Abstract
Background
Pancreatic cancer (PC) is one of the most lethal solid malignancies in the world due to its excessive cell proliferation and aggressive metastatic features. Emerging evidences revealed the importance of posttranscriptional modifications of RNAs in PC progression. However, knowledge about the 5-methylcytosine (m5C) RNA modification in PC is still extremely limited. In this study, we attempted to explore the expression changes and clinical significances of 12 known m5C-related genes among PC patients.
Methods
A total of 362 normal and 382 tumor specimens from PC patients were examined for candidate m5C-related gene and protein expression by using quantitative PCR (qPCR) and immunohistochemistry (IHC). The proliferation rate of PC cells was detected by MTS assay. Xenograft mouse models were used to assess the role of NSUN6 in PC tumor formation.
Findings
Through analyzing the four Gene Expression Omnibus (GEO) databases, six m5C-related genes shown significant and consistent alterations were selected for further examination in our 3 independent PC cohorts. Finally, we identified the reduction of NSUN6 as a common feature of all PC sample sets examined. NSUN6 expression correlated with clinicopathologic parameters including T stage, and Ki67+ cell rate. Further assessing the transcriptional profiles of 50 PC tissues, we found biological processes associated with cell proliferation like cell cycle and G2M checkpoint were enriched in NSUN6 lower expression group. Helped by in vitro PC cell lines and in vivo xenograft mouse models, we confirmed the role of NSUN6 in regulating cell proliferation and PC tumor growth. Last but also importantly, we also show the good performance of NSUN6 in evaluating tumor recurrence and survival among PC patients.
Interpretation
Our data suggested that NSUN6 is an important factor involved in regulating cell proliferation of PC, and highlights the potential of novel m5C-based clinical modalities as a therapeutic approach in PC patients.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81803014, 81802424, and 81802911).
Keywords: Pancreatic cancer, 5-methylcytosine, NSUN6, Cell proliferation
Abbreviations: PC, pancreatic cancer; m5C, 5-methylcytosine; GEO, Gene Expression Omnibus; NSUN6, NOP2/Sun domain family, member 6; FFPE, formalin-fixed paraffin-embedded; OS, overall survival; TCGA, The Cancer Genome Atlas; GSEA, gene set enrichment analysis; NES, normalized enrichment scores; FDR, false discovery rate; qPCR, quantitative PCR; IHC, immunohistochemical; SEM, standard error of mean; ROC, receiver operating characteristics curve; AUC, area under the curve; HR, Hazard ratios; CI, confidence interval; KPC, KrasG12D/+&Trp53R172H/+&Pdx1-Cre; PanIN, pancreatic intraepithelial neoplasia; DFS, disease free survival; KEGG, Kyoto Encyclopaedia of Genes and Genomes; CGN, Cancer Gene Neighborhoods; GO, gene ontology; CDK, cyclin-dependent kinase; CDKN, cyclin-dependent kinase inhibitor; m6A, N6-methyladenosine; NSUN, NOL1/NOP2/SUN domain
Research in context.
Evidence before this study
Numerous RNA modifications have been identified and reported to be involved in regulating diverse cellular functions. 5-methylcytosine (m5C), a novel discovered modification in RNA, has been confirmed to play essential role in regulating mRNA stabilization. It has also been reported that m5C-related genes were closely related to the tumorigenesis and tumor progression. However, the function of m5C-related genes in pancreatic cancer (PC) and the underlying mechanism remain unclear.
Added value of this study
The specific gene signatures and prognostic values of m5C-related genes human PC are still lacking. To address this question, a set of 12 m5C-related genes expression were assessed using 4 Gene Expression Omnibus (GEO) PC databases, and further verified in 3 independent PC cohorts. Results showed that NOP2/Sun domain family, member 6 (NSUN6) expression was closely correlated with the prognosis of PC patients. In addition, silencing of NSUN6 promoted cancer cell proliferation through elevating MKI67 levels and subsequently activating cell cycle related pathways.
Implications of all the available evidence
Our study provides new insights into understanding the impact of m5C-related gene NSUN6 on the prognosis of PC patients. These findings also imply that NSUN6 may be considered for future prognostic biomarker of patients with malignant tumors.
Alt-text: Unlabelled box
1. Introduction
Pancreatic cancer (PC) is a highly aggressive solid tumor with a 5-year survival of less than 10%, and with incidence rates increased during the past decade [1]. Despite the knowledge about PC still increasing, mechanisms involving in PC development remain unclear, and more effective therapies await discovery [2], [3], [4]. Therefore, figuring out the regulating mechanisms inside and thus identifying novel targets for effective treatments to improve the prognosis of PC patients were highly desirable.
Numerous RNA modifications have been identified and reported to be involved in regulating diverse cellular functions under physiological and/or pathological conditions [5, 6]. 5-methylcytosine (m5C), a long-standing DNA modification, is also found in RNAs (rRNA, tRNA and mRNA) and received considerable attention in recent years. It has been reported that RNA m5C modification plays essential roles in regulating RNAs alternative splicing, stabilization, localization, transportation, and translation [7], [8], [9], [10], [11], [12]. Until now, the m5C modification in RNAs have been found involve a series of regulators, including m5C methyltransferases (NSUN1-7, DNMT1, DNMT2, DNMT3A, and DNMT3B) and demethylases (TET2) [13]. More recently, ALYREF and YBX1 was also characterized as mRNA m5C binding protein (reader) in the nucleus, which could facilitate the transportation or maintain the stability of its target mRNA respectively [12, 14].
Dysregulation of m5C related genes have been implicated in cell differentiation, haematopoiesis, metabolic disorders, cancer initiation, metastasis and drug responses [14], [15], [16], [17], [18], [19], [20]. However, these studies were mainly focused on a limited number of genes identified in the m5C profiling of certain normal tissues and cancer cells. The specific gene signatures and prognostic values of m5C-related genes in cancers, especially in human PC, are still lacking. To address this question, a set of 12 m5C-related genes expression were assessed using 4 Gene Expression Omnibus (GEO) PC databases, and further verified in 3 independent PC cohorts. We found that NOP2/Sun domain family, member 6 (NSUN6) expression is closely correlated with tumor cell proliferation and prognosis of PC patients. These findings might be helpful in developing new biomarker and therapeutic rationale for targeting the m5C-related gene NSUN6 in PC.
