Abstract
Relative to conventional two-dimensional (2-D) culture, three-dimensional (3-D) suspension culture of epithelial cells more closely mimics the in vivo cell microenvironment regarding cell architecture, cell to matrix interaction, and osmosis exchange. However, primary normal human keratinocytes (NHKc) rapidly undergo terminal differentiation and detachment-induced cell death (anoikis) upon disconnection from the basement membrane, thus greatly constraining their use in 3-D suspension culture models. Here, we examined the 3-D anchorage-free growth potential of NHKc isolated from neonatal skin explants of 59 different individuals. We found that 40% of all isolates naturally self-assembled into multicellular spheroids within 24 h in anchorage-free culture, while 60% did not. Placing a single spheroid back into 2-D monolayer culture yielded proliferating cells that expressed elevated levels of nuclear P63 and basal cytokeratin 14. These cells also displayed prolonged keratinocyte renewal and a gene expression profile corresponding to cellular heterogeneity, quiescence, and de-differentiation. Notably, spheroid-derived (SD) NHKc were enriched for a P63/K14 double-positive population that formed holoclonal colonies and reassembled into multicellular spheroids during 3-D suspension subculture. This study reveals marked phenotypic differences in neonatal keratinocyte suspension cultures isolated from different individuals and present a model system that can be readily employed to study epithelial cell behavior, along with a variety of dermatological diseases.
Keywords: Anoikis, Epidermal stem cells, Spheroids, 3-D culture, Human keratinocytes, Cell-based model
1. Introduction
Three dimensional (3-D) culture systems provide unique platforms to study complex biological processes in vitro, particularly cell proliferation, differentiation, motility, and stress response. While 3-D spheroid cultivation methods are well established in certain tissue types such as mammary and colorectal epithelium, efficient 3-D epithelial spheroid systems for primary human skin epidermis are lacking. This challenge lies in keratinocytes’ sensitivity to surface-detachment, as basal keratinocyte renewal is tightly regulated by cell attachment to the basement membrane. In fact, normal human keratinocytes (NHKc) initiate terminal differentiation programs upon cell-surface detachment, eventually resulting in suspension-induced cell death (anoikis) (Green, 1977; Wakita and Takigawa, 1999; Guo and Jahoda, 2009).
While several reports have indicated that skin spheroid cultures possess enhance regenerative capacities (Zong et al., 2012; Toma et al., 2005), and that epidermosphere culture can improve skin cells’ potency (Borena et al., 2014; Higgins et al., 2010; Kang et al., 2012; Vollmers et al., 2012), most of these studies have been conducted in animals or with human dermal fibroblasts, with virtually no reports thoroughly characterizing the regenerative effects of 3-D spheroid culture using NHKc. Here, we describe a 3-D spheroid growth method to cultivate NHKc stem cells from neonatal foreskin tissue and use this system to investigate the spheroid forming capacity of NHKc isolates derived from 59 individual donors. By thoroughly characterizing skin epidermal spheroid cultures in 3-D regenerative states, we observed that most strains (60%) fail to autonomously form spheroids and readily undergo cell death when seeded in 3-D suspension. However, approximately 40% of NHKc strains can autonomously aggregate into multicellular epidermospheres and retain prolonged cellular growth in culture. When 3-D epidermospheres are placed into 2-D monolayer culture, the adherent spheroids expressed a transcriptome signature corresponding to ectoderm commitment and epidermal reconstitution. Strikingly, spheroid-derived NHKc (SD-NHKc) expressed marked induction of stem cell reprogramming factors and readily restored K14/P63 double-positive colonies that maintained enhanced regenerative properties. Our findings present NHKc 3-D spheroid cultivation and spheroid re-plating as an effective method for epidermal skin stem/progenitor cell enrichment from neonatal human skin and can be employed as an excellent cell-based model system to study complex cellular processes, such as tissue repair and cancer, as well as a variety of dermatological ailments in primary ex-vivo cultures.
