particular bacterial species on disease progression, using both human patient samples and mouse models of lupus.
Objective
The aim of this review is to summarize human and mouse studies that describe changes in the bacterial microbiome in lupus, the role of a leaky gut in stimulating inflammation, identification of specific bacterial species associated with lupus, and the potential roles of certain common bacterial infections in promoting lupus progression.
Methods
Information was collected using searches of the Pubmed database for articles relevant to bacterial infections in lupus and to microbiome changes associated with lupus.
Results
The reviewed studies demonstrate significant changes in the bacterial microbiome of lupus patients as compared to control subjects and in lupus-prone mice compared to control mice. Furthermore, there is evidence supporting the existence of a leaky gut in lupus patients and in lupus-prone mice. This leaky gut may allow live bacteria or bacterial components to enter the circulation and cause inflammation. Invasive bacterial infections are more common and often more severe in lupus patients. These include infections caused by Staphylococcus aureus, Salmonella enterica, Escherichia coli, Streptococcus pneumoniae and mycobacteria. These bacterial infections can trigger increased immune activation and inflammation, potentially stimulating activation of autoreactive lymphocytes and leading to worsening of lupus symptoms.
Conclusions
Together, the evidence suggests that lupus predisposes to infection, while infection may trigger worsening lupus, leading to a feedback loop that may reinforce autoimmune symptoms.
Keywords: Systemic lupus erythematosus, Microbiome, Leaky gut, Infection, Trigger, Feedback
Highlights
-
•
Severe bacterial infections are common in lupus patients and lead to morbidity and mortality.
-
•
A leaky gut syndrome is found in many lupus patients and mouse models of lupus.
-
•
There are changes to the bacterial microbiome in lupus.
-
•
Specific bacterial infections can stimulate development of lupus symptoms in mouse models.
-
•
There may be feedback loops between bacterial infection and autoimmunity such that they mutually amplify each other.
1. Introduction
Systemic lupus erythematosus (SLE or lupus) is a chronic multisystem autoimmune disorder that is characterized by severe immune dysregulation. As a result of loss of central and/or peripheral mechanisms of immune tolerance, self-reactive T and B lymphocytes persist in lupus patients and promote systemic immune activation [1]. The underlying causes of lupus are incompletely understood, but include both genetic and environmental risk factors. The result of these genetic and environmental factors is a significantly changed immune environment commonly characterized by reduced regulatory T cells, increased effector T cells and enhanced B cell activation [[2], [3], [4], [5], [6]]. Aberrant autoreactive B cell activation in response to self-antigen and the provision of T cell help by autoreactive T cells results in production of autoantibodies against various self-antigens including double-stranded DNA, ribonucleoproteins, connective tissues, and immunoglobulins [[7], [8], [9]]. The immune complexes formed can deposit in tissues and mediate antibody-dependent mechanisms of immune activation, such as complement activation, leading to inflammation and organ damage [10]. In addition, chronic activation of innate and adaptive immune cells results in a greatly altered cytokine milieu, with many cytokines found at increased levels [[11], [12], [13]].
The prevalence of lupus varies widely by geographical region, ethnicity, and sex with the highest rates among women and individuals of African descent. Women develop the disease at a 9:1 ratio as compared to men [14]. The altered immune state in SLE results in inflammation and damage to a variety of organs including the kidneys, lungs, cardiovascular system and brain, which contribute to mortality in patients with SLE. Lupus nephritis (LN), a form of glomerulonephritis, is a common manifestation that presents in approximately 50–75% of SLE patients and can develop into chronic kidney disease and severely impaired kidney function [[15], [16], [17], [18]]. Neuropsychiatric lupus, a result of chronic immune activation in the brain, can contribute to cognitive dysfunction as well as increased risk of stroke [16,[19], [20], [21], [22], [23]]. Patients with SLE have also been shown to develop increased rates of cardiovascular disease, with significantly increased rates of atherosclerosis and myocardial infarction as well as pericarditis [24,25]. These physical manifestations and the resulting end stage organ failures are common causes of death for SLE patients. However, studies over time have also identified infection as a major cause for hospitalization and mortality (Table 1) [[26], [27], [28], [29], [30], [31], [32], [33], [34]]. For instance, of 1000 European SLE patients followed prospectively over a 5-year period, 27% developed infections and among those that died during the study period, 29% died from infection [26]. As treatment for autoimmune pathology has improved, resulting in lower death rates due to organ damage caused by SLE, the percent of patients who die due to infection has increased. For instance, in a study from China, from 1986 to 1995 approximately 25% of deaths of SLE patients were due mainly to infection, while from 2006 to 2012 approximately 50% of deaths were due to infections [31]. Infections in SLE patients can be caused by bacterial, viral or fungal pathogens. In this review, we first focus on descriptions of the microbiome in lupus patients and mouse models of lupus, with an emphasis on mechanisms by which changes to the microbiome might influence lupus progression. Disruptions in the microbiome can allow pathogenic bacteria to invade the tissues and result in infections. In the second part of the review, we focus on specific bacterial infections in lupus patients describing their prevalence and relevance to morbidity and mortality.
Table 1.
Data supporting high rates of bacterial infection in lupus patients.
| Cervera et al., 1999 | Kang et al., 2011 | Jacobsen et al., 1999 | Fei et al., 2014 | Tsai et al., 2020 | Bharath et al., 2019 | Oud et al., 2019 | Ruiz-Irastorza et al., 2009 | |
|---|---|---|---|---|---|---|---|---|
| Location(s) | Seven countries in Europe | South Korea | Denmark | China | Taiwan | India | USA | Spain |
| SLE Cohort Size | 1000 | 1010 | 513 | 3831 | 427 | 53 | 94,338 | 249 |
| Clinical Outcome | Mortality | Mortality | Mortality | Mortality | Mortality | Mortality | Hospitalization due to sepsis | Hospitalization |
| Percent with infection | 24.4% (Bacterial) | 37.3% | 20.49% | 1986–1995: 25.7% 2006–2012: 53.6% |
72.47% | 47.2% | 18.1% | 29% |
| Infection sites | Respiratory Abdominal Urinary Tract |
Respiratory Sepsis Abscess Meningitis Peritonitis Infectious Colitis |
Respiratory Bacteremia Meningitis Brain Abscess Endocarditis Hepatitis |
Respiratory Generalized Bacteremia Extra-pulmonary tuberculosis Peritonitis Meningitis |
Respiratory Bacteremia Urinary Tract Skin |
Respiratory Urinary tract |
Respiratory Urinary Tract Abdominal Skin and Soft Tissue Device Related Other Unknown |
Respiratory Bacteremia Cellulitis and skin abscess Meningitis-encephalitis Pyelonephritis Abdominal Other |
| Bacterial Genus Identified |
None Identified | None Identified |
Staphylococcus Streptococcus Klebsiella |
Staphylococcus Klebsiella Pseudomonas Acinetobacter Enterococcus Escherichia Salmonella Pseudomonas Nocardia |
Staphylococcus Escherichia |
Acinetobacter Klebsiella Pseudomonas |
None Identified |
Escherichia Staphylococcus Mycobacterium Streptococcus Salmonella Pseudomonas Neisseria Campylobacter Legionella Serratia |
| Risk Factors Identified | Nephropathy | Irreversible organ damage related to SLE; cyclophosphamide therapy; glucocorticoid dosage | Early onset of disease; disease duration of 5–10 years | None Identified | Higher SLEDAI score; short disease duration; female Sex | Lung involvement; nephritis | Age 65+ years; High Deyo comorbidity index; multiple organ dysfunction | Lung disease; nephritis; leukopenia; anti-phospholipid autoantibodies; prednisone treatment |
2. Potential role of a leaky gut in driving lupus symptoms
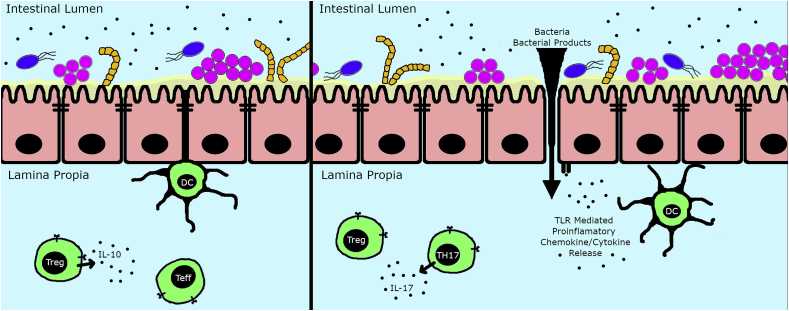
The majority of bacteria that colonize humans are found in the large intestine. Normally these bacteria remain within the intestinal tract, but damage to the lining of the intestine can lead to leakage of bacteria or their products (such as LPS) into the circulation. Bacteria leaving the intestine can colonize other organs such as liver or mesenteric lymph nodes. The leakage of bacteria or their products from the intestine is known as ‘leaky gut syndrome’ and has been identified in a number of human autoimmune diseases including rheumatoid arthritis, multiple sclerosis and Type I diabetes [[35], [36], [37]]. Fig. 1 shows the changes associated with leaky gut syndrome. Lupus patients also have a leaky gut as shown by detection of circulating LPS and (1 → 3)-β-D-glucan (a component of fungal cell walls) in their serum [38,39] and by the detection of human serum proteins (e.g., albumin and calprotectin) in the feces of lupus patients [40]. Albumin and calprotectin are normally absent from feces, but a breakdown of the intestinal barrier allows them to leak from the bloodstream into the intestine.
Fig. 1.
Leaky Gut Syndrome: On the left is shown a depiction of the normal gut, with colonization by varies bacterial species that remain in the lumen. Gut epithelial cells (shown in pink) are tightly attached to each other with tight junctions that prevent penetration of bacteria or their soluble products such as LPS into the submucosal tissues and thence into the bloodstream. Dendritic cells (DC) sample the gut contents to obtain bacterial antigens that are presented to T cells. Homeostasis is maintained by Tregs that prevent excess inflammation resulting from over-activation of the immune response by commensal microbes. This effect of Tregs is driven in part by their secretion of the immunoregulatory cytokine IL-10, which acts to suppress inappropriate activation by effector T cells (Teff). On the right is a depiction of a leaky gut. In the leaky gut, tight junctions between gut epithelial cells are partially disrupted, allowing bacteria and their products to penetrate into the submucosal tissue and then travel via the bloodstream to other tissues. The presence of bacteria and their products activates DCs and intestinal epithelial cells via TLR receptors. DCs can then drive formation of Th17 cells that secrete the pro-inflammatory cytokine IL-17. Treg activity in a leaky gut is insufficient to suppress immune activation due to the overwhelming proinflammatory signals. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
There are also significant data in mouse models of lupus indicating a leaky gut syndrome (Table 2). There is an increase in the levels of LPS in the bloodstream of the lupus-prone mouse strains MRL/lpr and NZBWF1, suggesting impaired gut barrier function [41,42]. This was confirmed by gavaging the mice with FITC-labeled dextran, which leaked from the gut to the bloodstream. Similarly, in lupus-prone MRL/lpr mice, (NZW × BXSB)F1 mice and TLR7.1 transgenic increased FITC-dextran leaked into the bloodstream when compared to non-lupus-prone control mice [[40], [41]]. Even wild-type C57BL/6 mice treated epicutaneously with the TLR7 ligand imiquimod, which triggers development of lupus-like disease, developed an impaired gut barrier [43]. Autoimmune damage to the gut wall may lead to leaky gut syndrome. Evidence supporting this comes from lupus-prone Fcgr2b−/− mice, which do not have a leaky gut in the young, pre-diseased state (8-24 weeks), but develop a leaky gut later in life when lupus is established (40 weeks) [39]. In pristane-induced lupus, there does not seem to be an intestinal barrier defect, unless mice are fed dextran sulfate solution (DSS) [44] and therefore lupus disease symptoms do not always lead to a leaky gut. DSS also triggers a leaky gut syndrome in young Fcgr2b−/− mice, when in the absence of DSS, Fcgr2b−/− mice only develop leaky gut later in life after lupus is established [39].
Table 2.