2. Method
2.1. Clinical specimens
A total of 362 para-cancer normal pancreatic tissues and 382 PC tissues were collected in this study. One part of them were obtained from the Department of Biliary-Pancreatic Surgery at Renji hospital (Cohort 1: 58-paired PC and normal pancreatic fresh tissues; Cohort 3: 224-paired PC and normal pancreatic formalin-fixed paraffin-embedded (FFPE) tissues), and the other part were come from Shanghai OutdoBiotech Ltd (Cohort 2: 100 PC and 80 normal pancreatic FFPE tissues). Before the tumors were resected, all PC patients did not undergo any neoadjuvant therapies. The definition of overall survival (OS) is the interval between the dates of surgery and last follow up or death. The clinical pathological features of all PC patients are shown in Supplemental Table 1.
2.2. Bioinformatics analysis
The data sets of GSE62452, GSE28735, GSE15471, and GSE16515 were downloaded from the public source GEO data repository (http://www.ncbi.nlm.nih.gov/geo/). We obtained 189 PC and 161 normal pancreatic tissues mRNA transcriptome data for differential m5C-related genes expression analysis. The NSUN6 and MKI67 mRNA expression data for PC tissue and the corresponding prognostic data were downloaded from The Cancer Genome Atlas (TCGA) (https://gdc-portal.nci.nih.gov/). Gene set enrichment analysis (GSEA), which is available in Java, was used to determine which gene sets were associated with NSUN6 expression in our published gene microarray data (GSE102238) [21]. The results are shown using normalized enrichment scores (NES), accounting for the size and degree to which a gene set in overrepresented at the top or bottom of the ranked list of genes with NES > 1, P < 0.05, and false discovery rate (FDR) < 0.25.
2.3. Cell culture
Human PC cell lines (PANC-1, BXPC-3, Capan-2, SW1990 and ASPC-1), human pancreatic ductal epithelial cell line (HPDE), and HEK293T were purchased from Cell Resource Centre of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China), and cultured in RPMI-1640 and DMEM (21870076 & 11054020, Gibco, NY, USA), which was supplemented with 10% fatal bovine serum in a humidified atmosphere of 5% CO2 at 37 °C. The length of time between cell line thawing and use in experiment did not exceed 4 weeks (two or more passages). The two PC cell lines used in this study were authenticated every four months, and the length of time between cell line thawing and use did not exceed two months.
2.4. Cell transfection
NSUN6-targeting short hairpin RNA (sh-NSUN6) and non-specific control shRNA (sh-Con) used in this study were obtained from Biochemistry and Molecular Cell Biology, Shanghai Jiao Tong University School of Medicine. The open reading frame (ORF) sequences of NSUN6 was cloned into the pLenti-CDH-IRES-Puromycin vector (Addgene, USA). For lentivirus production and infection, HEK293T cell with 80–90% confluency was co-transfected with 4.44 μg of the required plasmids, 3.3 μg of psPAX and 2.2 μg of pMD2.G with 30 μl of polyethylenimine in 100-mm dishes according to the procedure described previously [22]. After transfection for 6 h at 37 °C, the medium was replaced, and the lentivirus-containing medium was collected 72 h later. Then the target PC cells were infected by the packaged lentivirus for 24 h, and the positively stable cell lines were selected by using 2.5 μg/ml puromycin. The sh-NSUN6 sequence was listed in Supplementary Table 1.
2.5. Cell proliferation, colony formation and cell apoptosis assays
PC cells were seeded in 96-well plates (1000 cells per well) and cultured at 37 °C for 0–96 h. Then the MTS solution (G3581, Promega, WI, USA) was added to each well of the plate at the different time points, followed by incubating at 37 °C for 1 h. The absorbance at 490 nm was measured in a Synergy 2 microplate reader (Biotek, VT, USA). For colony formation, PC cells were seeded in 6-well plates (200 cells per well) for 2 weeks until colonies were visible. The colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Each experiment with three replicates was repeated three times. PC cell apoptosis rate was detected using an Annexin V-FITC/PI Apoptosis Detection Kit (556547, BD Biosciences, Heidelberg, Germany) according to the manufacturer's protocol. The cells were collected by trypsinization without EDTA and doubly stained with FITC conjugated Annexin V and propidium iodide (5 mg/ml) for 30 min, and measured by flow cytometry (BD Biosciences). The data were analyzed using FlowJo 9.1 software (Treestar, CA, USA).
2.6. RNA extraction and quantitative PCR (qPCR) assays
Total RNA was extracted from cells or tissues or using TRI Reagent (AM9738, Sigma-Aldrich, MO, USA), and 1 μg of total RNA was reverse transcribed using the PrimerScript RT Reagent Kit (RR037A, Takara Bio, Beijing, China) into cDNA. qPCR was performed by the SYBR Premix Ex Taq (RR420A, Takara Bio) in Applied Biosystems ViiATM 7 Real-Time PCR System (Applied Biosystems, CA, USA). The relative mRNA expression level was calculated with 2−ΔΔCT method and normalized to internal reference gene GAPDH. The primers were designed in the software of Primer3 Input (version 0.4.0) and purchased from Sangon Biotech (Shanghai, China). All primer sequences are listed in Supplementary Table 5.
2.7. Western blot assays
Total proteins were extracted from PC cells using RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, Dreieich, Germany), and were quantified using a Micro BCA Protein Assay Kit (Thermo Fisher Scientific). Standard immunoblotting procedures, and the Bio-Rad ChemiDoc MP imaging system (Hercules, CA, USA) were used according to the procedure described previously [22]. The images were analyzed using ImageJ software. GAPDH served as an internal control for the whole-cell lysates. The NSUN6 (HPA045902, 1:1000), Ki67 (ZRB1007, 1:2000) and GAPDH (HPA040067, 1:5000) antibody were purchased from Sigma-Aldrich. The secondary antibody was purchased from Cell Signaling Technology Inc (MA, USA).
2.8. Tissue microarrays immunohistochemical (IHC) staining assays
IHC staining was performed as described in our previous study [22]. In brief, paraffin sections of 5 μm thickness were prepared from tissue microarrays or xenograft tumors. The sections were deparaffinized, treated with 3% H2O2 for 10 min, autoclaved in 10 mM citric sodium (pH 6.0) for 30 min for heat-induced antigen retrieval, and then incubated with primary antibodies at 4 °C overnight, followed by incubation with biotinylated secondary antibody for 1 h at room temperature. Finally, 3,3-diaminobenzidine tetrahydrochloride was used as coloring reagent, and haematoxylin was used as a counterstain for nuclei. The stained fields were photographed using Olympus camera. Antibody against NSUN6 (HPA045902, 1:200) and Ki67 (ZRB1007, 1:200) were obtained from Sigma-Aldrich. Quantification of IHC staining was based upon the staining intensity (I score: negative, 0; weak, 1; moderate, 2; and intense, 3) and the percentage of positive stained cells (P score: 0–5%, score of 0; 6–35%, score of 1; 36–70%, score of 2; and >70%, score of 3) to obtain a final score (Q score = I score × P score). Sample with Q score of ≥ 4 were considered to be high expression, and < 4 were considered to be low expression. Two senior pathologists performed the scorings independently in a blinded manner.