2. Materials and methods
2.1. Cell culture
NHKc were isolated from neonatal foreskin as previously described (Akerman et al., 2001) and cultured according to Woappi et al. (Woappi et al., 2018). Briefly, KSFM media (Thermo Fisher Scientific Waltham, MA) was supplemented with 1% PenStrep/10 mg/ml gentamycine, 50 μg/mL bovine pituitary extract, 20 ng/ml EGF, 10 ng/ml basic fibroblast growth factor, 0.4% bovine serum albumin (BSA), and 4 μg/ml insulin. This cell culture medium is referred to as KSFM-stem cell medium (KSFM-scm). The normal adult human skin cell line L5Ep was obtained from the BWH Cell Culture Core.
2.2. 3D spheroid formation assay
Cultured NHKc (2x104 cells) were seeded into a 96-well roundbottom plate coated with a polymerized mixture of agarose (1.4%) and KSFM-scm. Cells were maintained in suspension for 24–96 h until they self-aggregated into spheroids. Multicellular epidermal spheroids were plated back into 2-D monolayer culture after 48 h and spheroid-derived (SD) cells were left to proliferate. Proliferating SD-NHKc were trypsinized, tested again for spheroid formation, then newly formed spheroids were again placed into 2-D monolayer culture to produce secondary SD-NHKc cultures. Cell size and morphology was determined using Lumenera Infinity 1 software (Lumenera Corporation, Ottawa, ON).
2.3. 3D organotypic cell culture assay
The method for organotypic culture was adapted from Dongari-Bagtzoglou and Kashleva (Dongari-Bagtzoglou and Kashleva, 2006). Briefly, seeding rafts were assembled by mixing Culturex 3-D Matrix (Sigma-Aldrich St. Louis, MO) with 4 × 105 NIH 3 T3 fibroblasts (1:3, v/v) in 200 μL/well of DMEM/10% FBS, in 6 well plates with Transwell culture inserts (VWR) and incubated at 37oC for 2 h. Dermal gel rafts were equilibrated after 24 h using 4 mL complete DMEM/10% FBS at 37oC until gels contracted. Medium was then withdrawn and 1 × 106 keratinocytes in 200 μL of complete KSFM were seeded on top of the raft and incubated for 2 h at 37 °C. Subsequently, skin equivalent on top of the insert was covered with complete KSFM, while DMEM/10% FBS filled the bottom of the well, coming in contact with the bottom surface of the skin equivalent. Plates were cultivated for 7 days—changing the medium every 2 days. At day 7, medium was withdrawn from the top and skin equivalents lifted to air–liquid interface, with KSFM/5% FBS/ 1mM calcium chloride at the bottom of the inserts, for 16 more days at 37 °C, changing the medium every other day. At day 16, skin equivalents were processed for H&E staining.
2.4. Low density growth assay
Cells were plated at low density (10,000–20,000 cells/dish) into duplicate 100-mm dishes, and fed KSFM-scm every 4 days, until ~ 25% confluence, then fed every 2 days. Cultures were serially passaged (1:100) in 100-mm dishes until cell proliferative capacity was exhausted. Cell numbers and viability were determined using Countess Automated Cell Counter (Invitrogen, CA). Cumulative population doublings were calculated according to the formula: population doublings = log(N/N0) / log2, where N represents the total cell number obtained at each passage and N0 represents the number of cells plated at the beginning of the experiment (Ma et al., 2015). Duplicate dishes were stained with 10% Giemsa to assess colony size and morphology.
2.5. Colony-forming efficiency assay
Spheroid non-forming NHKc (NF-NHKc) and spheroid forming NHKc (SF-NHKc) cultures were plated at 10,000 cells/dish in duplicate wells of a 6-well plate and fed once with 8 mL of medium, then incubated for 10–20 days. Cells were fixed with methanol, then stained with 10% Giemsa. Colony forming efficiencies (C.F.E) were calculated as colony number per dish divided by the original number of cells seeded. For 2-D-attached spheroid culture, C.F.E was determined by obtaining the ratio of colonies generated from each seeded spheroid to the original number of cells contained within the seeded spheroid.