Mouse models of lupus used in the studies described.
| Mouse model | Type of animal model | Shown to have a leaky gut? |
|---|---|---|
| MRL/pr | Spontaneous lupus – mutation in Fas gene with multigenic contribution from MRL background strain | Yes [41] |
| NZBWF1 | Spontaneous lupus - multigenic | Yes [42] |
| (NZW × BXSB)F1 | Spontaneous lupus = multigenic but with over-expressed TLR7 in males | Yes [40] |
| TLR7.1 transgenic | Transgenic model over-expressing TLR7 | Yes [40] |
| Fcgr2b−/− | Knockout of the inhibitory Fc RIIb receptor | Yes [39,44] |
| Pristane-induced lupus | Wildtype mice induced to have lupus by injection of pristane intraperitoneally | No, unless fed DSS∗ [44] |
| B6/lpr | Wildtype but carries the Fas lpr mutation leading to immune activation and mild lupus | Not tested |
| SNF1 | Spontaneous lupus - multigenic | Not tested |
∗ DSS – dextran sulfate solution.
To address whether there might be changes in the microbiota of lupus-prone strains that induce a leaky gut, Zegarra-Ruiz and colleagues transferred gut microbiota from lupus-prone TLR7.1 transgenic mice to wild-type C57BL/6 littermates by co-housing or gavage. This transfer resulted in an intestinal barrier defect in the wild-type littermate mice [43]. One bacterial species that seems to play a role in inducing gut leakiness is Enterococcus gallinarum, because monocolonizing germ-free wild-type C57BL/6 mice with E. gallinarum leads to a weakened intestinal barrier and translocation of bacteria to the liver and mesenteric lymph nodes [40]. Together, studies in both humans and mice indicate that an impaired gut barrier may allow leakage of bacteria or their products into the bloodstream, thereby stimulating immune cell responses, including those of autoreactive lymphocytes.
3. Microbiome studies
3.1. The bacterial microbiome in human lupus
Mice and humans contain a very diverse and abundant microbiota in the gastrointestinal tract as well as the skin, eyes, nose and genitourinary systems. Bacterial species found in these tissues can both promote inflammation (by releasing immune activating compounds such as TLR ligands) and inhibit inflammation (by promoting the development of regulatory immune cells). For instance, microbial-derived short chain fatty acids, such as butyrate, have been shown to promote development of regulatory T cells [45]. Microbial dysbiosis, an altered balance of the bacterial microbiota, is common in immune disorders and can lead to expansion of pathogenic and/or opportunistic bacterial species [[46], [47], [48]]. It is important to note that the composition of the gut microbiome can vary with age and sex of those tested and this must be taken into account in microbiome studies [49,50]. Furthermore, human and murine intestinal microbiota have substantial differences and thus care must be taken in interpreting mouse studies and their relevance to human lupus [51].
Loss of certain protective functions provided by the normal microbiota can lead to over-colonization by bacterial species that promote a stronger inflammatory response. Bacterial dysbiosis been well-documented in inflammatory diseases such as inflammatory bowel disease and ulcerative colitis [52,53]. A number of different studies have also described bacterial dysbiosis in the microflora of the skin, oral cavity and gut of patients with SLE (summarized in Table 3). As can be appreciated from the Table, different studies have identified different changes in the microbiome of lupus patients versus healthy controls. Differences at various taxonomic ranks (phylum, class, order, family, genus and species) have been described in lupus patients. Genetic differences between the populations studied and environmental differences (such as diet) can influence the microbial composition. Thus, changes that are identified in the microbiome must be further studied to determine if such changes are biologically relevant to lupus development. The microbiota changes can affect immune differentiation. For instance, in vitro stimulation of CD4+ T cells with dendritic cells plus fecal microbiota from SLE patients resulted in significantly reduced differentiation of FoxP3+ Tregs as compared to stimulation with fecal microbiota from healthy controls [54]. This impaired development of immunosuppressive Tregs in response to the SLE gut microbiome could contribute to chronic immune activation in the lupus patients. In this section, we summarize some of the changes identified in the human lupus microbiome and below we summarize changes identified in the mouse lupus microbiome and how mouse studies can approach the question of whether the changes identified may contribute to lupus progression.
Table 3.
Bacterial dysbiosis in lupus patients.
| Study | Hevia et al., 2014 | He et al., 2016 | Luo et al., 2018 | Azzouz et al., 2019 | Li et al., 2020 | Bellocchi et al., 2019 | Wei et al., 2019 | van der Meulen et al., 2019 | Huang et al., 2020 | Li et al., 2019 | Pessoa et al., 2019 | Correa et al., 2017 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organ system | Gut | Gut | Gut | Gut | Gut | Gut | Gut | Gut and Oral Cavity | Skin | Oral cavity | Oral cavity | Oral cavity |
| Patient population ethnicity | Spanish | Chinese | American | American | Chinese | Italian | Chinese | Dutch | Chinese | Chinese | Brazilian | Brazilian |
| Methods used | 16S rRNA Sequencing |
Whole genome sequencing | 16S rRNA Sequencing |
16S rRNA Sequencing | 16S rRNA Sequencing |
16S rRNA Sequencing |
16S rRNA Sequencing |
16S rRNA Sequencing |
16S rRNA Sequencing |
16S rRNA Sequencing | DNA-DNA checkerboard hybridization | 16S rRNA Sequencing |
| Overall bacterial species richness/diversity | Similar | Similar or decreased depending on measure used | Decreased | Decreased | Decreased | Similar | Similar or decreased depending on measure used | Decreased | Decreased | Decreased | Decreased | Decreased |
| Bacterial phyla over-represented in lupus | Bacteriodetes | Actinobacteria, proteobacteria, and Bacteriodetes | Proteobacteria | Not defined | Cyanobacteria | Not defined | Proteobacteria | Bacteriodetes and Proteobacteria | Firmicutes | Fusobacteria | Not defined | Not defined |
| Bacterial phyla under-represented in lupus | Firmicutes | Firmicutes | None | Not defined | Actinobacteria and Tenericutes | Not defined | None | Firmicutes | Acidobacteria, Gemmatimonadetes, and Tenericutes | Tenericutes | Not defined | Not defined |
| Species correlated with higher SLEDAI | None identified | None identified | None identified | Ruminococcus gnavus | None identified | None identified | None identified | None identified | None identified | Streptococcus anginosus | None identified | None identified |
In general, most studies with human SLE patients have demonstrated a reduced bacterial species diversity in patients as compared to healthy controls. In Table 3, we have listed changes in the microbiome of lupus patients at the phylum level. Many changes at other taxonomic levels (class, order, family and genus) have been described. Due to space constraints, we don’t describe all these changes in detail and the reader is referred to references in Table 3 for more details. The gut microbiome is dominated by species in two bacterial phyla, Firmicutes and Bacteriodetes, which together include ~90% of all bacterial species in the gut [55]. Three studies have identified a relative decrease in bacteria of the phylum Firmicutes and an increase in bacteria of the phylum Bacteroidetes (a decreased Firmicutes/Bacteriodetes ratio) in the gut of lupus patients [[56], [57], [58]], while two other studies did not observe this change [50,59]. The next most common bacterial phyla in the gut are Actinobacteria and Proteobacteria. Several studies have found an over-representation of Proteobacteria in the gut of lupus subjects [50,57,58,60]. One study also found an over-representation of Actinobacteria [57], but a different study showed Actinobacteria to be decreased [61]. In addition, there may be changes in bacterial phyla that are typically relatively minor constituents of the intestinal flora, such as increased Cyanobacteria and decreased Teniricutes [61]. The skin and oral cavity tend to have different bacterial composition compared to the gut with bacteria in the phyla Actinobacteria, Firmicutes and Proteobacteria being common. On the skin, Huang et al. found that the phylum Firmicutes was over-represented, while certain phyla that are less common constituents of the skin microbiome (Acidobacteria, Gemmatimonadetes, and Tenericutes) were under-represented [62]. The phyla Actinobacteria, Firmicutes, Bacteriodetes, Fusobacteria and Proteobacteria are common in the oral cavity. Li et al. found an increase in Fusobacteria in the oral cavity of lupus patients, while the less common phylum Tenericutes was reduced [59].
Many of the microbiome studies were conducted on lupus patients in remission with low SLEDAI scores. However, the studies by Azzouz et al. [63] and Li et al. [59] included patients both in remission and in active phases of the disease. Note that while the Azzouz study and Li study both compared active versus inactive lupus patients, they did not find the same changes to the microbiome to be associated with active disease. The reasons for this difference could include differences in genetic makeup of the populations as well as differences in environmental factors like different diets or differences in drug therapies. In the Azzouz study, the bacterial families Veillonellaceae and Ruminococcaceae showed a statistically significant increase in lupus patients with active disease versus those with low disease activity. As described in more detail below, the Ruminococcaceae includes the bacterial species Ruminococcus gnavus and the study by Azzouz linked this to lupus progression. In the Li et al. study, the phylum Firmicutes was increased in patients with active lupus, while the phylum Actinobacteria was reduced. Among the Firmicutes, the order Lactobacillales and the family Streptococcaceae and genus Streptococcus were most increased. Among the Actinobacteria, the order Actinomycetales and Bifidobacterales were most reduced. Although the overall prevalence of the phylum Proteobacteria was not statistically changed in active lupus, the class Epsilonproteobacteria within this phylum was increased. Within the Epsilonproteobacteria, the order Campylobacterales and family Campylobacteraceae were increased. The best-studied representative of Campylobacteraceae in the human gut is Campylobacter jejuni, which is a common cause of food poisoning. C. jejuni is also associated with Guillain-Barre syndrome and reactive arthritis [64]. Immunization of Balb/c mice with formalin-fixed C. jejuni in the presence of Freund’s adjuvant induces a lupus-like syndrome [65]. It is possible that immunosuppressive therapies given to lupus patients with active disease result in increased susceptibility to infections with C. jejuni or other Campylobacter species. Indeed, the level of Campylobacter was positively correlated with SLEDAI scores [59]. These infections might contribute to certain lupus symptoms and merit further study.
3.2. Specific bacterial species associated with human lupus
One species of bacteria that has been linked to lupus pathogenesis in humans is Enterococcus gallinarum. As described above, E. gallinarum has been linked to a leaky gut syndrome in mice [40]. A similar leakiness also appears in human SLE patients. Liver biopsies of lupus patients revealed the presence of E. gallinarum in the liver tissue, while biopsies of liver from normal donors did not find E. gallinarum, although other species in the genus Enterococcus were present in four of six normal liver donors using PCR with primers specific to E. gallinarum or all species in the Enterococcus genus [40]. Co-culturing E. gallinarum with primary hepatocytes induced production of type I interferon, a cytokine with a major role in driving lupus pathogenesis. Therefore, a weak intestinal barrier may allow E. gallinarum to escape the gut and colonize the liver, where it can promote an inflammatory state and induce production of immune-stimulating cytokines.
Two studies have found an enrichment of members of the Ruminococcus genus in SLE patients [60,63]. One of those studies showed that the presence of the species Ruminococcus gnavus was correlated with worse disease in lupus patients as represented by high SLEDAI scores [63]. Interestingly, some anti-dsDNA antibodies seem to crossreact with antigens found in the R. gnavus strain RG2, suggesting the R. gnavus might stimulate anti-DNA autoantibody production [63]. R. gnavus has also been found to be enriched in patients with the inflammatory bowel disorder Crohn’s Disease [[66], [67], [68]]. This suggests the possibility that R. gnavus has a unique role in stimulating inflammatory immune responses. However, in a different analysis, R. gnavus was found to be reduced in the gut microbiome of lupus patients with active disease as compared to disease in remission [59]. Li et al. found that bacteria in the genus Streptococcus, including S. anginosus, were more prevalent in lupus patients, especially those with active disease. The level of Streptococcus and S. anginosus was also positively correlated with higher SLEDAI values and negatively correlated with the levels of complement C3 [59].
Together, the studies described above show that there are typically alterations in the bacterial microbiome between lupus patients and control subjects and these studies have pinpointed some specific species of bacteria that may play a pathogenic role. However, it is still not clear if the differences identified are critical for lupus progression or are instead a consequence of lupus-associated damage to the organs being tested or to differences in environmental factors [69]. Mouse studies can help address mechanistically the role of various bacterial species in lupus progression.