2.9. Xenograft model
To evaluate PC cell proliferation in vivo, NSUN6-overexpressing, NSUN6-silencing and negative control PC cells (2 × 106) were subcutaneously injected into the hind footpads of the 4-week-old male BALB/c nude mice (Shanghai Laboratory Animal Research Centre), and then housed in laminar flow cabinets under specific pathogen free conditions with food and water provided ad libitum. Tumor growth curves were measured weekly with a calliper from the 1st week to the 5th week. Tumor volume analyses were calculated using the equation: V = (length × width2)/2. All the mice were killed at the 5th week, and subcutaneous tumors were collected and weighed. The tumor volume and weight were presented as the mean ± standard error of mean (SEM) (n = 5).
2.10. Statistical analysis
All the data in this study were presented as mean ± SEM. Group comparisons of normally distributed measurement data and categorical data were performed using unpaired Student's t test and χ2 test respectively. Receiver operating characteristic (ROC) curve analysis was performed to evaluate the sensitivity and specificity of NSUN6 levels for PC diagnosis, and the area under the curve (AUC) value was calculated and used to designate the ROC effect. For survival analysis, the Kaplan–Meier method with the log-rank test (univariate analysis) and Cox proportional hazards regression model (multivariate analysis) were used to analysis the prognostic factors of PC patients. Hazard ratios (HR) for disease prognosis and 95% confidence intervals (CI) were calculated by the Cox risk proportion model. Pearson correlation coefficient were used to analyzed the correlation between NSUN6 and MKI67 or CDK11 mRNA expression. SPSS 17.0 software (IBM, IL, USA) was used for all statistical analysis, and a P < 0.05 was considered to be statistically significant.
2.11. Ethics statement
Humans: This study was approved by the Ethical Committee of Renji hospital, School of Medicine, Shanghai Jiao Tong University (approved protocol number: RA-2019-116). All of the subjects were provided with written informed consent before enrolment. All experimental protocols and procedures were carried out following the ethical standards of the Renji hospital at which the studies were conducted. PC and normal pancreatic FFPE tissues were collected in the pathology department of Renji hospital, with the approval of the Renji hospital Ethical Committee and according to local legal and ethical regulations. Animals: Procedures involving animals and their care were conducted according to the guidelines of the Animal Research Committee of Renji hospital, School of Medicine, Shanghai Jiao Tong University.
2.12. Role of funding sources
Funders had no role in study design, data collection, data analyses, interpretation, or the writing of this report.
3. Results
3.1. Differential expression analysis of 12 m5C-related genes in PC patients
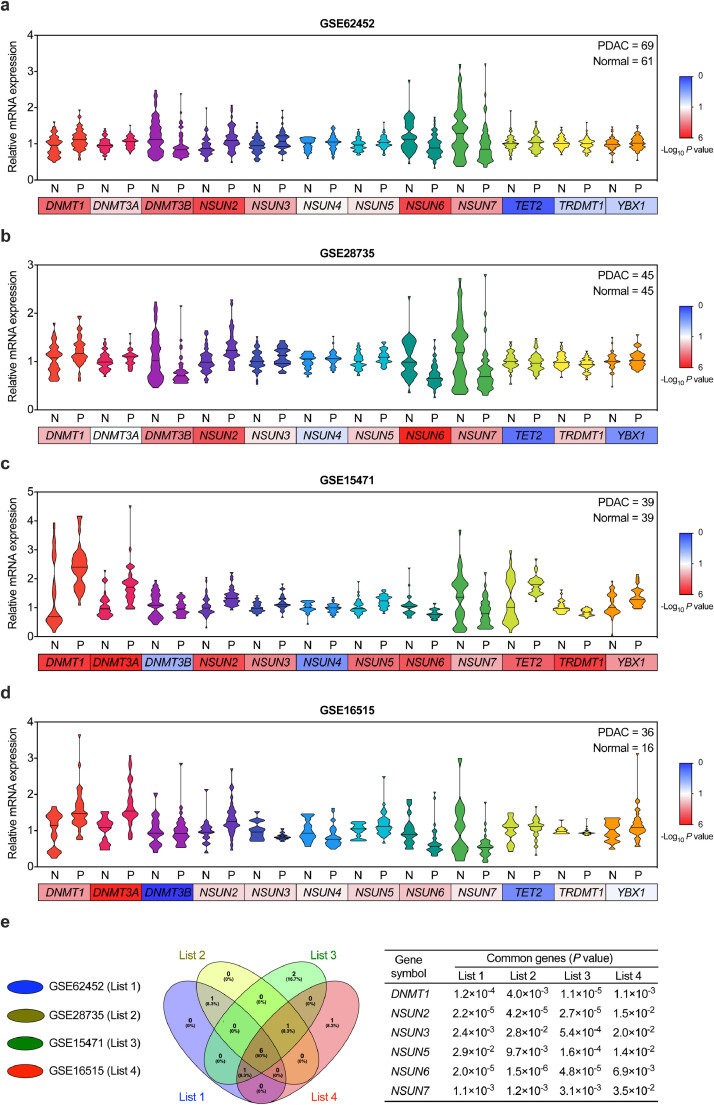
To determine the possible implications of m5C-related genes in PC tumorigenesis and progression, the expression levels of 12 m5C-related genes were analyzed based on the 4 GEO databases (GSE62452, GSE28735, GSE15471, and GSE16515) (Fig. 1a–d). Among them, six genes shown consistent elevations or reductions in all four databases were identified (Fig. 1e). As compared with normal pancreatic tissues, DNMT1, NSUN2, NSUN3 and NSUN5 were increased in PC tissues, otherwise NSUN6 and NSUN7 were significantly reduced (Fig. 1a–e).
Fig. 1.
m5C modification-related genes expression in PC. (a–d) Violin plot of m5C-related gene expression profiles in PC and para-cancer normal pancreatic tissues from the 4 GEO databases (A: GSE62452; B: GSE28735; C: GSE15471; D: GSE16515). (e) Left is the Venn diagram show the overlapping m5C-related genes differentially expressed between PC and para-cancer normal pancreatic tissues identified in the 4 GEO databases. Right form shows the P values of the identified 6 common altered genes.