2.6. Real-time PCR
Total RNA was isolated from cells using the All Prep DNA/RNA Mini Kit (Qiagen, CA) according to the manufacturer’s protocol. Reverse transcription was carried out with 1 μg of total RNA using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad) following the manufacturer’s instructions. Amplicon products were validated by agarose gel electrophoresis (2% v/v). GAPDH was used as an internal control. All samples were assayed in triplicate. The primer sequences used for real-time PCR are shown in Supplementary Table 1. We considered “Edge” cells to be all cells contained within the compact spheroid ring and “outside” cells those floating around the compact spheroid ring, completely detached from the spheroid structure. Each group was isolated separately for PCR analysis.
2.7. Immunohistochemistry
Cells were grown on coverslips coated with poly-lysine until 75% confluent. The cells were then washed twice in ice-cold phosphate buffered saline (PBS), fixed with 4% paraformaldehyde for 20 min at room temperature, permeabilized with 0.5% Triton in 1% glycine and then blocked using 0.5% BSA and 5% goat serum for 30 min at room temperature. Samples were then incubated with antibodies against P63 (Thermo Scientific, catalog # 703809, 1:200 dilution), and cytokeratin 14 (Santa Cruz Biotechnology, catalog #sc-53253, 1:200 dilution) in blocking solution overnight at 4 °C. Samples were then washed three times with PBS containing Tween 20 (PBST), followed by incubation with FITC-and Alexa 568- conjugated secondary antibodies (Invitrogen, 1:1000 dilution). Nuclei were also stained with 4', 6-diamidino-2-phenylindole (DAPI) (Invitrogen, 1:5000 dilution) before cells were mounted. Cells were observed using a Nikon Eclipse E600 microscope and a Zeiss confocal laser-scanning microscope.
2.8. Cell lineage tracing assay
2-D monolayer cultures were transfected with 1 μg the pMSCV-IRES-EGFP plasmid vector carrying the enhanced green fluorescent protein (eGFP) gene using the Promega TransFast kit (Madison, WI). Cells were trypsinized and seeded (2x104 cells) as 3-D spheroid suspension cultures. After 24 h eGFP-expressing spheroids were plated into monolayer 2-D cell culture. Cells were tracked with a Zeiss Axionvert 135 fluorescence microscope using Axiovision Rel. 4.5 software. The number of eGFP-expressing cells was determined by quantifying a calibrated pixel-by-pixel ratio between the green fluorescent image channel and the phase contrast image using ImageJ software.
2.9. Fluorescent activated cell sorting analysis
Cells (2–4 × 106 cells/ml) were stained with FITC-conjugated anti-integrinα6 (Abcam, Cambridge, MA; catalog number ab30496) and PE conjugated anti-EGFR (BD Pharmingen San Jose, CA; catalog number 555997) both at 1:50 dilutions. Flow cytometry analysis was performed using a BD FACSAria II flow cytometer (BD Biosciences, San Jose, CA).
2.10. Microarray processing and analysis
Total RNA was isolated from mass cultures of a single NHKc donor and spheroids derived from the same donor, in triplicate, using the Qiagen RNeasy Plus Micro Kit, according to the manufacturer’s protocol. RNA quality was assessed using an Agilent 2100 Bioanalyzer and RNA Integrity Numbers (RIN) ranged from 9.0 to 9.1. Microarray experiments were performed using the Affymetrix platform. Total RNA samples were amplified and biotinylated using GeneChip WT PLUS Reagent Kit (Affymetrix/Thermo Fisher Scientific). Briefly, 100 ng of total RNA per sample was reverse transcribed into ds-cDNA using NNN random primers containing a T7 RNA polymerase promoter sequence. T7 RNA polymerase was then added to cDNA samples to amplify RNA, and then RNA was copied to ss-cDNA and degraded using RNase H. ss-cBNA molecules were then fragmented and labelled with biotin. Amplified and labeled samples were hybridized to GeneChip HuGene 2.0 ST Arrays (Affymetrix/Thermo Fisher Scientific) for 16 h at 45 °C using a GeneChip Hybridization Oven 640 and a GeneChip Hybridization, Wash, and Stain Kit (Affymetrix/Thermo Fisher Scientific). Hybridized arrays were washed and stained using GeneChip Fluidics Stations 450 (Affymetrix/Thermo Fisher Scientific). Arrays were scanned using a GeneChip Scanner 3000 7G system and computer workstation equipped with GeneChip Command Console 4.0 software (Affymetrix/Thermo Fisher Scientific). Following completion of array scans, probe cell intensity (CEL) files were imported into Expression Console Software (Affymetrix/Thermo Fisher Scientific) and processed at the gene-level using Affymetrix’s HuGene-2_0-st library file and Robust Multichip Analysis (RMA) algorithm to generate CHP files. After confirming data quality within Expression Console, CHP files containing log2 expression signals for each probe were then imported into Transcriptome Analysis Console Software version 3.0.0.466 (Affymetrix/Thermo Fisher Scientific) to analyze cell type specific transcriptional responses using one-way between-subject analysis of variance.