3.3. The bacterial microbiome in mouse lupus
Several studies have described the gut microbiome of lupus-prone mice compared to non-lupus-prone control animals. Zhang et al. first described a reduced prevalence of the bacterial family Lactobacillaceae in 5 week old lupus-prone MRL/lpr mice compared to non-lupus-prone MRL mice [49]. There was a corresponding increase in the prevalence of the bacterial family Lachnospiraceae, as well as other gut bacteria including members of the families Ruminococcaceae and Rikenellaceae. Lymphadenopathy and glomerulonephritis correlated negatively with the abundance of Lactobacillaceae and positively with Lachnospiraceae in the gut of MRL/lpr mice. A reduction in bacteria from the order Lactobacillales (which includes the Lactobacillaceae family) was confirmed in another study with MRL/lpr mice [70]. B6/lpr mice, which carry the same Fas lpr mutation as MRL/lpr, but on the C57BL/6 genetic background, also showed an alteration in gut microbiota compared to wild-type C57BL/6 controls [49]. In these B6/lpr mice and in normal C57BL/B6 controls, the abundance of Lactobacillaceae varied over time, but did not show a distinct correlation with the presence of lupus. Lactobacillaceae was also identified as a taxon that was changed in human lupus, but only when comparing lupus patients with active versus inactive SLE [59]. However, Lactobacillaceae was increased in active SLE, while in mouse models of lupus Lactobacillaceae are reduced. So far, no studies have explored the microbiome in mouse tissues other than the gut. Therefore, we do not know how the microbiome of the skin or oral cavity of lupus-prone mice compares to the microbiome of human lupus patients in these locations.
To test roles of the microbiota in driving lupus disease in mouse models, lupus-prone MRL/lpr mice were derived under gnotobiotic (germ-free) conditions [71]. Germ-free MRL/lpr mice developed an overall similar degree of autoimmunity as MRL/lpr mice housed under conventional conditions, including the extent of lymphoproliferation and presence of kidney disease. In 5-month-old mice, there was reduced production of anti-single-stranded DNA and anti-chromatin autoantibodies in the germ-free MRL/lpr mice. However, anti-Smith antigen autoantibodies were actually elevated in 5-month-old germ-free MRL/lpr mice. These data would tend to support the idea that microbiota may have a minimal effect in promoting stronger autoimmune responses. However, in another mouse model of lupus, TLR7.1 transgenic mice, maintaining the animals under germ-free conditions led to a reduction in lupus symptoms [43]. Furthermore, transfer of cecal bacteria from non-lupus-prone MRL mice into lupus-prone MRL/lpr mice resulted in reduced autoimmune symptoms, indicating the changes to the microbiome can influence lupus disease course in MRL/lpr mice [41]. Complete loss of the microbiota results in the removal of bacteria that have a protective role in stimulating development of regulatory cells, such as Tregs [72], as well as removal of more pathological species that stimulate immune activation. Therefore, results with gnotobiotic mice must be interpreted cautiously.
Treatment of MRL/lpr mice with a combination of antibiotics that kills both Gram-positive and Gram-negative bacterial species (a combination of ampicillin, neomycin, metronidazole and vancomycin) results in reduction of lupus symptoms [70]. Oral treatment with vancomycin alone, which selectively kills gram-positive bacteria, also results in reduced lupus [70]. Similar results have been obtained in TLR7.1 transgenic mice where treatment with a combination of antibiotics (ampicillin, neomycin, metronidazole and vancomycin) reduced lupus symptoms [43]. Treatment of (NZW × BXSB)F1 hybrid mice with vancomycin or ampicillin alone (which are most active against Gram-positive bacteria) also leads to reduced lupus symptoms [40]. On the other hand, treatment of (NZW × BXSB)F1 hybrid mice with metronidazole alone (most active against Gram-negative anaerobic bacteria) or neomycin alone (also most active against Gram-negative bacteria), does not reduce lupus symptoms. These results suggest that Gram-positive bacteria may be especially important in triggering lupus development. Firmicutes and Actinobacteria are the most prominent bacterial phyla in the gut that are Gram-positive. Thus, it is possible that one or more species in these phyla are particularly important in driving lupus pathogenesis. Altogether, the results of the antibiotic treatment regimens indicate that the microbiota does contribute to development of lupus disease and further suggests that in MRL/lpr gnotobiotic mice the absence of bacteria that promote development of regulatory cells may explain the fact that total lack of bacteria did not significantly alter the lupus phenotype.
3.4. Lactobacillaceae and lupus
As described above, several studies have shown a relative deficiency of bacteria in the family Lactobacillaceae in lupus-prone mouse models [49,70]. A study by Johnson et al. showed that lupus-prone SNF1 mice maintained on acidified water (AW) had a slower progression of lupus than SNF1 mice maintained on neutral pH water (NW) [73]. In SNF1 mice that had already developed nephritis, there was an increase in Lactobacillus reuteri and Turicibacter species, while in pre-nephritic SNF1 mice there was an increase of bacteria in the families Rikenellaceae and Christensenellaceae when the mice were maintained on AW. Transfer of microbiota from AW-treated mice to NW mice resulted in reduced lupus progression. The importance of bacteria in the Lactobacillaceae family was tested by transferring a mixture of 5 different Lactobacillus species (Lactobacillus oris, Lactobacillus rhamnosus, Lactobacillus reuteri, Lactobacillus johnsonii, and Lactobacillus gasseri) into MRL/lpr mice that are deficient in Lactobacillaceae [70]. Transfer of Lactobacillus species into MRL/lpr mice pre-treated with antibiotics for 2 days resulted in reduced lupus symptoms, which was associated with reduced leakiness of the gut. Treatment with Lactobacilli was more effective if begun early (3 weeks of age) rather than after disease onset. L. reuteri was the only transferred Lactobacillus species that was recovered in fecal pellets, suggesting that among the species transferred only L. reuteri survives well in the environment of the mouse gut [70]. This also suggests that L. reuteri may be the most important Lactobacillus species involved in suppressing lupus.
Several other studies suggest a role for L. reuteri in reducing lupus-associated inflammation in NZBWF1 mice [74,75]. On the other hand, L. reuteri has been shown to be over-represented in TLR7.1 transgenic mice and to promote lupus symptoms, while another Lactobacillus species L. johnsonii was protective against lupus development in this model [43]. Therefore, the role of L. reuteri in promoting immune suppression remains ill-defined and may be influenced by the mouse model studied. In addition to L. reuteri, several other Lactobacillus species (L. fermentum, L. paracasei, L. delbrueckii or L. rhamnosus) have been shown to suppress lupus inflammation in either NZBWF1 mice or in pristane-induced lupus models [42,[76], [77], [78]]. However, Luo et al. showed that an uncharacterized Lactobacillus species was increased in NZBWF1 lupus-prone mice after disease onset and that it was associated with more severe symptoms [50]. Enterococcus gallinarum, which was identified as a potential pathogenic bacterium in lupus, is a member of the order Lactobacillales and therefore genetically related to the Lactobacilli. In summary, the potential roles of Lactobacillus species in either protecting from lupus or driving lupus symptoms remains unclear. Differences in the species of Lactobacillus as well as differences in mouse age, sex, severity of lupus symptoms, diet or other environmental factors could play a role in these sometimes contradictory findings and further study is warranted to clear up these inconsistencies.
Data supporting roles for Lactobacillus in human lupus are still very preliminary. An increased prevalence of Lactobacillus species was found in the fecal microbiota of lupus patients when compared to normal healthy individuals [43]. Furthermore, as described above, bacteria in the order Lactobacillales (which includes the genus Lactobacillus) are increased in lupus patients with active disease versus those with lupus in remission [59]. Therefore, it’s possible that Lactobacilli play a pathogenic role in human lupus. Alternatively, they simply colonize the gut of lupus patients more readily than normal controls.
3.5. Specific bacterial species associated with mouse lupus
In human lupus patients several bacterial species have been associated with lupus severity, including Ruminococcus gnavus and Enterococcus gallinarum. Potential roles for these bacteria have also been studied in mouse models of lupus. Mouse studies concerning the role of R. gnavus are contradictory making it difficult to confidently ascribe a role to this species in pathogenesis. R. gnavus is a member of the bacterial family Lachnospiraceae. A study of MRL/lpr lupus-prone mice showed an enrichment of Lachnospiraceae family bacteria in the gut of female lupus-prone mice as compared to the non-lupus-prone mice [49]. This difference was not noted in male MRL/lpr mice that tend to have a somewhat delayed and less severe lupus phenotype. NZBWF1 lupus-prone mice also showed an increase in Lachnospiraceae species when comparing microbiota from diseased versus pre-diseased mice [50]. Although these two studies found an increased prevalence of the family Lachnospiraceae, they did not report any specific increase in R. gnavus Itself. However, another study in lupus-prone MRL/lpr mice did examine R. gnavus prevalence and found no increase in either Lachnospiraceae or R. gnavus in the intestinal microbiota, although this study did not separately analyze male and female mice [79]. In lupus-prone SNF1 mice, provision of acidified water was shown to reduce lupus symptoms, but this was associated with an increase in R. gnavus levels [73]. Altogether, the data are confusing as to the prevalence and role of R. gnavus in mouse lupus.
Enterococcus gallinarum is a Gram-positive bacterium that has been shown to be elevated in (NZW × BXSB)F1 lupus prone mice, with colonization present in the mesenteric veins, mesenteric lymph nodes, liver, and spleen [40]. Germ-free mice that were monocolonized with E. gallinarum significantly downregulated gut barrier genes such as occludin, claudins, and mucin-2 indicating that the bacterium is capable of weakening gut barrier function and inducing a leaky gut. Importantly, vaccination against E. gallinarum prolonged survival of mice with lupus indicating that the bacterium may play an important role in development of autoimmunity in the mice [40].
3.6. Segmented filamentous bacteria and Th17 cells
Segmented filamentous bacteria (SFB also referred to as ‘Candidatus Arthromitus’ or ‘Candidatus Savagella’) are a type of bacteria found in the intestine of mice and other organisms and they are closely associated with and attached to gut epithelial cells. They cannot be cultured and hence have been studied mainly in germ-free mice monocolonized with SFB. Bacteria from these monocolonized animals have been sequenced and represent a single species in the order Clostridiales within the phylum Firmicutes [80]. These bacteria have a greatly reduced genome typical of microbes that depend on the host for providing part of their nutritional needs. Introduction of SFB into germ-free mice stimulates development of Th17 cells [81]. IL-17 has a prominent role in inducing inflammation in a variety of autoimmune diseases, including lupus [82]. Therefore, SFB might potentially contribute to the pathogenesis of lupus by inducing development of inflammatory Th17 cells. In addition to their roles in inducing Th17 cells, SFB also induce other T cell responses [83]. SFB-like bacteria are rarely found in adult humans, but have been shown to be more common in children 3 years old and younger [84]. Hence, the role of SFB in development of Th17 cells in humans is debatable. However, the bacteria Bifidobacterium adolescentis isolated from human gut was also able to induce Th17 cells when transferred into germ-free mice [85]. Like SFB, B. adolescentis is closely associated with and bound to gut epithelial cells.
The role of SFB in lupus development remains unclear. SFB can stimulate anti-nuclear autoantibody production in mice with a deletion of LTβR (which is required for generation of lymph nodes) or a combined deletion of Hox11 (which is required for generation of the spleen) and LTβR [86]. Mice lacking either LTβR alone or both Hox11 and LTβR develop anti-nuclear autoantibodies, similar to those found in lupus and this was shown to be due to enhanced colonization of SFB in the intestine leading to increased Th17 cells [86]. However, SFB was shown to have no effect on development of lupus nephritis in SNF1 mice maintained either on acidified water (AW) on neutral pH water [73]. Further studies will be required to understand how SFB might contribute to lupus development in various mouse models.
3.7. Summary of microbiome studies
Studies have begun to tease out contributions of the bacterial microbiome to lupus progression. It is clear that the microbiome of lupus patients and lupus-prone mice often differ substantially from controls. However, it is less clear how these changes impact disease progression. This is further complicated because different studies have identified different changes in the microbiome and these alterations can be influenced by sex, age, genetics and various environmental factors of the populations studied. A number of bacterial species have been implicated as potentially important in either stimulating or suppressing lupus (Table 4). These include Enterococcus gallinarum, Ruminococcus gnavus, segmented filamentous bacteria and Lactobacillus species. A leaky gut is present in many human lupus patients and in mouse models of lupus, indicating the bacteria or bacterial products can leak into the bloodstream and potentially contribute to immune activation. Future studies will be needed to better address how these microbiome changes impact lupus progression.
Table 4.