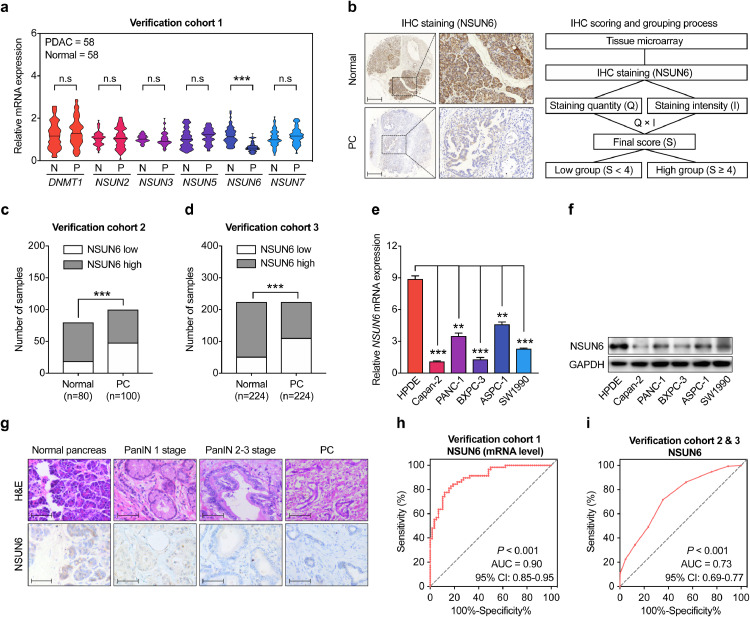
For verifying the expression changes of the six genes in PC tissues, we then examined their expressions in our 58-paired fresh PC and their corresponding para-cancer normal pancreatic tissues by qPCR (Cohort 1). However, we found that only NSUN6, which was again shown significantly reduction in PC tissues (P < 0.001). DNMT1 and NSUN5 were elevated in PC tissues, but which were not achieved the significance level (Fig. 2a). Further assessing the protein level of NSUN6 in two other independent PC cohorts by tissue microarrays IHC staining, we confirmed the reductions of NSUN6 in PC tissue as compared with normal controls (cohort 2: P < 0.001; cohort 3: P < 0.001) (Fig. 2b–d). Interestingly, we found that as compared with normal human pancreas duct cell line (HPDE), both the mRNA and protein levels of NSUN6 were obviously lower in all five PC cell lines examined (Fig. 2e–f).
Fig. 2.
Validation of expression levels of candidate m5C-related genes in PC cell lines and tissues. (a) qPCR assessment of the expression of the 6 candidate m5C-related genes in 58 paired PC and para-cancer normal pancreatic tissues from verification cohort 1. (b) Left is the representative IHC staining of NSUN6 in PC and normal pancreatic tissue. Right is the flow chart show the evaluation method for NSUN6 IHC scoring and grouping process. Scale bar = 200 μm. (c–d) Statistical quantification of the number of samples classified by NSUN6 expression level in normal and PC group from verification cohort 2 and 3. (e–f) qPCR and western blot examination of the protein and mRNA levels of NSUN6 in normal pancreatic epithelial cell (HPDE) and five PC cell lines. (g) H&E and NSUN6 IHC staining for KPC mice pancreatic tissue with lesions at different stage. (h–i) ROC analysis show the sensitivity and specificity for the NSUN6 expression in predicting PC of verification cohort 1 and 2 combined 3. ***P < 0.001; n.s, not significant; Student's t test.
The KrasG12D/+&Trp53R172H/+&Pdx1-Cre (KPC) mice, characterized by highly accelerated development of pancreatic intraepithelial neoplasia (PanIN) and well differentiated PC [23], was used for exploring the mechanism of pancreatic tumorigenesis in our previous study [24]. We then measured the levels of NSUN6 protein in pancreas from KPC mice and found gradually reduced NSUN6 levels in normal pancreas, pancreatic intraepithelial neoplasia (PanIN), and PC tissues (Fig. 2g). As the prominent reduction of NSUN6 in PC identified, we then performed a ROC curve analysis to evaluate its sensitivity and specificity for the diagnosis of PC. Indeed, NSUN6 achieved high AUC value in both cohort 1 and cohort 2 combined 3 by using qPCR (AUC: 0.90) and IHC (AUC: 0.73) respectively, implying that NSUN6 harbor high sensitivity and specificity for PC diagnosis (Fig. 2h–i). Hence, above results showed that both the mRNA and protein levels of NSUN6 were significantly decreased in PC tissues, and the capacity of NSUN6 as biomarker for PC diagnosis.
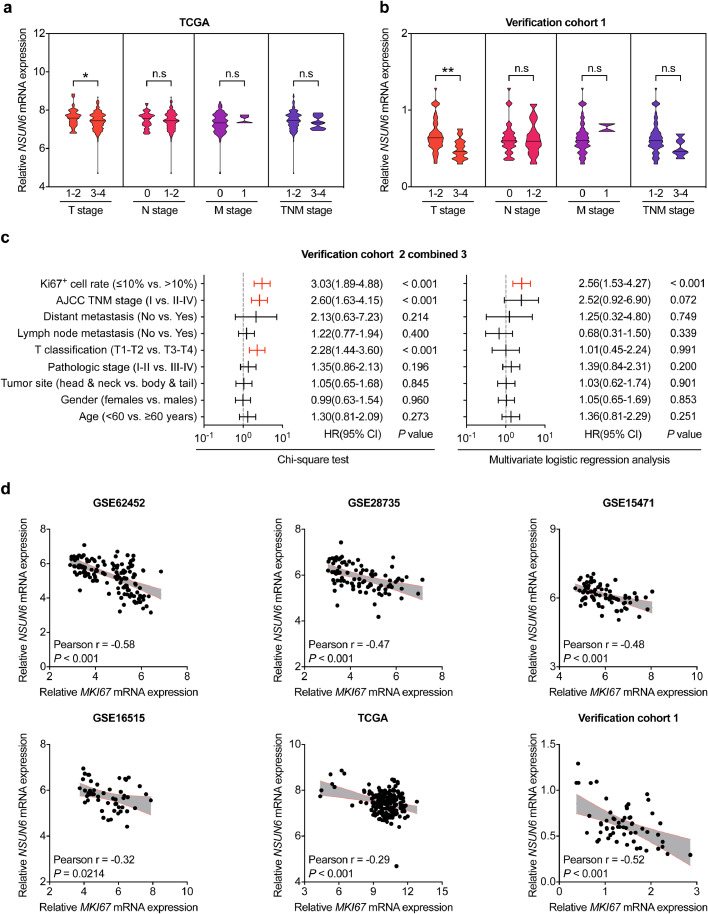
3.2. Correlations between the NSUN6 and the clinical features of PC patients
In order to find out the clinical importance of NSUN6 in PC tissues, the relationship between NSUN6 levels and various important clinicopathologic parameters were assessed in the verification cohort and TCGA database. We found that lower NSUN6 carriers are prone to have advanced T stages in our PC verification cohort 1 and TCGA-PC cohort (Fig. 3a–b). Otherwise, no obvious relationships between NSUN6 and N stage or M stage were observed. Furthermore, in our PC verification cohort 2 and 3, both univariate and multivariate logistic regression analyses show that NSUN6 expression was tightly correlated with Ki67+ cell rate in PC tissue (Fig. 3c, Supplemental Fig. 1a–b, and Supplemental Tables 2–4). Interestingly, an inverse relationship between NSUN6 and MKI67 mRNA expression were also identified in the 4 GEO databases, TCGA database and our PC verification cohort 1 (Fig. 3d). Taken together, these results suggest that NSUN6 might influence tumor size through regulating cellular proliferation.