2.11. Statistical analysis
Data were expressed as the mean ± standard deviation (SD). Differences between mean values were analyzed using Student’s t-test. P < 0.05, P < 0.01 or P < 0.001 were considered statistically significant and indicated in the figures by *, ** or *** respectively.
3. Results
3.1. NHKc isolates derived from neonatal skin epidermis display autonomous spheroid self-assembling ability in 3-D suspension culture
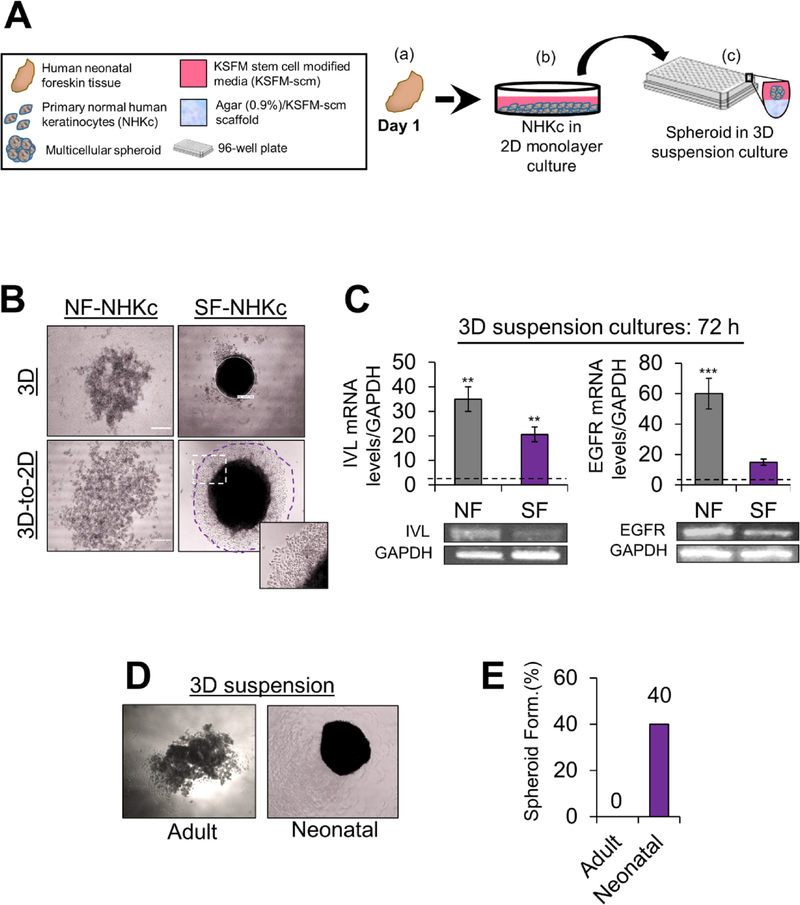
To investigate the anchorage-independent growth potential of NHKc isolates, NHKc cultures derived from skin explants of various donors were seeded into 96-well round-bottomed plates coated with a semisolid polymerized mixture of agarose and KSFM-scm (Fig. 1A). In this 3-D condition, cells grow as self-aggregating suspension spheroids of reproducible sizes. Intriguingly, we observed that 3-D suspension cultures obtained from some NHKc isolates displayed an ability to aggregate into multicellular spheroids -spheroid-forming NHKc (SF-NHKc) - while other isolates consistently failed to self-assemble into spheroids throughout their time in suspension culture - spheroid non-forming NHKc (NF-NHKc) (Fig. 1B). Moreover, when transferred back to 2D monolayer culture, SF-NHKc spheroids generated proliferating keratinocytes, while transferring of cells from NF-NHKc suspensions did not yield any proliferating cells (Fig. 1B). To assess the status of cellular differentiation in suspension cultures, we measured mRNA levels of the late-differentiation gene involucrin (IVL) as well as the EGF receptor (EGFR) in SF-NHKc and NF-NHKc isolates cultivated in 3-D suspension for 72 h. We found that suspension cultures from NF-NHKc and SF-NHKc exhibited a 35-fold and 21-fold respective increase of IVL mRNA, relative to adherent monolayer mass cultures (Fig. 1C). This marked upregulation of involucrin mRNA expression indicate that suspension culture considerably stimulated terminal differentiation in both NF-NHKc and SF-NHKc. In contrast, EGFR mRNA levels exhibited a 45-fold greater increase in NF-NHKc compared to SF-NHKc, relative to levels found in monolayer mass cultures (Fig. 1C). These results indicate that 3-D cultures from SF-NHKc were more resistant to suspension-induced terminal differentiation than 3-D cultures from NF-NHKc.. We also determined that NHKc from an adult skin cell line (4 technical replicates) were incapable of self-aggregating