Bacterial species associated with lupus in humans and mice.
| Bacterial species | Associations noted in humans | Associations noted in mice |
|---|---|---|
| Enterococcus gallinarum | Present in the liver (indicating leaky gut) and promotes type I interferon production [40] | Elevated in (NZW x BXSB)F1 mice and vaccination against E. gallinarum associated with reduced disease symptoms |
| Ruminococcus gnavus | Differing results in various studies - two studies showed correlation with worse disease (higher SLEDAI score) [60,63], but a third study showed R. gnavus is reduced in active disease [59] | Not increased in prevalence in MRL/lpr mice [79]. Increased in prevalence in SNF1 mice on acidified water (which reduces symptoms) [73]. |
| Streptococcus anginosus | Presence correlated with worse disease (SLEDAI score) [59] | Not tested in mouse lupus. |
| Lactobacillus species | Increased in human lupus patients [43] and in active disease [59] | Confusing results that depend on the species of Lactobacillus studied and the mouse model used – Lactobacillus is associated with worse disease in some studies [43,50,73], but associated with reduced symptoms in other studies [42,43,69,[74], [75], [76], [77], [78]]. |
| Candidatus Arthromitus (segmented filamentous bacteria, SFB) | Role in humans unclear; bacteria of this genus typically not present in adult humans [84] | Stimulates development of Th17 cells that may contribute to inflammation |
4. Bacterial infections in lupus
4.1. Increased bacterial infection rates in lupus
The burden of infection in patients with SLE is great resulting in hospitalization rates 12 times greater than the general populace by some estimates [87]. In addition to this greater prevalence of hospitalization, high rates of serious infection contribute significantly to increased mortality as compared to the general populace [17,32,[88], [89], [90], [91], [92]]. Bacterial pathogens have been identified as a major contributor to infection in SLE patients, comprising 60–75% of all infections reported [17,26,27,[29], [30], [31], [32], [33], [34],90,[93], [94], [95], [96]]. The risk of opportunistic infection, including bacterial infections, is elevated in SLE patients compared to the general populace [97]. Lupus nephritis (LN) is a severe manifestation of SLE in which kidney function is impaired due to deposition of autoantibodies and complement factors resulting in tissue inflammation and damage [18]. The resulting glomerulonephritis can cripple normal activity of the kidney contributing to proteinuria and eventually if untreated renal failure [98]. Retrospective analysis of hospital admissions of SLE patients has consistently identified LN as a risk factor in bacterial infection [62,95,[99], [100], [101], [102], [103], [104], [105], [106]]. Patients with LN exhibit significant rates of bacteremia, which in turn can contribute to systemic immune activation and flare development [107,108].
The susceptibility of SLE patients to severe bacterial infections is consistent with the use of immunosuppressive drugs such as glucocorticoids or cyclophosphamide. However, SLE patients often have immunological abnormalities that might also contribute to susceptibility. These immunological abnormalities include lymphopenia, neutropenia and/or low levels of complement proteins and they may lead to an immunodeficiency that precedes use of immunosuppressive drugs. For instance, in LN patients there is a depletion of complement proteins resulting from binding to immune complexes deposited in the glomeruli. Murine models of lupus also sometimes show an immunodeficiency to certain bacterial infections under basal conditions where no immunosuppressive drugs are used. MRL/lpr mice exhibit an enhanced susceptibility to infection with Mycobacterium leprae and B6/lpr mice exhibit a deficiency in Haemophilus influenzae clearance [113,114]. On the other hand, there is also some evidence that the increased immune activation that occurs in lupus can lead to better clearance of some infections. For instance, mice carrying the lupus susceptibility locus Sle3 have enhanced antibacterial responses to pneumonia caused by Klebsiella pneumoniae and to polymicrobial sepsis caused by cecal ligation and puncture [115]. Lupus-prone Fcgr2b−/− or TLR7.1 transgenic mice are resistant to cerebral malaria [116]. While the malaria parasite is not a bacterium, but rather a unicellular eukaryotic pathogen, this result supports the idea that excess immune activation in lupus may sometimes be protective against infection.
Retrospective studies conducted in North America, India, Africa, Europe, and Asia have identified particular pathogens to be extremely prevalent in SLE including Staphylococcus aureus, Salmonella enterica, Escherichia coli, Streptococcus pneumoniae and various mycobacterial species. These species, while also prevalent in the general population, often lead to more serious and invasive infections in SLE patients. Colonization with these bacteria may enhance autoimmune responses to potentially precipitate development of SLE. Below we summarize the evidence linking specific bacterial species to lupus pathogenesis.
4.2. Staphylococcus aureus
Staphylococcus aureus is a Gram-positive bacteria and common commensal organism on human epithelial surfaces such as the skin and nasal mucosa. In the event of microbial dysbiosis or breaches in epithelial integrity, S. aureus can become pathogenic [117,118]. The burden of infection of S. aureus in populations with SLE is significant comprising around 15–35% of all infections, with pneumonia, urinary tract infections, and septicemia as the most common manifestations [32,93,95,119,120]. Analysis of the skin microbiome in lupus patients showed enhanced colonization by S. aureus and other Staphylococcal species (S. epidermidis and S. hominis) [62]. S. aureus can be recovered from ~50% of cutaneous lupus erythematosus lesions, while it was not found in the skin rashes of patients with the autoimmune disease psoriasis [121]. S. aureus colonization of skin lesions was associated with a higher Cutaneous Lupus Disease Area and Severity Index (CLASI) score. This high rate of lupus discoid rash colonization by S. aureus may be the result of the elevated type I interferons in lupus patients. Keratinocytes isolated from nonlesional lupus skin, cultured in vitro and then treated with IFNα significantly downregulated skin barrier genes such as filaggrin and loricrin, while also increasing expression of adhesion factors such as fibronectin, integrin α5, integrin β1 and fibrinogen, resulting in increased S. aureus binding to keratinocytes [121]. Like IFNα, IFNγ also reduces expression of barrier genes in keratinocytes. However, unlike IFNα, IFNγ did not upregulate adhesion factors or increase S. aureus attachment to the cells [121]. Another immune mechanism that may play a role in skin infections by S. aureus is the formation of neutrophil extracellular traps (NETs). In wild-type C57BL/6 mice, skin inflammation caused by tape-stripping results in recruitment of neutrophils and secretion of NETs. The presence of these NETs was shown to enhance the interactions of S. aureus with keratinocytes [122]. This enhancement of skin colonization may be increased in SLE due to anti-NET autoantibodies impairing degradation of the NET structure by DNAse 1 [123].
S. aureus can colonize the nasal cavity in addition to the skin and is found in the nose of about 25–30% of people tested [124]. Carriage of S. aureus intranasally by lupus patients has been shown to correlate with significantly elevated anti-dsDNA, anti-RNP, anti-SSA, and anti-SSB autoantibody titers and increased rates of kidney disease, furthering a link between S. aureus and lupus [125]. In another study of Egyptian patients, nasal carriage of S. aureus was associated with low levels of complement in the bloodstream and with the occurrence of flares of the disease [126].
Murine models of lupus have indicated that infection by S. aureus can influence the autoimmune phenotype. MRL/lpr and NZBWF1 lupus-prone mice have been shown to develop arthritis upon active or accidental infection with S. aureus [127,128]. Staphylococcal enterotoxin B (SEB) is a superantigen produced by some but not all strains of S. aureus. SEB polyclonally activates mouse T cells carrying specific Vβ8 TCRs in an antigen-independent manner [129]. Lupus-like autoimmunity develops in non-lupus-prone mice carrying a transgene for human HLA-DQ8 (which binds with higher affinity to SEB than mouse MHC molecules) were exposed to a constant low-level of purified SEB protein [130]. On the other hand, lupus nephritis is reduced in MRL/lpr mice subjected to one or multiple intravenous injections with SEB [[131], [132], [133]]. However, this effect of SEB in reducing lupus may be due to its ability to trigger anergy of memory T cells resulting from excessive TCR signaling [134,135]. Together, these results implicate S. aureus in the induction and progression of autoimmunity. Failure to effectively clear S. aureus leading to chronic colonization may promote disease progression and trigger flares in patients with SLE.
4.3. Salmonella enterica
Salmonella bacteria are Gram-negative organisms that are common causes of infection in lupus patients [112,[136], [137], [138]]. Salmonella can cause disseminated infections in lupus patients leading to bacteremia, septic arthritis, pneumonia and soft-tissue infections [136,138,139]. In fact, Salmonella is the most common cause of septic arthritis in younger SLE patients [140]. Among all patients with Salmonella bloodstream infections, lupus was the most frequent underlying comorbidity [141].
Lupus-prone NZBWF1 mice are relatively resistant to intravenous S. Typhimurium infection at 4 months of age (prior to overt lupus development), but are susceptible at 8 months of age when lupus is established [142]. Intravenous infection of 2-month-old pre-diseased lupus-prone NZBWF1 mice with a single dose of an attenuated strain of S. Typhimurium strain resulted in reduced proteinuria symptoms over time and reduced anti-DNA autoantibodies after 8.5 months of age [142]. Intravenous infection of older NZBWF1 mice at the onset of lupus disease (6 months) also showed a reduction in proteinuria, but not anti-DNA autoantibodies. These results indicate Salmonella can be protective if given early in the disease course.
In a different study, a virulent strain of S. Typhimurium was injected intraperitoneally every other week for 8 weeks into NZBWF1 mice (6 weeks of age at the beginning of the study) [143]. Under these experimental conditions, Salmonella stimulated production of anti-DNA and anti-chromatin autoantibodies [143]. In this study, expression of the bacterial protein curli, which is the major component of the biofilm produced by Salmonella bacteria, was required for maximal stimulatory effect of Salmonella on autoimmune responses. The curli biofilm can bind and trap bacterial DNA and the DNA/curli complex is highly immunogenic. The different roles of S. Tymphimurium in the two studies [142,143] may be due to differences in age of mice infected (2 month old versus 6 week old), differences in route of infection (intravenous versus intraperitoneal), differences in the frequency of infection (a single dose or repeated doses) or differences in the virulence of the Salmonella strains used (attenuated versus virulent). Given the contrasting findings described above, it is difficult to know whether Salmonella is protective against lupus or stimulates development of disease symptoms.
4.4. Escherichia coli
Escherichia coli is a Gram-negative bacterial organism and normal commensal present in the gut microflora [144]. Approximately, 5–20% of infection-related hospitalizations in SLE patients are the result of E. coli infections, including approximately to 25–40% of bloodstream infections [[91], [92], [93],95,99,119,145]. The role of E. coli in stimulating inflammation or autoimmunity in human lupus has not been studied in detail. However, the presence of anti-heat shock protein autoantibodies in lupus patients is correlated with the presence of antibodies that recognize the E. coli heat shock protein groEL [146,147]. The correlation suggests the possibility that molecular mimicry by E. coli proteins may trigger autoimmune responses.
E. coli extracts given orally were reported to reduce autoimmune symptoms in MRL/lpr mice [148]. This was attributed to their effects on inducing regulatory T cells [146]. Like Salmonella, E. coli produces the amyloid protein curli as a part of its biofilm. When E. coli expressing curli protein were injected intraperitoneally into young pre-diseased lupus-prone NZBWF1 mice, the mice rapidly developed anti-DNA and anti-chromatin autoantibodies, while mice injected with a mutant E. coli strain lacking curli failed to show these responses [143]. Therefore, E. coli infection is capable of promoting autoimmunity and may precipitate the development of SLE in individuals that are genetically predisposed.
4.5. Streptococcus pneumoniae
Streptococcus pneumoniae is a Gram-positive species of bacteria that is known to be the most common causative agent of bacterial pneumonia. S. pneumoniae infection is common in lupus patients [145,149]. SLE patients are more likely to have a pneumococcal infection at a younger age and more likely to have severe and invasive infections with a higher need for intensive care unit (ICU) admission [150]. Lupus patients often have a reduction in serum levels of complement proteins, including complement component C3 and C4. Serum isolated form lupus patients exhibited a significant reduction in C3 deposition on S. pneumoniae cells [151]. This deficiency in complement factor deposition could result in increased susceptibility and impaired clearance of S. pneumoniae.
Despite the frequency and importance of S. pneumoniae infection in lupus patients, there are very limited studies in mice to address potential roles for S. pneumoniae in lupus. Interestingly, non-lupus prone complement C4 knockout mice (C4−/−) infected with S. pneumoniae via the lung developed IgA anti-DNA autoantibodies, suggesting that S. pneumoniae might induce production of lupus autoantibodies. Deletion of the complement component C4 from lupus-prone MRL/lpr mice enhances the autoimmune disease that develops [152]. More work needs to be done to understand the role of S. pneumoniae in mouse models of lupus.
4.6. Mycobacterial species
Mycobacterium tuberculosis and Mycobacterium leprae are the bacterial causative agents of tuberculosis and leprosy [153,154]. Retrospective analyses of hospitalization data have identified an increased rate of infection Mycobacterium tuberculosis in SLE patients, indicating there may be a susceptibility to infection or impairment of clearance [104,[155], [156], [157], [158]]. Lupus-prone MRL/lpr mice have also been shown to be susceptible to infection with M. leprae, while non-obese diabetic (NOD) mice were resistant [113]. Infection by either Mycobacterium tuberculosis or Mycobacterium leprae in humans can mimic autoimmune diseases like lupus and can result in significantly increased titers of autoantibodies [[159], [160], [161]]. Due to this phenotype, mycobacterial infection in SLE patients is often mistaken for a lupus flare, resulting in delay in treatment and uncontrolled infection.