Fig. 3.
Correlations between NSUN6 expression and the clinicopathologic features of PC patients. (a–b) The comparison of mRNA expression of NSUN6 in PC tissue at different TNM stages from TCGA database and verification cohort 1. (c) Univariate and multivariate analysis of the associations between NSUN6 protein levels and the clinicopathologic features of PC patients from verification cohort 2 combined 3. Red lines represent clinicopathologic features significantly related with NSUN6. P < 0.05. (d) Correlation analysis between the NSUN6 and MKI67 mRNA expression from the 4 GEO databases, TCGA database, and verification cohort 1. *P < 0.05, **P < 0.01; n.s, not significant; Student's t test.
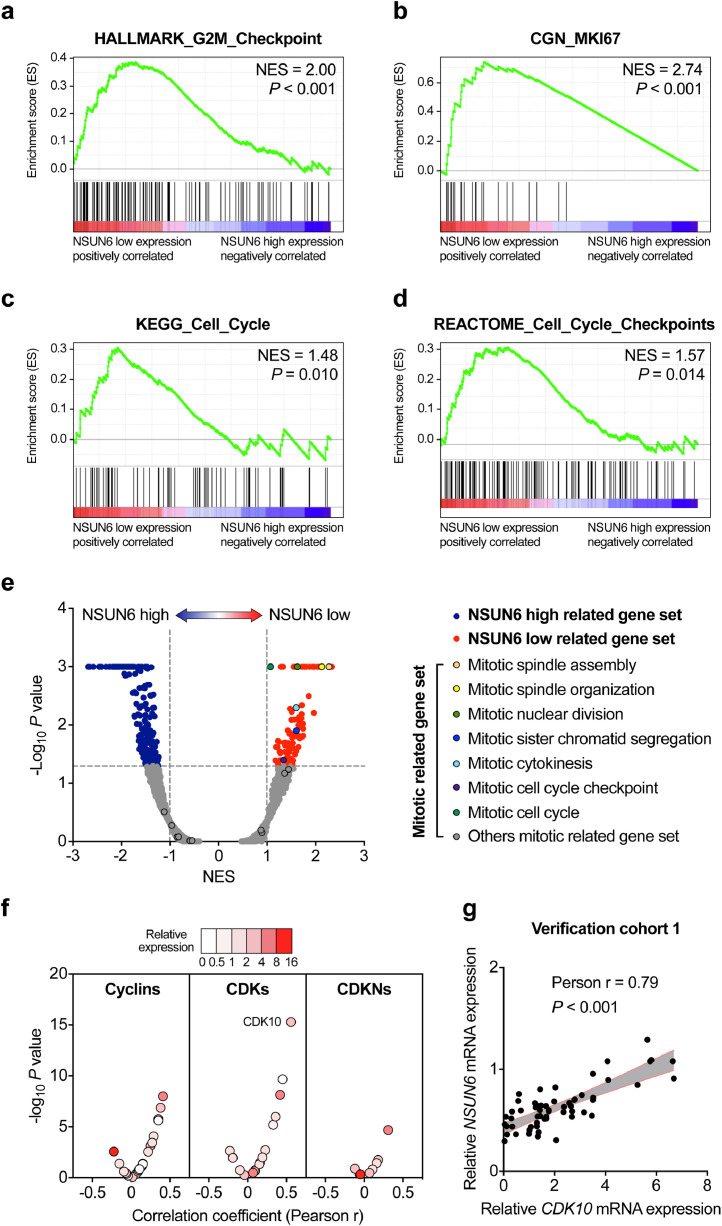
3.3. Functional analysis of NSUN6 in PC
To investigate the significant altered gene sets associated with NSUN6, we performed GSEA analysis of transcriptional profiles of PC samples from our published gene microarray data (GSE102238), of which 50 paired PC and corresponding para-cancer normal pancreatic tissues were included [21]. The PC samples were divided into high NSUN6 group (n = 25) and low NSUN6 group (n = 25). Hallmark, Kyoto Encyclopaedia of Genes and Genomes (KEGG), Reactome, and Cancer Gene Neighborhoods (CGN) gene sets analysis revealed that pathways associated with cell proliferation like G2M checkpoint, cell cycle were enriched in PC tissues with lower NSUN6 expression (Fig. 4a–d). Gene ontology (GO) analysis also revealed obviously elevated mitotic related genes involved in mitotic spindle assembly, mitotic spindle organization in samples with lower NSUN6 expression (Fig. 4e).
Fig. 4.
GSEA analysis of NSUN6-related biological features in PC tissues. (a–d) Cell proliferation-related pathway gene sets were enriched and positively correlated with PC tissues with low NSUN6 expression. (e) Volcano plot show GO analysis of NSUN6 related gene sets in PC tissues. Red plots represent NSUN6 low expression group related gene set with P < 0.05. Blue plots represent NSUN6 high expression group related gene set with P < 0.05. (f) Correlation analysis between the NSUN6 and Cyclins, or CDKs, or CDKNs expression from the GSE102238 databases. (g) Correlation analysis between the NSUN6 and CDK10 mRNA expression in verification PC cohort 1.
Cyclins, cyclin-dependent kinases (CDKs) and cyclin-dependent kinase inhibitors (CDKNs) are well-known markers of cell proliferation [25]. The association between NSUN6 levels and cellular proliferation markers including 26 Cyclins, 20 CDKs and 8 CDKNs were analyzed based on our PC gene microarray data. We found that CDK10, a tumor suppressor gene which reported to inhibit cell proliferation [26], were positively correlated with NSUN6 expression (Fig. 4f). This correlation was also identified in our verification PC cohort 1 (Fig. 4g). These results indicate that changes in cell cycle progression genes like CDK10 might be one of the main effectors associated with NSUN6 in PC tissues.