into spheroids during 3-D suspension culture, while 40% (24 of 59 isolates) of neonatal foreskin derived NHKc readily formed spheroids in 3-D culture conditions (Fig. 1D, E and Table 1). These findings indicate that some neonatal NHKc isolates can better resist detachment-induced differentiation and detachment-induced cell death (anoikis), and that this phenotype is linked to the developmental stage of the host tissue. These intriguing findings compelled us to conduct more thorough molecular phenotypic characterization of 3-D human neonatal spheroid cultures and their corresponding spheroid-derived cell populations.
Fig. 1. Spheroid formation assay in NHKc isolates derived from different individuals.
(A) Schematic overview of the epidermal spheroid formation assay. NHKc were isolated from (a) fresh neonatal foreskin explants and (b) cultivated in 100-mm dishes, then passaged into (c) 96-well round-bottomed plates coated with a polymerized mixture of agarose and KSFM-scm. Seeded cells then generate single suspension cultures of reproducibly sized spheroids in each well. (B) 2 × 104 NHKc cultured in stem cell media were seeded onto soft agarose cushions coating individual wells of a 96-well plate. The ability for spheroid formation was assessed over the course of 144 h. Aggregation of NHKc suspensions from (left) spheroid non-forming (NF-NHKc) and (right) spheroid-forming (SF-NHKc) isolates occurred in 3-D suspension. (Bottom) 3-D cell suspensions were then transferred to 2-D monolayer culture after 48 h. Purple circle demarks propagating spheroid-derived cells captured by Nikon TMS phase microscope using Infinity 1 Analyze Software. (C) Expression levels of (Left) involucrin and (Right) EGF receptor (EGFR) mRNA, in NF-NHKc and SF-NHKc cells after 72 h in suspension culture as determined by reverse transcriptase real-time PCR relative to monolayer mass cultures. Dotted lines represent mRNA expression levels in monolayer mass cultures. (Bottom) amplicon products of each real time PCR reaction was run on a 2% agarose gel and observed by ethidium bromide staining. Data were normalized to GAPDH mRNA levels expression. Bars indicate standard deviation, and *, **, and *** indicate statistically significant P values ≤ 0.05, 0.01 and 0.001, respectively. (D) 3-D suspension cultures of NHKc isolated from (left) adult skin and (right) neonatal skin and (E) their respective spheroid forming efficiencies.