Recent bioinformatics-based analyses have identified a number of M. tuberculosis peptides that share sequence homology with self-peptides, suggesting that T cell responses to these mycobacterial peptides could initiate anti-self responses [162,163]. Two mycobacterial proteins show sequence similarity to human HSP-60 and isoleucyl-tRNA synthase, which can serve as self-antigens in autoimmune disorders such as SLE [164,165]. Supporting a role for mycobacterial proteins in inducing lupus symptoms, injection of HSP65 derived from M. leprae accelerates autoimmunity and increases mortality in NZBWF1 lupus-prone mice [166]. The mice injected with the recombinant mycobacterial protein exhibited increased IFNγ production, elevated anti-dsDNA titers and increased necrosis/apoptosis in immune cells. Therefore, molecular mimicry may be an important aspect to altered immune activation in lupus patients infected with mycobacteria.
4.7. Summary of bacterial infections in lupus
Lupus patients show in increased susceptibility to severe and life-threatening infections. A number of factors likely contribute to this susceptibility including development of lymphopenia, neutropenia and complement deficiencies as well as treatment with immunosuppressive drugs. It is also possible that genetic factors that result in a propensity to developing lupus may skew the immune response in a way that impairs bacterial clearance. Infections that are common in lupus patients are also common in other patient populations and include infections caused by Staphylococcus aureus, Salmonella enterica, Escherichia coli, Streptococcus pneumoniae and mycobacteria. This is not a complete list of all bacteria that cause infections in lupus patients and infections have been reported with less common species such as Nocardia [167]. Mouse models of lupus have been used to investigate the roles of some bacterial species in lupus disease progression. At this time, S. aureus, E. coli and S. enterica are the best studied bacterial organisms in mouse lupus and evidence suggests that all of these species can stimulate development of lupus symptoms (Table 5).
Table 5.
Bacterial species that have been mechanistically studied in mouse lupus.
| Bacterial species | Effects on Lupus |
|---|---|
| Staphylococcus aureus | Subcutaneous infection of MRL/lpr worsens arthritis [127]; accidental introduction of S. aureus into MRL/lpr colony results in worse arthritis [128] |
| S. aureus-derived superantigen SEB | Chronic exposure of HLA-DQ8 transgenic mice to SEB results in lupus development [126]; bi-weekly intravenous injection of SEB into MRL/lpr mice reduces disease [131]; single injection of SEB into young pre-diseased MRL/lpr mice prevents disease [132]; intraperitoneal injection of SEB into MRL/lpr mice that were neonatally tolerized to this superantigen results in dramatically worse lupus disease symptoms [133] |
| Salmonella enterica | Intravenous injection of attenuated Salmonella into 2-month pre-diseased NZBWF1 mice results in reduced proteinuria and reduced anti-DNA autoantibody [142]; four intraperitoneal infections of virulent Salmonella into young pre-diseased NZBWF1 mice results in increased autoantibody titers [143] |
| Escherichia coli | Four intraperitoneal infections of E. coli into young pre-diseased NZBWF1 mice results in increased autoantibody titers [143] |
| Streptococcus pneumoniae | No direct studies on S. pneumoniae in lupus-prone strains of mice |
| Mycobacterium leprae | MRL/lpr lupus-prone mice were shown to be more susceptible to M. leprae footpad infection than non-obese diabetic (NOD) mice [113], but the effects of the infection on lupus symptoms were not tested. Injection of Hsp65 protein derived from M. leprae accelerates autoimmunity and increases mortality in NZBWF1 lupus-prone mice [166] |
5. Conclusions
In this review, we’ve summarized published studies that cover aspects of microbial colonization and infection in systemic lupus erythematosus including aspects of the bacterial microbiome and bacterial species that are prevalent in causing infections in lupus patients. Various studies have shown that the gut microbiome is different in lupus patients versus control subjects. Similarly, the oral and skin microbiome also appear different in lupus, suggesting a generalized dysbiosis. Many of the infections that are common in lupus patients, such as infections caused by S. aureus, S. enterica, E. coli and S. pneumoniae, are also common in the general population. But lupus patients are more susceptible to severe and invasive infections with these organisms. Evidence from mouse models suggests that bacterial infection with a number of different bacterial species can trigger increased immune activation and can promote autoimmune progression. The high rate of bacterial infections in lupus patients is likely caused both by inherent deficiencies in the immune response as well as immunosuppressive therapies. Therefore, there may be a feedback loop in which SLE patients become infected with bacteria, due to immunodeficiencies caused by immunological abnormalities or immunosuppressive therapies, and the bacterial infection stimulates further immune activation thereby worsening the autoimmune symptoms. Further studies will be needed to more fully understand the alterations in bacterial colonization and infection in lupus patients and how they contribute to disease progression.
Author contributions
Michael Battaglia, Conceptualization, Writing – original draft. Lee Ann Garrett-Sinha, Conceptualization, Funding acquisition, Project administration, Writing – review & editing.
Declarations of competing interest
None.
Acknowledgements
This work was supported by grants from the National Institutes of Health (AI085127) and the Lupus Research Alliance.
References
- 1.Long H., Yin H., Wang L., Gershwin M.E., Lu Q. The critical role of epigenetics in systemic lupus erythematosus and autoimmunity. J. Autoimmun. 2016;74:118–138. doi: 10.1016/j.jaut.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 2.Legorreta-Haquet M.V., Chavez-Rueda K., Chavez-Sanchez L., Cervera-Castillo H., Zenteno-Galindo E., Barile-Fabris L. Function of treg cells decreased in patients with systemic lupus erythematosus due to the effect of prolactin. Medicine (Baltim.) 2016;95 doi: 10.1097/MD.0000000000002384. e2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleczynska W., Jakiela B., Plutecka H., Milewski M., Sanak M., Musial J. Imbalance between Th17 and regulatory T-cells in systemic lupus erythematosus. Folia Histochem. Cytobiol. 2011;49:646–653. doi: 10.5603/fhc.2011.0088. [DOI] [PubMed] [Google Scholar]
- 4.Streicher K., Morehouse C.A., Groves C.J., Rajan B., Pilataxi F., Lehmann K.P. The plasma cell signature in autoimmune disease. Arthritis Rheum. 2014;66:173–184. doi: 10.1002/art.38194. [DOI] [PubMed] [Google Scholar]
- 5.Shah K., Lee W.W., Lee S.H., Kim S.H., Kang S.W., Craft J. Dysregulated balance of Th17 and Th1 cells in systemic lupus erythematosus. Arthritis Res. Ther. 2010;12 doi: 10.1186/ar2964. R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S.J., Lee K., Diamond B. Follicular helper T cells in systemic lupus erythematosus. Front. Immunol. 2018;9:1793. doi: 10.3389/fimmu.2018.01793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q.Z., Zhou J., Wandstrat A.E., Carr-Johnson F., Branch V., Karp D.R. Protein array autoantibody profiles for insights into systemic lupus erythematosus and incomplete lupus syndromes. Clin. Exp. Immunol. 2007;147:60–70. doi: 10.1111/j.1365-2249.2006.03251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witte T., Hartung K., Sachse C., Matthias T., Fricke M., Kalden J.R. Rheumatoid factors in systemic lupus erythematosus: association with clinical and laboratory parameters. SLE study group. Rheumatol. Int. 2000;19:107–111. doi: 10.1007/s002960050112. [DOI] [PubMed] [Google Scholar]
- 9.Han S., Zhuang H., Shumyak S., Yang L., Reeves W.H. Mechanisms of autoantibody production in systemic lupus erythematosus. Front. Immunol. 2015;6:228. doi: 10.3389/fimmu.2015.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truedsson L., Bengtsson A.A., Sturfelt G. Complement deficiencies and systemic lupus erythematosus. Autoimmunity. 2007;40:560–566. doi: 10.1080/08916930701510673. [DOI] [PubMed] [Google Scholar]
- 11.Grondal G., Gunnarsson I., Ronnelid J., Rogberg S., Klareskog L., Lundberg I. Cytokine production, serum levels and disease activity in systemic lupus erythematosus. Clin. Exp. Rheumatol. 2000;18:565–570. [PubMed] [Google Scholar]
- 12.Lourenco E.V., La Cava A. Cytokines in systemic lupus erythematosus. Curr. Mol. Med. 2009;9:242–254. doi: 10.2174/156652409787847263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howe H.S., Leung B.P.L. Anti-cytokine autoantibodies in systemic lupus erythematosus. Cells. 2019:9. doi: 10.3390/cells9010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stojan G., Petri M. Epidemiology of systemic lupus erythematosus: an update. Curr. Opin. Rheumatol. 2018;30:144–150. doi: 10.1097/BOR.0000000000000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anders H.J., Saxena R., Zhao M.H., Parodis I., Salmon J.E., Mohan C. Lupus nephritis. Nat Rev Dis Primers. 2020;6:7. doi: 10.1038/s41572-019-0141-9. [DOI] [PubMed] [Google Scholar]
- 16.Mok C.C., Tse S.M., Chan K.L., Ho L.Y. Effect of immunosuppressive therapies on survival of systemic lupus erythematosus: a propensity score analysis of a longitudinal cohort. Lupus. 2018;27:722–727. doi: 10.1177/0961203317739129. [DOI] [PubMed] [Google Scholar]
- 17.Bharath G., Kumar P., Makkar N., Singla P., Soneja M., Biswas A. Mortality in systemic lupus erythematosus at a teaching hospital in India: a 5-year retrospective study. J. Fam. Med. Prim. Care. 2019;8:2511–2515. doi: 10.4103/jfmpc.jfmpc_362_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Fang X., Li Q.Z. Biomarker profiling for lupus nephritis. Dev. Reprod. Biol. 2013;11:158–165. doi: 10.1016/j.gpb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu C.C., Huang C.C., Chan W.L., Chung C.M., Huang P.H., Lin S.J. Increased risk of ischemic stroke in patients with systemic lupus erythematosus: a nationwide population-based study. Intern. Med. 2012;51:17–21. doi: 10.2169/internalmedicine.51.6154. [DOI] [PubMed] [Google Scholar]
- 20.Tsukamoto E., Tanei T., Senda J., Kato T., Naito T., Ishii K. Subarachnoid hemorrhage after ischemic stroke associated with systemic lupus erythematosus and antiphospholipid syndrome. World Neurosurg. 2020;136:248–252. doi: 10.1016/j.wneu.2020.01.030. [DOI] [PubMed] [Google Scholar]
- 21.Mimori A., Suzuki T., Hashimoto M., Nara H., Yoshio T., Masuyama J.I. Subarachnoid hemorrhage and systemic lupus erythematosus. Lupus. 2000;9:521–526. doi: 10.1177/096120330000900708. [DOI] [PubMed] [Google Scholar]
- 22.J F. Baizabal Carvallo, C. Cantu Brito, B. Estanol, G. S. Garcia Ramos. Subarachnoid hemorrhage as a complication of systemic lupus erythematosus. Cerebrovasc. Dis. 2007;24:301–304. doi: 10.1159/000105684. [DOI] [PubMed] [Google Scholar]
- 23.Duarte-Delgado N.P., Vasquez G., Ortiz-Reyes B.L. Blood-brain barrier disruption and neuroinflammation as pathophysiological mechanisms of the diffuse manifestations of neuropsychiatric systemic lupus erythematosus. Autoimmun. Rev. 2019;18:426–432. doi: 10.1016/j.autrev.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Tselios K., Urowitz Cardiovascular M.B., Manifestations Pulmonary. Of systemic lupus erythematosus. Curr. Rheumatol. Rev. 2017;13:206–218. doi: 10.2174/1573397113666170704102444. [DOI] [PubMed] [Google Scholar]