3.4. NSUN6 inhibited PC cell proliferation in vitro and in vivo
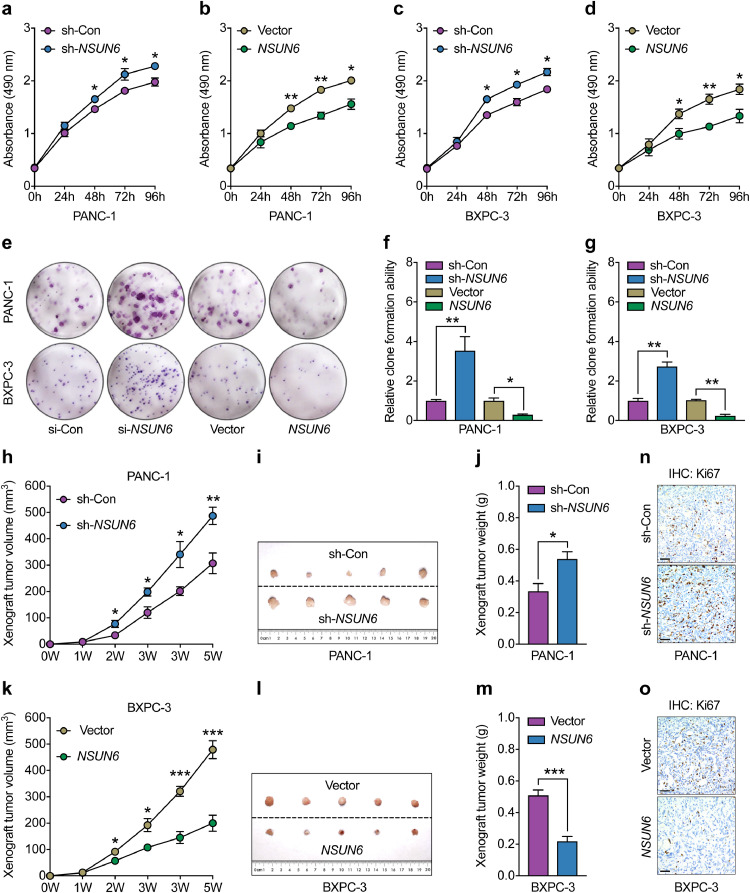
To study the potential influence of NSUN6 on the biological behavior of PC cells, MTS and colony formation assays were first conducted to determine PC cell proliferation rates. Compared with the group transfected with control shRNA, both PANC-1 and BXPC-3 cells transfected with NSUN6 shRNA show increased proliferation rates and enhanced cloning formation capacity (Fig. 5a–g). Conversely, opposite effects on cellular proliferation rates were observed in PC cells with NSUN6 overexpression (Fig. 5a–g). However, cell apoptosis rates were not affected by altering NSUN6 expression levels in PANC-1 and BXPC-3 cells (Supplemental Fig. 2a–b).
Fig. 5.
NSUN6 influenced PC cell proliferation in vitro and in vivo. (a–d) MTS analysis of PANC-1 and BXPC-3 cells transfected with NSUN6 shRNA (sh-NSUN6), control shRNA (sh-Con), NSUN6 overexpression vector (NSUN6), or control vector (Vector). (e–g) Representative images and statistical analysis of colony formation ability among PANC-1 and BXPC-3 cells transfected with NSUN6 shRNA, control shRNA, NSUN6 overexpression vector, or control vector. (h–j) Xenograft tumour growth curves, shape and weight of PANC-1 cells transfected with NSUN6 shRNA or control shRNA. (k–m) Xenograft tumor growth curves, shape and weight of BXPC-3 cells transfected with NSUN6 overexpression vector or control vector. (n–o) Representative IHC images for Ki67 staining of tumours formed in sh-Con, sh-NSUN6, Vector, and NSUN6 groups. All in vitro experiment, n = 3; All in vivo experiment n = 5; bar, S.E.M., *P < 0.05; **P < 0.01; ***P < 0.001; Student's t-test.
We then used PC xenograft mouse models to determine the function of NSUN6 in vivo. Consistent with the in vitro findings, PC cells with NSUN6 silencing had faster tumor growth rates and higher tumor weights than the control group (Fig. 5h–j). In contrast, overexpression NSUN6 in BXPC-3 cells slowed tumor growth rates and thus reduced tumor weights (Fig. 5k–m). Through IHC staining, we found that NSUN6 deficiency increased Ki67+ cell rate, while NSUN6 overexpression reduced Ki67+ cell rate in xenograft tumors respectively (Fig. 5n–o). Indeed, both the mRNA transcripts and protein levels of Ki67 were increased in NSUN6 knock-down PANC-1 and BXPC-3 cells, and otherwise reduced in NSUN6 overexpression cells (Supplemental Fig. 2c–d). In addition, we found that CDK10 were decreased in NSUN6 deficient PANC-1 and BXPC-3 cells, and otherwise increased in NSUN6 overexpression cells. Therefore, our data suggested that CDK10 might be one of the important factors mediating the function of NSUN6 in inhibiting PC cell proliferation.