Table 1.
Individual NHKc strains assayed for spheroid-forming ability in 3-D culture.
| SF-NHKc | NF-NHKc | |
|---|---|---|
| Number of samples | 24 | 35 |
| Percent | 40% | 60% |
n = 59.
3.2. Suspension spheroid cultures display orchestrated cell kinetics during 3D growth
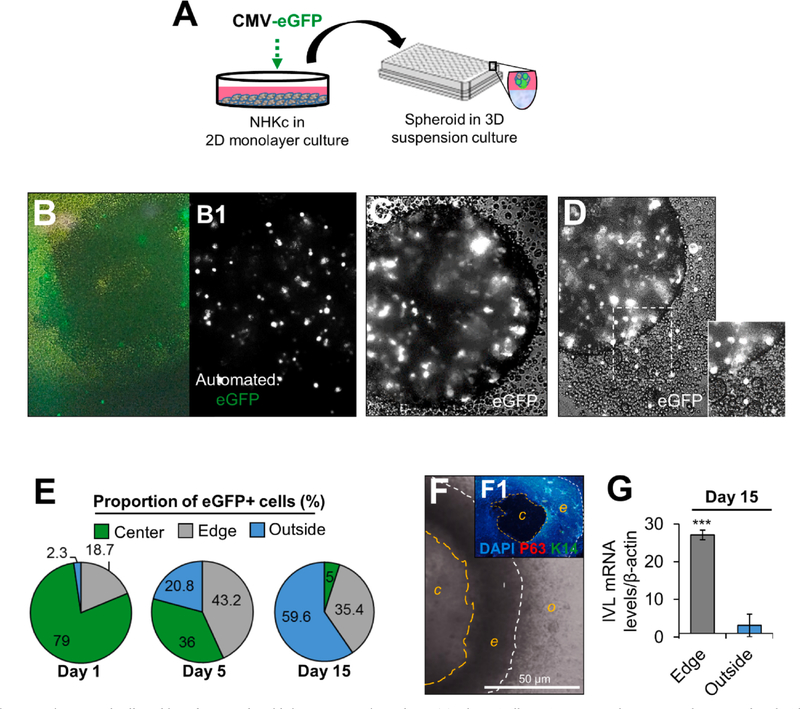
To determine the movement kinetics of spheroid-forming cells during their cultivation in suspension, monolayer-cultured SF-NHKc were transfected with a plasmid transiently expressing enhanced green fluorescent protein (eGFP) and seeded in 3-D agar suspension (Fig. 2A–B1). Interestingly, we observed that most cells progressively aligned along the spheroid edge within 6 h, then some gradually began migrating out of the aggregated spheroid (Fig. 2C, D). By tracking the movement of eGFP-expressing spheroid cells, we quantified cell proportions present in different regions of the spheroid throughout its time in suspension. We observed that while the spheroid is full of cells at the start of the assay, the center is progressively hollowed over time, leaving behind a compact aggregate of cells in the shape of a ring. However, cells located outside of the spheroid increased in number by 10-fold. By day 15, over 59% of all eGFP-labelled spheroid cells were located outside of the spheroid (Fig. 2E), while cells remaining within the spheroid formed a dense hollow ring of cells (Fig. 2F, F1; Supplemental Fig. 1). The basal markers P63 and cytokeratin 14 (K14) were undetectable within the dense hollow ring (Fig. 2F, F1). mRNA for the terminal differentiation marker IVL was increased 27.1-fold in cells of the dense hollow ring, while cells present outside the ring expressed reduced levels of IVL (Fig. 2G). These results indicate that the dense ring of cells was largely composed of terminally differentiated progenies, while cells that traveled outside were more resistant to differentiation.
Fig. 2. Spatiotemporal cell tracking of NHKc spheroids in 3-D suspension culture.