- 25.Munguia-Realpozo P., Mendoza-Pinto C., Sierra Benito C., Escarcega R.O., Garcia-Carrasco M., Mendez Martinez S. Systemic lupus erythematosus and hypertension. Autoimmun. Rev. 2019;18:102371. doi: 10.1016/j.autrev.2019.102371. [DOI] [PubMed] [Google Scholar]
- 26.Cervera R., Khamashta M.A., Font J., Sebastiani G.D., Gil A., Lavilla P. Morbidity and mortality in systemic lupus erythematosus during a 5-year period. A multicenter prospective study of 1,000 patients. European Working Party on Systemic Lupus Erythematosus. Medicine (Baltim.) 1999;78:167–175. doi: 10.1097/00005792-199905000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Kim W.U., Min J.K., Lee S.H., Park S.H., Cho C.S., Kim H.Y. Causes of death in Korean patients with systemic lupus erythematosus: a single center retrospective study. Clin. Exp. Rheumatol. 1999;17:539–545. [PubMed] [Google Scholar]
- 28.Kang K.Y., Kwok S.K., Ju J.H., Park K.S., Cho C.S., Kim H.Y. The causes of death in Korean patients with systemic lupus erythematosus over 11 years. Lupus. 2011;20:989–997. doi: 10.1177/0961203311402245. [DOI] [PubMed] [Google Scholar]
- 29.Jacobsen S., Petersen J., Ullman S., Junker P., Voss A., Rasmussen J.M. Mortality and causes of death of 513 Danish patients with systemic lupus erythematosus. Scand. J. Rheumatol. 1999;28:75–80. doi: 10.1080/030097499442522. [DOI] [PubMed] [Google Scholar]
- 30.Adwan M.H., Qasem U., Mustafa K.N. In-hospital mortality in patients with systemic lupus erythematosus: a study from Jordan. Rheumatol. Int. 2020:2002–2017. doi: 10.1007/s00296-020-04538-z. [DOI] [PubMed] [Google Scholar]
- 31.Fei Y., Shi X., Gan F., Li X., Zhang W., Li M. Death causes and pathogens analysis of systemic lupus erythematosus during the past 26 years. Clin. Rheumatol. 2014;33:57–63. doi: 10.1007/s10067-013-2383-3. [DOI] [PubMed] [Google Scholar]
- 32.Tsai P.H., Jang S.S., Liou L.B. Septicaemia is associated with increased disease activity and mortality in systemic lupus erythematosus: a retrospective analysis from Taiwan. Lupus. 2020;29:191–198. doi: 10.1177/0961203319899162. [DOI] [PubMed] [Google Scholar]
- 33.Liang H., Pan H.F., Tao J.H., Ye D.Q. Causes and factors associated with frequent hospitalization in Chinese patients with systemic lupus erythematosus: an ambispective cohort study. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2019;25:8061–8068. doi: 10.12659/MSM.919381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anastasiou C., Dulai O., Baskaran A., Proudfoot J., Verhaegen S., Kalunian K. Immunosuppressant use and hospitalisations in adult patients with systemic lupus erythematosus admitted to a tertiary academic medical centre. Lupus Sci Med. 2018;5 doi: 10.1136/lupus-2017-000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taneja V. Arthritis susceptibility and the gut microbiome. FEBS Lett. 2014;588:4244–4249. doi: 10.1016/j.febslet.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdelhamid L., Luo X.M. Retinoic acid, leaky gut, and autoimmune diseases. Nutrients. 2018:10. doi: 10.3390/nu10081016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters A., Wekerle H. Autoimmune diabetes mellitus and the leaky gut. Proc. Natl. Acad. Sci. U. S. A. 2019;116:14788–14790. doi: 10.1073/pnas.1909224116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L., Zhang Z., Yu A.M., Wang W., Wei Z., Akhter E. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PloS One. 2014;9 doi: 10.1371/journal.pone.0093846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Issara-Amphorn J., Surawut S., Worasilchai N., Thim-Uam A., Finkelman M., Chindamporn A. The synergy of endotoxin and (1-->3)-beta-D-glucan, from gut translocation, worsens sepsis severity in a lupus model of fc gamma receptor IIb-deficient mice. J Innate Immun. 2018;10:189–201. doi: 10.1159/000486321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manfredo Vieira S., Hiltensperger M., Kumar V., Zegarra-Ruiz D., Dehner C., Khan N. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359:1156–1161. doi: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mu Q., Zhang H., Liao X., Lin K., Liu H., Edwards M.R. Control of lupus nephritis by changes of gut microbiota. Microbiome. 2017;5:73. doi: 10.1186/s40168-017-0300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toral M., Robles-Vera I., Romero M., de la Visitacion N., Sanchez M., O’Valle F. Lactobacillus fermentum CECT5716: a novel alternative for the prevention of vascular disorders in a mouse model of systemic lupus erythematosus. Faseb. J. 2019;33:10005–10018. doi: 10.1096/fj.201900545RR. [DOI] [PubMed] [Google Scholar]
- 43.Zegarra-Ruiz D.F., El Beidaq A., Iniguez A.J., Lubrano Di Ricco M., Manfredo Vieira S., Ruff W.E. A diet-sensitive commensal Lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe. 2019;25:113–127. doi: 10.1016/j.chom.2018.11.009. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thim-Uam A., Surawut S., Issara-Amphorn J., Jaroonwitchawan T., Hiengrach P., Chatthanathon P. Leaky-gut enhanced lupus progression in the Fc gamma receptor-IIb deficient and pristane-induced mouse models of lupus. Sci. Rep. 2020;10:777. doi: 10.1038/s41598-019-57275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amoroso C., Perillo F., Strati F., Fantini M.C., Caprioli F., Facciotti F. The role of gut microbiota biomodulators on mucosal immunity and intestinal inflammation. Cells. 2020:9. doi: 10.3390/cells9051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haase S., Haghikia A., Wilck N., Muller D.N., Linker R.A. Impacts of microbiome metabolites on immune regulation and autoimmunity. Immunology. 2018;154:230–238. doi: 10.1111/imm.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee N., Kim W.U. Microbiota in T-cell homeostasis and inflammatory diseases. Exp. Mol. Med. 2017;49 doi: 10.1038/emm.2017.36. e340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H., Liao X., Sparks J.B., Luo X.M. Dynamics of gut microbiota in autoimmune lupus. Appl. Environ. Microbiol. 2014;80:7551–7560. doi: 10.1128/AEM.02676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo X.M., Edwards M.R., Mu Q., Yu Y., Vieson M.D., Reilly C.M. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl. Environ. Microbiol. 2018:84. doi: 10.1128/AEM.02288-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hugenholtz F., de Vos W.M. Mouse models for human intestinal microbiota research: a critical evaluation. Cell. Mol. Life Sci. 2018;75:149–160. doi: 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan I., Ullah N., Zha L., Bai Y., Khan A., Zhao T. Alteration of gut microbiota in inflammatory bowel disease (IBD): cause or consequence? IBD treatment targeting the gut microbiome. Pathogens. 2019:8. doi: 10.3390/pathogens8030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen Z.H., Zhu C.X., Quan Y.S., Yang Z.Y., Wu S., Luo W.W. Relationship between intestinal microbiota and ulcerative colitis: mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J. Gastroenterol. 2018;24:5–14. doi: 10.3748/wjg.v24.i1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez P., de Paz B., Rodriguez-Carrio J., Hevia A., Sanchez B., Margolles A. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci. Rep. 2016;6:24072. doi: 10.1038/srep24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tremaroli V., Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 56.Hevia A., Milani C., Lopez P., Cuervo A., Arboleya S., Duranti S. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio. 2014;5:14. doi: 10.1128/mBio.01548-14. e01548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He Z., Shao T., Li H., Xie Z., Wen C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog. 2016;8:64. doi: 10.1186/s13099-016-0146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Meulen T.A., Harmsen H.J.M., Vila A.V., Kurilshikov A., Liefers S.C., Zhernakova A. Shared gut, but distinct oral microbiota composition in primary Sjogren’s syndrome and systemic lupus erythematosus. J. Autoimmun. 2019;97:77–87. doi: 10.1016/j.jaut.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Wang H.F., Li X., Li H.X., Zhang Q., Zhou H.W. Disordered intestinal microbes are associated with the activity of Systemic Lupus Erythematosus. Clin. Sci. (Lond.) 2019;133:821–838. doi: 10.1042/CS20180841. [DOI] [PubMed] [Google Scholar]
- 60.Wei F., Xu H., Yan C., Rong C., Liu B., Zhou H. Changes of intestinal flora in patients with systemic lupus erythematosus in northeast China. PloS One. 2019;14 doi: 10.1371/journal.pone.0213063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li B.Z., Zhou H.Y., Guo B., Chen W.J., Tao J.H., Cao N.W. Dysbiosis of oral microbiota is associated with systemic lupus erythematosus. Arch. Oral Biol. 2020;113:104708. doi: 10.1016/j.archoralbio.2020.104708. [DOI] [PubMed] [Google Scholar]
- 62.Huang C., Yi X., Long H., Zhang G., Wu H., Zhao M. Disordered cutaneous microbiota in systemic lupus erythematosus. J. Autoimmun. 2020;108:102391. doi: 10.1016/j.jaut.2019.102391. [DOI] [PubMed] [Google Scholar]
- 63.O A., Azzouz D., Heguy A., Schwudke D., Gisch N. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 2019:947–956. doi: 10.1136/annrheumdis-2018-214856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riddle M.S., Guerry P. Status of vaccine research and development for Campylobacter jejuni. Vaccine. 2016;34:2903–2906. doi: 10.1016/j.vaccine.2016.02.080. [DOI] [PubMed] [Google Scholar]
- 65.Jiang L., Wang Z., Zhu H.W., Di H.Y., Li H., Zhang Y.Y. Beneficial effect of Eucommia polysaccharides on systemic lupus erythematosus-like syndrome induced by Campylobacter jejuni in BALB/c mice. Inflammation. 2011;34:402–411. doi: 10.1007/s10753-010-9247-7. [DOI] [PubMed] [Google Scholar]
- 66.Henke M.T., Kenny D.J., Cassilly C.D., Vlamakis H., Xavier R.J., Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. U. S. A. 2019;116:12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joossens M., Huys G., Cnockaert M., De Preter V., Verbeke K., Rutgeerts P. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 68.Hall A.B., Yassour M., Sauk J., Garner A., Jiang X., Arthur T. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9:103. doi: 10.1186/s13073-017-0490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez P., Sanchez B., Margolles A., Suarez A. Intestinal dysbiosis in systemic lupus erythematosus: cause or consequence? Curr. Opin. Rheumatol. 2016;28:515–522. doi: 10.1097/BOR.0000000000000309. [DOI] [PubMed] [Google Scholar]
- 70.Mu Q., Tavella V.J., Kirby J.L., Cecere T.E., Chung M., Lee J. Antibiotics ameliorate lupus-like symptoms in mice. Sci. Rep. 2017;7:13675. doi: 10.1038/s41598-017-14223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maldonado M.A., Kakkanaiah V., MacDonald G.C., Chen F., Reap E.A., Balish E. The role of environmental antigens in the spontaneous development of autoimmunity in MRL-lpr mice. J. Immunol. 1999;162:6322–6330. [PubMed] [Google Scholar]
- 72.Pandiyan P., Bhaskaran N., Zou M., Schneider E., Jayaraman S., Huehn J. Microbiome dependent regulation of Tregs and Th17 cells in mucosa. Front. Immunol. 2019;10:426. doi: 10.3389/fimmu.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson B.M., Gaudreau M.C., Al-Gadban M.M., Gudi R., Vasu C. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clin. Exp. Immunol. 2015;181:323–337. doi: 10.1111/cei.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsu T.C., Huang C.Y., Liu C.H., Hsu K.C., Chen Y.H., Tzang B.S. Lactobacillus paracasei GMNL-32, Lactobacillus reuteri GMNL-89 and L. reuteri GMNL-263 ameliorate hepatic injuries in lupus-prone mice. Br. J. Nutr. 2017;117:1066–1074. doi: 10.1017/S0007114517001039. [DOI] [PubMed] [Google Scholar]