3.5. Comprehensive analysis the role of NSUN6 in PC prognosis prediction
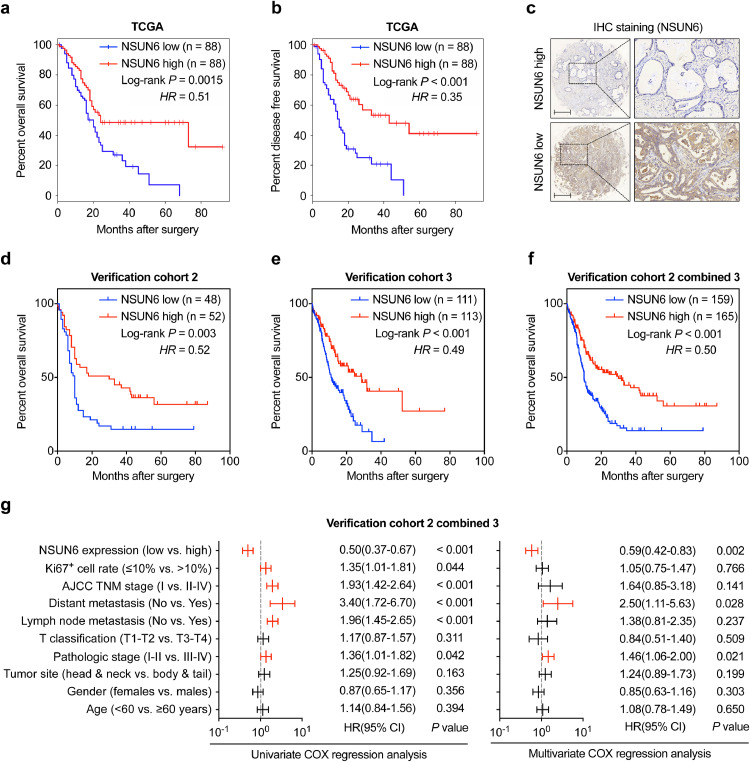
In order to investigate whether the expression levels of NSUN6 could be used for predicting the prognosis of PC patients, we first explored the correlations between NSUN6 and the OS as well as the DFS rate using data from the TCGA database. Our analysis results revealed that the mRNA level of NSUN6 significantly associated with OS and DFS time in TCGA dataset, of which patients with higher NSUN6 expression harbor obviously improved outcomes (Fig. 6a–b). Consistently, analysis of data from our PC verification cohort 2 and 3 confirmed reduced cumulative OS rates among those patients with lower tissue NSUN6 expression (Fig. 6c–f). In addition, our univariate COX regression analysis displayed that NSUN6 expression, along with Ki67+ cell rate, TNM stage, lymph node metastasis, distant metastasis and pathological stage are all important prognostic factors in predicting the OS rate of our PC patients (Fig. 6g). Meanwhile, multivariate COX regression analysis suggested that NSUN6 expression is an independent predictor of OS in our PC cohorts (Fig. 6g and Supplemental Fig. 3a–b). Taken together, these data demonstrated that NSUN6 level might be a potential marker for predicting the prognosis of PC patients.
Fig. 6.
Prognostic prediction value of NSUN6 for clinical PC patients. (a–b) Kaplan–Meier OS and DFS curves of PC patients with high or low NSUN6 expression analyzed from TCGA database. (c) Representative IHC staining image of NSUN6 high and low expression by PC tissue microarray. (d–f) Kaplan–Meier OS curves assessed for PC patients with high or low NSUN6 expression from verification cohort 2, 3, as well as cohort 2 and 3 combined. (g) Univariable and multivariable COX regression analyses of the associations between the prognostic parameters and OS among the PC patients in verification cohort 2 combined 3. Red lines represent the factors significantly related with PC prognosis with P < 0.05.
4. Discussion
PC is a highly aggressive disease, and although recent decades have seen gaining advancement in technological treatment of PC, the 5-year survival rate (8.2%) remain low [27]. Therefore, identification new factors underlying PC progression and novel therapeutic strategies are urgently needed. Until now, over a hundred of modifications of RNA have been found, with majority of these modifications seen on rRNA and tRNA and still increasing modification in mRNA and in less abundant non-coding RNA. Correspondingly increased are the interests in elucidating the nature and functions of those “epitransciptomic” modifications in RNA. Increasing evidences shown that RNA methyltransferases are aberrantly expressed and play critical roles in cancer development and pathogenesis [28, 29]. Among these, N6-methyladenosine (m6A) is the best understood and frequent mark of mRNA, and the functions of which ranges from pre-mRNA processing, translation, miRNA biogenesis to mRNA decay [30]. The importance of METTL3 and ALKBH5 in the development and progression of pancreatic cancer have pointed out [31, 32]. By contrast, much less research has been conducted on m5C, which was detected in tRNAs and rRNAs and more recently in poly(A)RNAs. In mammalian cells, the addition of m5C to RNA cytosines is carried out by enzymes of the NOL1/NOP2/SUN domain (NSUN) family as well as the DNA methyltransferase homologue DNMT2. One of the m5C RNA modifier NSUN2 has been found highly expressed in various tumors and linked with the oncogene activation [33, 34]. However, little knowledge was known about the expression and functions of m5C related genes in PC. Through systematically examine the expression of 12 m5C-related genes in 4 GEO databases, we found that while DNMT1, NSUN2, NSUN3 and NSUN5 were consistently elevated, NSUN6 and NSUN7 were reduced in PC tissues as compared with normal pancreatic tissues. This differential expression pattern might imply different biological significance existed between m5C-related modulators. Obviously decreased NSUN6 were verified in our 58-paired fresh PC and their corresponding para-cancer normal pancreatic tissues by qPCR and two other independent PC cohorts by using tissue microarrays IHC staining. Importantly, the reduction of NSUN6 expression in PC patients were tightly correlated with poor OS and DFS rates using data from both the TCGA database and our two PC verification cohort. NSUN6 expression, along with Ki67+ cell rate, TNM stage, lymph node metastasis, distant metastasis and pathological stage are all important prognostic factors in predicting the OS rate of our PC patients. Meanwhile, multivariate COX regression analysis also suggested that NSUN6 expression was an independent predictor of OS in our PC cohorts. Taken together, these data for the first time show that NSUN6 level be a potential marker tightly correlated with the prognosis of PC patients.
The functions of m5C RNA methyltransferases regulators in cellular processes are versatile including cellular differentiation and proliferation. Helped by analysing the GSEA of transcriptional profiles of our gene microarray data (GSE102238), we found an interestingly phenomenon that cell proliferation and cell cycle related signatures genes were profoundly increased in patients with lower NSUN6 expression as compared with those with higher NSUN6. The capacity of NSUN6 in regulating cellular proliferation were verified by in vitro PC cell lines and in vivo mouse xenograft models. NSUN6 overexpression indeed suppressed tumor growth and increased CDK10. These observations imply that NSUN6 possibly affect PC proliferation status through regulating CDK10 involved in mitotic spindle assembly, mitotic nuclear division and so on. However, our investigation was still limited as how NSUN6 involved in regulating mitotic related genes expression unclarified. Human NSUN6 was found localize in the cytoplasm of HEK293 cells and catalyze methylation at C72 of tRNA Cys and tRNA Thr isoacceptors [35, 10]. The physiological role of tRNA fragments have been proposed, as which could be processed to smaller RNAs by the Dicer RNase and affect RNA silencing pathways [36], [37], [38]. DNMT2, another RNA m5C methyltransferases catalyse methylation at C38 in the anticodon stem loop of tRNA. The methylation mark at C38 modified by DNMT2 could inhibit stress-induced cleavage of tRNAs [39]. Whether the tRNA modification by NSUN6 influenced proliferation related genes through similar mechanisms needed to be proven. In fact, the altered proliferation related genes by NSUN6 could be directly influenced by their tRNA methylation status or indirectly. Of course, the m5Cs modifications of the mRNAs changed with NSUN6 could not be excluded. Further examination of NSUN6-dependent tRNA/mRNA methylation profiles in PC by ultra-performance liquid chromatography-mass spectrometry or RNA bisulphite sequencing would be helpful for us clarifying this. Finding out the regulatory mechanisms inside would provide valuable therapeutic targets for patients with pancreatic cancer.