- 75.Tzang B.S., Liu C.H., Hsu K.C., Chen Y.H., Huang C.Y., Hsu T.C. Effects of oral Lactobacillus administration on antioxidant activities and CD4+CD25+forkhead box P3 (FoxP3)+ T cells in NZB/W F1 mice. Br. J. Nutr. 2017;118:333–342. doi: 10.1017/S0007114517002112. [DOI] [PubMed] [Google Scholar]
- 76.Hu W.S., Rajendran P., Tzang B.S., Yeh Y.L., Shen C.Y., Chen R.J. Lactobacillus paracasei GMNL-32 exerts a therapeutic effect on cardiac abnormalities in NZB/W F1 mice. PloS One. 2017;12 doi: 10.1371/journal.pone.0185098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khorasani S., Mahmoudi M., Kalantari M.R., Lavi Arab F., Esmaeili S.A., Mardani F. Amelioration of regulatory T cells by Lactobacillus delbrueckii and Lactobacillus rhamnosus in pristane-induced lupus mice model. J. Cell. Physiol. 2019;234:9778–9786. doi: 10.1002/jcp.27663. [DOI] [PubMed] [Google Scholar]
- 78.Mardani F., Mahmoudi M., Esmaeili S.A., Khorasani S., Tabasi N., Rastin M. In vivo study: Th1-Th17 reduction in pristane-induced systemic lupus erythematosus mice after treatment with tolerogenic Lactobacillus probiotics. J. Cell. Physiol. 2018;234:642–649. doi: 10.1002/jcp.26819. [DOI] [PubMed] [Google Scholar]
- 79.Mu Q., Cabana-Puig X., Mao J., Swartwout B., Abdelhamid L., Cecere T.E. Pregnancy and lactation interfere with the response of autoimmunity to modulation of gut microbiota. Microbiome. 2019;7:105. doi: 10.1186/s40168-019-0720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sczesnak A., Segata N., Qin X., Gevers D., Petrosino J.F., Huttenhower C. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ivanov K., II, Atarashi N., Manel E.L., Brodie T., Shima U., Karaoz Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koga T., Ichinose K., Kawakami A., Tsokos G.C. The role of IL-17 in systemic lupus erythematosus and its potential as a therapeutic target. Expet Rev. Clin. Immunol. 2019;15:629–637. doi: 10.1080/1744666X.2019.1593141. [DOI] [PubMed] [Google Scholar]
- 83.Gaboriau-Routhiau V., Rakotobe S., Lecuyer E., Mulder I., Lan A., Bridonneau C. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 84.Yin Y., Wang Y., Zhu L., Liu W., Liao N., Jiang M. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 2013;7:615–621. doi: 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan T.G., Sefik E., Geva-Zatorsky N., Kua L., Naskar D., Teng F. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E8141–E8150. doi: 10.1073/pnas.1617460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Praet J.T., Donovan E., Vanassche I., Drennan M.B., Windels F., Dendooven A. Commensal microbiota influence systemic autoimmune responses. EMBO J. 2015;34:466–474. doi: 10.15252/embj.201489966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tektonidou M.G., Wang Z., Dasgupta A., Ward M.M. Burden of serious infections in adults with systemic lupus erythematosus: a national population-based study, 1996-2011. Arthritis Care Res. 2015;67:1078–1085. doi: 10.1002/acr.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yen E.Y., Shaheen M., Woo J.M.P., Mercer N., Li N., McCurdy D.K. 46-Year trends in systemic lupus erythematosus mortality in the United States, 1968 to 2013: a nationwide population-based study. Ann. Intern. Med. 2017;167:777–785. doi: 10.7326/M17-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gomez-Puerta J.A., Barbhaiya M., Guan H., Feldman C.H., Alarcon G.S., Costenbader K.H. Racial/Ethnic variation in all-cause mortality among United States medicaid recipients with systemic lupus erythematosus: a Hispanic and asian paradox. Arthritis Rheum. 2015;67:752–760. doi: 10.1002/art.38981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gladman D.D., Hussain F., Ibanez D., Urowitz M.B. The nature and outcome of infection in systemic lupus erythematosus. Lupus. 2002;11:234–239. doi: 10.1191/0961203302lu170oa. [DOI] [PubMed] [Google Scholar]
- 91.Marcos M., Fernandez C., Soriano A., Marco F., Martinez J.A., Almela M. Epidemiology and clinical outcomes of bloodstream infections among lupus patients. Lupus. 2011;20:965–971. doi: 10.1177/0961203311403345. [DOI] [PubMed] [Google Scholar]
- 92.Rua-Figueroa I., Lopez-Longo F.J., Del Campo V., Galindo-Izquierdo M., Uriarte E., Torre-Cisneros J. Bacteremia in systemic lupus erythematosus in patients from a Spanish registry: risk factors, clinical and microbiological characteristics, and outcomes. J. Rheumatol. 2020;47:234–240. doi: 10.3899/jrheum.180882. [DOI] [PubMed] [Google Scholar]
- 93.Dubula T., Mody G.M. Spectrum of infections and outcome among hospitalized South Africans with systemic lupus erythematosus. Clin. Rheumatol. 2015;34:479–488. doi: 10.1007/s10067-014-2847-0. [DOI] [PubMed] [Google Scholar]
- 94.Martinez-Martinez M.U., Sturbaum A.K., Alcocer-Varela J., Merayo-Chalico J., Gomez-Martin D., Gomez-Banuelos Jde J. Factors associated with mortality and infections in patients with systemic lupus erythematosus with diffuse alveolar hemorrhage. J. Rheumatol. 2014;41:1656–1661. doi: 10.3899/jrheum.130927. [DOI] [PubMed] [Google Scholar]
- 95.Ruiz-Irastorza G., Olivares N., Ruiz-Arruza I., Martinez-Berriotxoa A., Egurbide M.V., Aguirre C. Predictors of major infections in systemic lupus erythematosus. Arthritis Res. Ther. 2009;11 doi: 10.1186/ar2764. R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oud L. Epidemiology and outcomes of sepsis among hospitalizations with systemic lupus erythematosus admitted to the ICU: a population-based cohort study. J Intensive Care. 2020;8:3. doi: 10.1186/s40560-019-0424-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hsu C.-Y., Ko C.-H., Wang J.-L., Hsu T.-C., Lin C.-Y. Comparing the burdens of opportunistic infections among patients with systemic rheumatic diseases: a nationally representative cohort study. Arthritis Res. Ther. 2019;21:211. doi: 10.1186/s13075-019-1997-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mok C.C. Understanding lupus nephritis: diagnosis, management, and treatment options. Int J Womens Health. 2012;4:213–222. doi: 10.2147/IJWH.S28034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu Y., Shen H., Zhu C., Guo R., Gao Y., Lu L. Infections caused by extended-spectrum beta-lactamase producing Escherichia coli in systemic lupus erythematosus patients: prevalence, risk factors, and predictive model. BioMed Res. Int. 2018;2018:8296720. doi: 10.1155/2018/8296720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goldblatt F., Chambers S., Rahman A., Isenberg D.A. Serious infections in British patients with systemic lupus erythematosus: hospitalisations and mortality. Lupus. 2009;18:682–689. doi: 10.1177/0961203308101019. [DOI] [PubMed] [Google Scholar]
- 101.Horta-Baas G., Guerrero-Soto O., Barile-Fabris L. Central nervous system infection by Listeria monocytogenes in patients with systemic lupus erythematosus: analysis of 26 cases, including the report of a new case. Reumatol. Clínica. 2013;9:340–347. doi: 10.1016/j.reuma.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 102.Tobon G.J., Serna M.J., Canas C.A. Listeria monocytogenes infection in patients with systemic lupus erythematosus. Clin. Rheumatol. 2013;(Suppl 1):S25–S27. doi: 10.1007/s10067-010-1416-4. 32. [DOI] [PubMed] [Google Scholar]
- 103.Zhang L., Wang D.X., Ma L. [A clinical study of tuberculosis infection in systemic lupus erythematosus] Zhonghua Nei Ke Za Zhi. 2008;47:808–810. [PubMed] [Google Scholar]
- 104.Sayarlioglu M., Inanc M., Kamali S., Cefle A., Karaman O., Gul A. Tuberculosis in Turkish patients with systemic lupus erythematosus: increased frequency of extrapulmonary localization. Lupus. 2004;13:274–278. doi: 10.1191/0961203303lu529xx. [DOI] [PubMed] [Google Scholar]
- 105.Mitander A., Fei Y., Trysberg E., Mohammad M., Hu Z., Sakiniene E. Complement consumption in systemic lupus erythematosus leads to decreased opsonophagocytosis in. Vitro. J Rheumatol. 2018;45:1557–1564. doi: 10.3899/jrheum.171325. [DOI] [PubMed] [Google Scholar]
- 106.Jung J.Y., Yoon D., Choi Y., Kim H.A., Suh C.H. Associated clinical factors for serious infections in patients with systemic lupus erythematosus. Sci. Rep. 2019;9:9704. doi: 10.1038/s41598-019-46039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu P., Tan H.Z., Li H., Lim C.C., Choo J.C.J. Infections in hospitalized lupus nephritis patients: characteristics, risk factors, and outcomes. Lupus. 2018;27:1150–1158. doi: 10.1177/0961203318768881. [DOI] [PubMed] [Google Scholar]
- 108.Lim C.C., Liu P.Y., Tan H.Z., Lee P., Chin Y.M., Mok I.Y. Severe infections in patients with lupus nephritis treated with immunosuppressants: A retrospective cohort study. Nephrology. 2017;22:478–484. doi: 10.1111/nep.12809. [DOI] [PubMed] [Google Scholar]
- 112.Gencer S., Balkan Y.Y., Benzonana N., Ozer S. Undiagnosed systemic lupus erythematosus presenting with salmonella bacteremia: a case report and mini-review. Clin. Microbiol. Infect. 2003;9:572–573. doi: 10.1046/j.1469-0691.2003.00604.x. [DOI] [PubMed] [Google Scholar]
- 113.Yogi Y., Nakamura K., Suzuki A. The experimental inoculation with Mycobacterium leprae in autoimmune mice: results of MRL/lpr mice inoculated into the right hind foot (continued) Nippon. Rai Gakkai Zasshi. 1989;58:235–240. doi: 10.5025/hansen1977.58.235. [DOI] [PubMed] [Google Scholar]
- 114.Li W., Chen W., Huang S., Tang X., Yao G., Sun L. Infection with opportunistic bacteria triggers severe pulmonary inflammation in lupus-prone mice. Mediat. Inflamm. 2019;2019:1701367. doi: 10.1155/2019/1701367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mehrad B., Park S.J., Akangire G., Standiford T.J., Wu T., Zhu J. The lupus-susceptibility locus, Sle3, mediates enhanced resistance to bacterial infections. J. Immunol. 2006;176:3233–3239. doi: 10.4049/jimmunol.176.5.3233. [DOI] [PubMed] [Google Scholar]
- 116.Waisberg M., Tarasenko T., Vickers B.K., Scott B.L., Willcocks L.C., Molina-Cruz A. Genetic susceptibility to systemic lupus erythematosus protects against cerebral malaria in mice. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1122–1127. doi: 10.1073/pnas.1017996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Otto M. Staphylococci in the human microbiome: the role of host and interbacterial interactions. Curr. Opin. Microbiol. 2020;53:71–77. doi: 10.1016/j.mib.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 118.Oogai Y., Matsuo M., Hashimoto M., Kato F., Sugai M., Komatsuzawa H. Expression of virulence factors by Staphylococcus aureus grown in serum. Appl. Environ. Microbiol. 2011;77:8097–8105. doi: 10.1128/AEM.05316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Barrera-Vargas A., Gomez-Martin D., Merayo-Chalico J., Ponce-de-Leon A., Alcocer-Varela J. Risk factors for drug-resistant bloodstream infections in patients with systemic lupus erythematosus. J. Rheumatol. 2014;41:1311–1316. doi: 10.3899/jrheum.131261. [DOI] [PubMed] [Google Scholar]
- 120.Al-Rayes H., Al-Swailem R., Arfin M., Sobki S., Rizvi S., Tariq M. Systemic lupus erythematosus and infections: a retrospective study in Saudis. Lupus. 2007;16:755–763. doi: 10.1177/0961203307079943. [DOI] [PubMed] [Google Scholar]
- 121.Sirobhushanam S., Parsa N., Reed T.J., Berthier C.C., Sarkar M.K., Hile G.A. Staphylococcus aureus colonization is increased on lupus skin lesions and is promoted by IFN-mediated barrier disruption. J. Invest. Dermatol. 2019 doi: 10.1016/j.jid.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bitschar K., Staudenmaier L., Klink L., Focken J., Sauer B., Fehrenbacher B. Staphylococcus aureus skin colonization is enhanced by the interaction of neutrophil extracellular traps with keratinocytes. J. Invest. Dermatol. 2020;140:1054–1065. doi: 10.1016/j.jid.2019.10.017. e4. [DOI] [PubMed] [Google Scholar]
- 123.Hakkim A., Furnrohr B.G., Amann K., Laube B., Abed U.A., Brinkmann V. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mehraj J., Witte W., Akmatov M.K., Layer F., Werner G., Krause G. Epidemiology of Staphylococcus aureus nasal carriage patterns in the community. Curr. Top. Microbiol. Immunol. 2016;398:55–87. doi: 10.1007/82_2016_497. [DOI] [PubMed] [Google Scholar]
- 125.Conti F., Ceccarelli F., Iaiani G., Perricone C., Giordano A., Amori L. Association between Staphylococcus aureus nasal carriage and disease phenotype in patients affected by systemic lupus erythematosus. Arthritis Res. Ther. 2016;18:177. doi: 10.1186/s13075-016-1079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hajialilo M., Ghorbanihaghjo A., Khabbazi A., Valizadeh H., Raeisi S., Hasani A. Nasal carriage rate of Staphylococcus aureus among patients with systemic lupus erythematosus and its correlation with disease relapse. Egypt. Rheumatol. 2015;37:81–84. [Google Scholar]
- 127.Salinas-Carmona M.C., de la Cruz-Galicia G., Perez-Rivera I., Solis-Soto J.M., Segoviano-Ramirez J.C., Vazquez A.V. Spontaneous arthritis in MRL/lpr mice is aggravated by Staphylococcus aureus and ameliorated by Nippostrongylus brasiliensis infections. Autoimmunity. 2009;42:25–32. doi: 10.1080/08916930802228290. [DOI] [PubMed] [Google Scholar]
- 128.Bremell T., Lange S., Svensson L., Jennische E., Grondahl K., Carlsten H. Outbreak of spontaneous staphylococcal arthritis and osteitis in mice. Arthritis Rheum. 1990;33:1739–1744. doi: 10.1002/art.1780331120. [DOI] [PubMed] [Google Scholar]
- 129.Jenkinson E.J., Kingston R., Smith C.A., Williams G.T., Owen J.J. Antigen-induced apoptosis in developing T cells: a mechanism for negative selection of the T cell receptor repertoire. Eur. J. Immunol. 1989;19:2175–2177. doi: 10.1002/eji.1830191132. [DOI] [PubMed] [Google Scholar]
- 130.Chowdhary V.R., Tilahun A.Y., Clark C.R., Grande J.P., Rajagopalan G. Chronic exposure to staphylococcal superantigen elicits a systemic inflammatory disease mimicking lupus. J. Immunol. 2012;189:2054–2062. doi: 10.4049/jimmunol.1201097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim C., Siminovitch K.A., Ochi A. Reduction of lupus nephritis in MRL/lpr mice by a bacterial superantigen treatment. J. Exp. Med. 1991;174:1431–1437. doi: 10.1084/jem.174.6.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gonzalo J.A., Tarazona R., Schuurman H.J., Uytdehaag F., Wick G., Martinez C. A single injection of Staphylococcus aureus enterotoxin B reduces autoimmunity in MRL/lpr mice. Clin. Immunol. Immunopathol. 1994;71:176–182. doi: 10.1006/clin.1994.1069. [DOI] [PubMed] [Google Scholar]
- 133.Zhou T., Bluethmann H., Zhang J., Edwards C.K., Mountz J.D. Defective maintenance of T cell tolerance to a superantigen in MRL-lpr/lpr mice. J. Exp. Med. 1992;176:1063–1072. doi: 10.1084/jem.176.4.1063. 3rd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Janik D.K., Lee W.T. Staphylococcal enterotoxin B (SEB) induces memory CD4 T cell anergy in vivo and impairs recall immunity to unrelated antigens. J. Clin. Cell. Immunol. 2015;6:1–8. doi: 10.4172/2155-9899.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Watson A.R., Mittler J.N., Lee W.T. Staphylococcal enterotoxin B induces anergy to conventional peptide in memory T cells. Cell. Immunol. 2003;222:144–155. doi: 10.1016/s0008-8749(03)00117-5. [DOI] [PubMed] [Google Scholar]
- 136.Huang C.F., Chen P.L., Liu M.F., Lee C.C., Lee N.Y., Chang C.M. Nontyphoidal Salmonella bacteremia in patients with connective tissue diseases. J. Microbiol. Immunol. Infect. 2012;45:350–355. doi: 10.1016/j.jmii.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 137.Tsao C.H., Chen C.Y., Ou L.S., Huang J.L. Risk factors of mortality for salmonella infection in systemic lupus erythematosus. J. Rheumatol. 2002;29:1214–1218. [PubMed] [Google Scholar]
- 138.Gerona J.G., Navarra S.V. Salmonella infections in patients with systemic lupus erythematosus: a case series. Int J Rheum Dis. 2009;12:319–323. doi: 10.1111/j.1756-185X.2009.01440.x. [DOI] [PubMed] [Google Scholar]
- 139.Wu K.-C., Yao T.-C., Yeh K.-W., Huang J.-L. Osteomyelitis in patients with systemic lupus erythematosus. J. Rheumatol. 2004;31:1340–1343. [PubMed] [Google Scholar]
- 140.Huang J.L., Hung J.J., Wu K.C., Lee W.I., Chan C.K., Ou L.S. Septic arthritis in patients with systemic lupus erythematosus: salmonella and nonsalmonella infections compared. Semin. Arthritis Rheum. 2006;36:61–67. doi: 10.1016/j.semarthrit.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 141.Abramson S., Kramer S.B., Radin A., Holzman R. Salmonella bacteremia in systemic lupus erythematosus. Eight-year experience at a municipal hospital. Arthritis Rheum. 1985;28:75–79. doi: 10.1002/art.1780280112. [DOI] [PubMed] [Google Scholar]
- 142.Matsiota-Bernard P., Hentati B., Pie S., Legakis N., Nauciel C., Avrameas S. Beneficial effect of Salmonella typhimurium infection and of immunoglobulins from S. typhimurium-infected mice on the autoimmune disease of (NZB x NZW) F1 mice. Clin. Exp. Immunol. 1996;104:228–235. doi: 10.1046/j.1365-2249.1996.15724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gallo P.M., Rapsinski G.J., Wilson R.P., Oppong G.O., Sriram U., Goulian M. Amyloid-DNA composites of bacterial biofilms stimulate autoimmunity. Immunity. 2015;42:1171–1184. doi: 10.1016/j.immuni.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kaper J.B., Nataro J.P., Mobley H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 145.Luijten R.K., Cuppen B.V., Bijlsma J.W., Derksen R.H. Serious infections in systemic lupus erythematosus with a focus on pneumococcal infections. Lupus. 2014;23:1512–1516. doi: 10.1177/0961203314543918. [DOI] [PubMed] [Google Scholar]
- 146.Wendling U., Farine J.C. Oral administration of HSP-containing E. coli extract OM-89 has suppressive effects in autoimmunity. Regulation of autoimmune processes by modulating peripheral immunity towards hsp’s? Biotherapy. 1998;10:223–227. doi: 10.1007/BF02678300. [DOI] [PubMed] [Google Scholar]
- 147.Handley H.H., Yu J., Yu D.T., Singh B., Gupta R.S., Vaughan J.H. Autoantibodies to human heat shock protein (hsp)60 may be induced by Escherichia coli groEL. Clin. Exp. Immunol. 1996;103:429–435. doi: 10.1111/j.1365-2249.1996.tb08298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dhaher Y.Y., Khamashta M.A., Hartley B., Taub N., Farine J.C., Hughes G.R. The effect of OM-89 (Subreum) on the murine model of systemic lupus erythematosus MRL-lpr/lpr. Lupus. 1997;6:436–440. doi: 10.1177/096120339700600504. [DOI] [PubMed] [Google Scholar]
- 149.Garcia-Guevara G., Rios-Corzo R., Diaz-Mora A., Lopez-Lopez M., Hernandez-Flores J., Fragoso-Loyo H. Pneumonia in patients with systemic lupus erythematosus: epidemiology, microbiology and outcomes. Lupus. 2018;27:1953–1959. doi: 10.1177/0961203318799207. [DOI] [PubMed] [Google Scholar]
- 150.Schurder J., Goulenok T., Jouenne R., Dossier A., Van Gysel D., Papo T. Pneumococcal infection in patients with systemic lupus erythematosus. Joint Bone Spine. 2018;85:333–336. doi: 10.1016/j.jbspin.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 151.Goldblatt F., Yuste J., Isenberg D.A., Rahman A., Brown J. Impaired C3b/iC3b deposition on Streptococcus pneumoniae in serum from patients with systemic lupus erythematosus. Rheumatology. 2009;48:1498–1501. doi: 10.1093/rheumatology/kep289. [DOI] [PubMed] [Google Scholar]
- 152.Einav S., Pozdnyakova O.O., Ma M., Carroll M.C. Complement C4 is protective for lupus disease independent of C3. J. Immunol. 2002;168:1036–1041. doi: 10.4049/jimmunol.168.3.1036. [DOI] [PubMed] [Google Scholar]
- 153.Lastoria J.C., Abreu M.A. Leprosy: review of the epidemiological, clinical, and etiopathogenic aspects - part 1. An. Bras. Dermatol. 2014;89:205–218. doi: 10.1590/abd1806-4841.20142450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chai Q., Zhang Y., Liu C.H. Mycobacterium tuberculosis: an adaptable pathogen associated with multiple human diseases. Front Cell Infect Microbiol. 2018;8:158. doi: 10.3389/fcimb.2018.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lin Y.C., Liang S.J., Liu Y.H., Hsu W.H., Shih C.M., Sung F.C. Tuberculosis as a risk factor for systemic lupus erythematosus: results of a nationwide study in Taiwan. Rheumatol. Int. 2012;32:1669–1673. doi: 10.1007/s00296-011-1847-5. [DOI] [PubMed] [Google Scholar]
- 156.Mok M.Y., Lo Y., Chan T.M., Wong W.S., Lau C.S. Tuberculosis in systemic lupus erythematosus in an endemic area and the role of isoniazid prophylaxis during corticosteroid therapy. J. Rheumatol. 2005;32:609–615. [PubMed] [Google Scholar]
- 157.Erdozain J.G., Ruiz-Irastorza G., Egurbide M.V., Martinez-Berriotxoa A., Aguirre C. High risk of tuberculosis in systemic lupus erythematosus? Lupus. 2006;15:232–235. doi: 10.1191/0961203306lu2289xx. [DOI] [PubMed] [Google Scholar]
- 158.Gaitonde S., Pathan E., Sule A., Mittal G., Joshi V.R. Efficacy of isoniazid prophylaxis in patients with systemic lupus erythematosus receiving long term steroid treatment. Ann. Rheum. Dis. 2002;61:251–253. doi: 10.1136/ard.61.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Machado Ribeiro F., Goldenberg T. Mycobacteria and autoimmunity. Lupus. 2015;24:374–381. doi: 10.1177/0961203314559634. [DOI] [PubMed] [Google Scholar]
- 160.Hsieh T., Wu Y. Leprosy mimicking lupus erythematosus. Dermatol. Sin. 2014;32:47–50. [Google Scholar]
- 161.Karadeniz A., Lally L., Magro C., Levy R., Erkan D., Lockshin M.D. Lepromatous leprosy mimicking systemic lupus erythematosus: a clinical pathology conference held by the division of rheumatology at hospital for special surgery. HSS J. 2014;10:286–291. doi: 10.1007/s11420-014-9405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chodisetti S.B., Rai P.K., Gowthaman U., Pahari S., Agrewala J.N. Potential T cell epitopes of Mycobacterium tuberculosis that can instigate molecular mimicry against host: implications in autoimmune pathogenesis. BMC Immunol. 2012;13:13. doi: 10.1186/1471-2172-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pahari S., Chatterjee D., Negi S., Kaur J., Singh B., Agrewala J.N. Morbid sequences suggest molecular mimicry between microbial peptides and self-antigens: a possibility of inciting autoimmunity. Front. Microbiol. 2017;8:1938. doi: 10.3389/fmicb.2017.01938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Res P.C., Schaar C.G., Breedveld F.C., van Eden W., van Embden J.D., Cohen I.R. Synovial fluid T cell reactivity against 65 kD heat shock protein of mycobacteria in early chronic arthritis. Lancet. 1988;2:478–480. doi: 10.1016/s0140-6736(88)90123-7. [DOI] [PubMed] [Google Scholar]
- 165.Ohosone Y., Ishida M., Takahashi Y., Matsumura M., Hirakata M., Kawahara Y. Spectrum and clinical significance of autoantibodies against transfer RNA. Arthritis Rheum. 1998;41:1625–1631. doi: 10.1002/1529-0131(199809)41:9<1625::AID-ART13>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 166.Marengo E.B., de Moraes L.V., Faria M., Fernandes B.L., Carvalho L.V., Tambourgi D.V. Administration of M. leprae Hsp65 interferes with the murine lupus progression. PloS One. 2008;3 doi: 10.1371/journal.pone.0003025. e3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yagishita M., Tsuboi H., Tabuchi D., Sugita T., Nishiyama T., Okamoto S. Clinical features and prognosis of nocardiosis in patients with connective tissue diseases. Mod. Rheumatol. 2020:1–17. doi: 10.1080/14397595.2020.1823070. [DOI] [PubMed] [Google Scholar]