Overall, we discovered that a m5C-related gene NSUN6 was associated with clinicopathologic features and are useful in predicting the recurrence and survival of patients with PC. The influence of NSUN6 on suppressing cell proliferation were further demonstrated in vitro and in vivo. These findings suggest that the NSUN6 might be helpful for prognostic risk stratification among PC patients and facilitate personalized therapy decision making for them. However, a more detailed molecular mechanism analysis for NSUN6 will be carried out in the future to clarify their role in promoting PC progression.
Funding sources
This study was supported by the National Natural Science Foundation of China (grant nos. 81803014, 81802424, and 81802911). The funding sponsors played no role in study design, data collection, data analysis, interpretation, writing of the report, and the decision to submit the paper for publication.
Data sharing statement
Data, materials and software information supporting the conclusions of this article are included within the article and its additional file.
Declaration of Interests
The authors have declared no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103195.
Contributor Information
Linhua Yang, Email: yanglinhua1981@126.com.
Ming Zhan, Email: zhanming@shsmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Grossberg AJ, Chu LC, Deig CR, Fishman EK, Hwang WL, Maitra A. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J Clin. 2020;70(5):375–403. doi: 10.3322/caac.21626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15(6):333–348. doi: 10.1038/s41575-018-0005-x. [DOI] [PubMed] [Google Scholar]
- 3.Uzunparmak B, Sahin IH. Pancreatic cancer microenvironment: a current dilemma. Clin Transl Med. 2019;8(1):2. doi: 10.1186/s40169-019-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniel SK, Sullivan KM, Labadie KP, Pillarisetty VG. Hypoxia as a barrier to immunotherapy in pancreatic adenocarcinoma. Clin Transl Med. 2019;8(1):10. doi: 10.1186/s40169-019-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machnicka MA, Olchowik A, Grosjean H, Bujnicki JM. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014;11(12):1619–1629. doi: 10.4161/15476286.2014.992273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017;18(1):1. doi: 10.1186/s13059-016-1139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T. Transcriptome-wide mapping of RNA 5-methylcytosine in arabidopsis mRNAs and noncoding RNAs. Plant Cell. 2017;29(3):445–460. doi: 10.1105/tpc.16.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu RJ, Long T, Li J, Li H, Wang ED. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. 2017;45(11):6684–6697. doi: 10.1093/nar/gkx473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38(5):1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY. 5-methylcytosine promotes mRNA export – NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27(5):606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31(5):458–464. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21(8):978–990. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 15.Bohnsack KE, Hobartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m(5)C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes. 2019;10(2) doi: 10.3390/genes10020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21(3):261–266. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69(20):8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 18.Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015;34(18):2350–2362. doi: 10.15252/embj.201591382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol. 2018;20(5):535–540. doi: 10.1038/s41556-018-0087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Wang S, Xing Z, Lin A, Liang K, Song J. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19(2):106–119. doi: 10.1038/ncb3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang MW, Tao LY, Jiang YS, Yang JY, Huo YM, Liu DJ. Perineural invasion reprograms the immune microenvironment through cholinergic signaling in pancreatic ductal adenocarcinoma. Cancer Res. 2020;80(10):1991–2003. doi: 10.1158/0008-5472.CAN-19-2689. [DOI] [PubMed] [Google Scholar]
- 22.Xu S, Zhan M, Jiang C, He M, Yang L, Shen H. Genome-wide CRISPR screen identifies ELP5 as a determinant of gemcitabine sensitivity in gallbladder cancer. Nat Commun. 2019;10(1):5492. doi: 10.1038/s41467-019-13420-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 24.Jiang SH, Li J, Dong FY, Yang JY, Liu DJ, Yang XM. Increased serotonin signaling contributes to the Warburg effect in pancreatic tumor cells under metabolic stress and promotes growth of pancreatic tumors in mice. Gastroenterology. 2017;153(1) doi: 10.1053/j.gastro.2017.03.008. 277-91 e19. [DOI] [PubMed] [Google Scholar]
- 25.Campbell GJ, Hands EL, Van de Pette M. The role of CDKs and CDKIs in murine development. Int J Mol Sci. 2020;21(15) doi: 10.3390/ijms21155343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu JH, Zhong XY, Zhang WG, Wang ZD, Dong Q, Tai S. CDK10 functions as a tumor suppressor gene and regulates survivability of biliary tract cancer cells. Oncol Rep. 2012;27(4):1266–1276. doi: 10.3892/or.2011.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verma HK, Kampalli PK, Lakkakula S, Chalikonda G, Bhaskar L, Pattnaik S. A retrospective look at Anti-EGFR agents in pancreatic cancer therapy. Curr Drug Metab. 2019;20(12):958–966. doi: 10.2174/1389200220666191122104955. [DOI] [PubMed] [Google Scholar]
- 28.Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18(1):103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat Cell Biol. 2019;21(5):552–559. doi: 10.1038/s41556-019-0319-0. [DOI] [PubMed] [Google Scholar]
- 30.Shi H, Chai P, Jia R, Fan X. Novel insight into the regulatory roles of diverse RNA modifications: re-defining the bridge between transcription and translation. Mol Cancer. 2020;19(1):78. doi: 10.1186/s12943-020-01194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geng Y, Guan R, Hong W, Huang B, Liu P, Guo X. Identification of m6A-related genes and m6A RNA methylation regulators in pancreatic cancer and their association with survival. Ann Transl Med. 2020;8(6):387. doi: 10.21037/atm.2020.03.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 2020;19(1):91. doi: 10.1186/s12943-020-01158-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16(10):971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 34.Holgersson J, Breimer ME, Jacobsson A, Svensson L, Ulfvin A, Samuelsson BE. Glycolipid- and glycoprotein-based blood group A antigen expression in human thrombocytes. A1/A2 difference. Glycoconj J. 1990;7(6):601–608. doi: 10.1007/BF01189080. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Li H, Long T, Dong H, Wang ED, Liu RJ. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2019;47(4):2041–2055. doi: 10.1093/nar/gky1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15(12):2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S. Hidden layers of human small RNAs. BMC Genom. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311(5759):395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.