(A) Schematic illustrating 2-D monolayer NHKc cultures transfected with an eGFP reporter plasmid and employed for epidermal spheroid formation assay. (B) Manual image of eGFP-expressing SF-NHKc after 6 h in 3-D suspension culture. (B1) Automated image of eGFP-tagged NHKc suspensions (white dots) at 6 h. (C) Bright phase image of eGFP-expressing keratinocytes within a multicellular spheroid after 24 h in suspension culture. (D) eGFP-labeled cells are seen out of a spheroid after 36 h in suspension culture. (E) Fluorescent tracking of eGFP-labelled cells after 15 days in suspension culture. The proportion of eGFP-expressing cells in each spheroid compartment (i.e. center, edge, or outside) was quantified at distinct time points by ImageJ analysis. (F) Compact ring of cells generated from a single spheroid after 15 days in suspension. White contour lines delineate the outer edge of the spheroid (o); orange contour lines delineate the spheroid’s center (c) from the spheroid’s inner edge (e). (F1) Dense ring of cells immunostained with DAPI, antibodies against pan-tumor protein 63 targeting all TP63 isoforms (P63, red), and basal cytokeratin 14 (K14, green). Scale bar = 50 μM. (G) Expression levels of mRNAs encoding involucrin in cells within the dense floating ring compared to cells located outside the ring, relative to corresponding monolayer mass cultures as determined by Real-time RT-PCR. Data were normalized to GAPDH expression and reported as mean +/- standard deviation.
3.3. 2-D Time-lapse imaging of spheroid-derived cells reveals dynamic cellular behavior upon transfer to monolayer culture
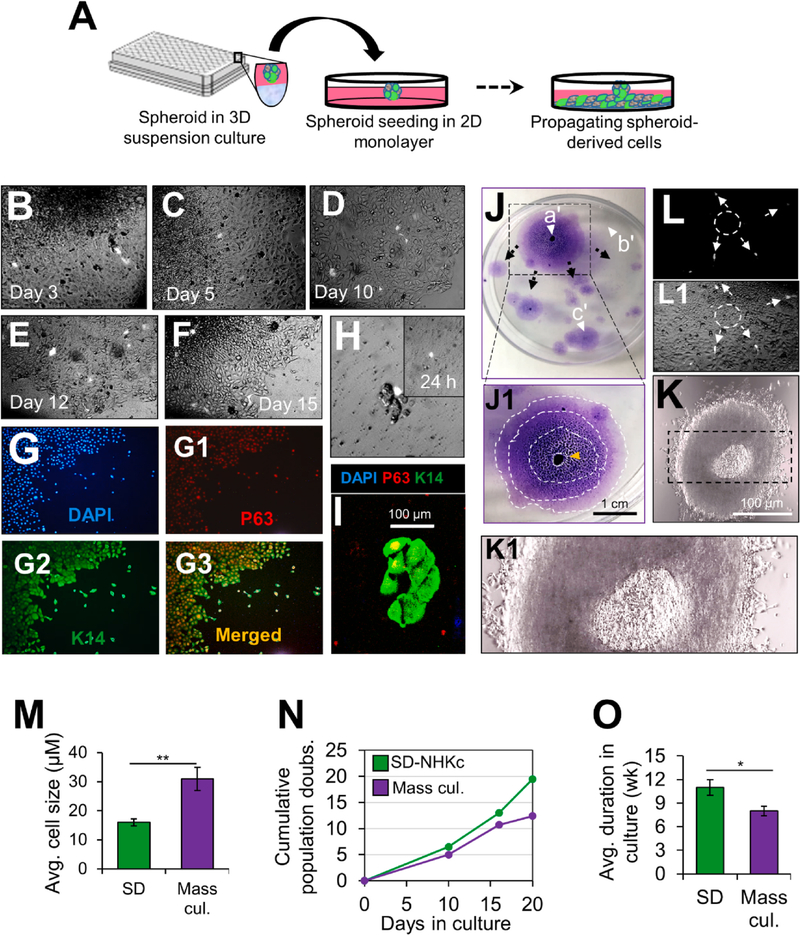
To gain further insight into the clonal fate of spheroid-derived cells, a single eGFP-expressing spheroid was transferred into an empty 2-D polystyrene cell culture dish and movement of eGFP-positive SD cells (SDGFP) was tracked over several days (Fig. 3A). We found that the center of the 2-D plated spheroid became necrotic (Fig. 3J–J1), while the surviving lower layer of epithelial cells grew out. Similar to patterns observed during suspension culture, SDGFP migrated outwardly into empty spaces around the dish, generating monolayer sheets along their paths (Fig. 3B–F). Notably, within 12 days in culture, SDGFP were aligned along the edge of colonies, on the side most distal to the adherent spheroid, and progressively expanded the colony’s surface area (Fig. 3E, F). Dual immunostaining of cells found along the colony edges revealed that they intensely expressed K14 and nuclear P63 (Fig. 3G–G3). When tracking a single SDGFP in culture, we found that it could regenerate a dividing basal K14+/P63+ colony (Fig. 3H, I). Given that SD progenies played such a direct role in colony renewal, we aimed to further characterize their colony regenerative patterns. Colony formation analysis demonstrated that SD cells migrated in a centrifugal configuration to form colonies around the distal spheroid focal point (Fig. 3J, J1). Notably, areas of the dish with confined space were void of colonies (Fig. 3J), suggesting that detection of empty open space triggered epidermal renewal by SD cells, typical of re-epithelization patterns following wound repair. Strikingly, the configuration of multicellular spheroids after their attachment in 2-D monolayer culture was reminiscent of the organization of the stratified epidermis, with the 2-D attached spheroid recapitulating features of a cornified envelope and the propagating epidermal cell sheets underneath behaving as the basal layers (Fig. 3J–K1). Moreover, secondary SD-NHKc cultures accumulated more population doublings (PD) and displayed small-sized cells that could be subcultivated for over 10 weeks as monolayers (Fig. 3M–O). These observations indicate that SD-NHKc are a proliferating population of basal keratinocytes, with characteristics of stem and transit-amplifying cells (TAC) of the skin epidermis.
Fig. 3. 2-D Time-lapse imaging of spheroid-derived cells in 2-D monolayer culture.
(A) Schematic illustrating an eGFP-positive NHKc spheroid plated into 2-D monolayer culture as part of the epidermal spheroid re-playing assay. (B-E) Monitoring eGFP-labelled spheroid-derived (SD) cells moving from a spheroid over the course of 12 days in monolayer 2-D culture. (F) SDEGFP cells positioned along the edge of monolayer sheets by day 15. (G) SD-NHKc located along the edge of monolayer sheets were immunostained against DAPI (blue) and antibodies against (G1) P63 (red), (G2) K14 (green), and (G3) overlaid image. (H) A single SDGFP cell forming a dividing colony after 10 days in 2-D adherent culture. (I) Immunostaining of a SD-NHKc colony with antibodies against K14 (green), P63 (red), and DAPI (blue). (J) Colony renewal visible by Giemsa staining 20 days after a single spheroid is plated back into 2-D monolayer culture. Black arrows depict the radial migratory patterns of SD cells as they travel from (a’) the adherent spheroid to (b’-c’) open spaces throughout the dish. (J1) Configuration mapping of progressively expanding epidermal sheets from the attached spheroid. Contour lines delineate each new sheet layer. Orange arrow indicates the location of the 2-D attached spheroid. (K) Bright-field microscopic image of an adherent spheroid 15 h after attachment in 2-D culture. (K1) Magnified image depicting epidermal sheets forming underneath the adherent spheroid. (L-L1) Automated image of eGFP-tagged SD cells (pointed by white arrows) after > 10 days in monolayer 2-D culture. The white circle demarks the original location of the 2D attached spheroid. (M) Mean cell size of SD-NHKc at passage 3 and their mass cultured counterparts at corresponding passages as quantified by a Nikon TMS phase microscope using Infinity 1 Analyze Software. (N) Low density growth assay assessing the cumulative population doublings of adherent SD-NHKc and their monolayer mass cultures over the course of 20 days. (O) Survival time of SD-NHKc cells and their corresponding mass cultures in monolayer cultures by weeks.
3.4. Spheroid re-plating assay activates regenerative gene programs
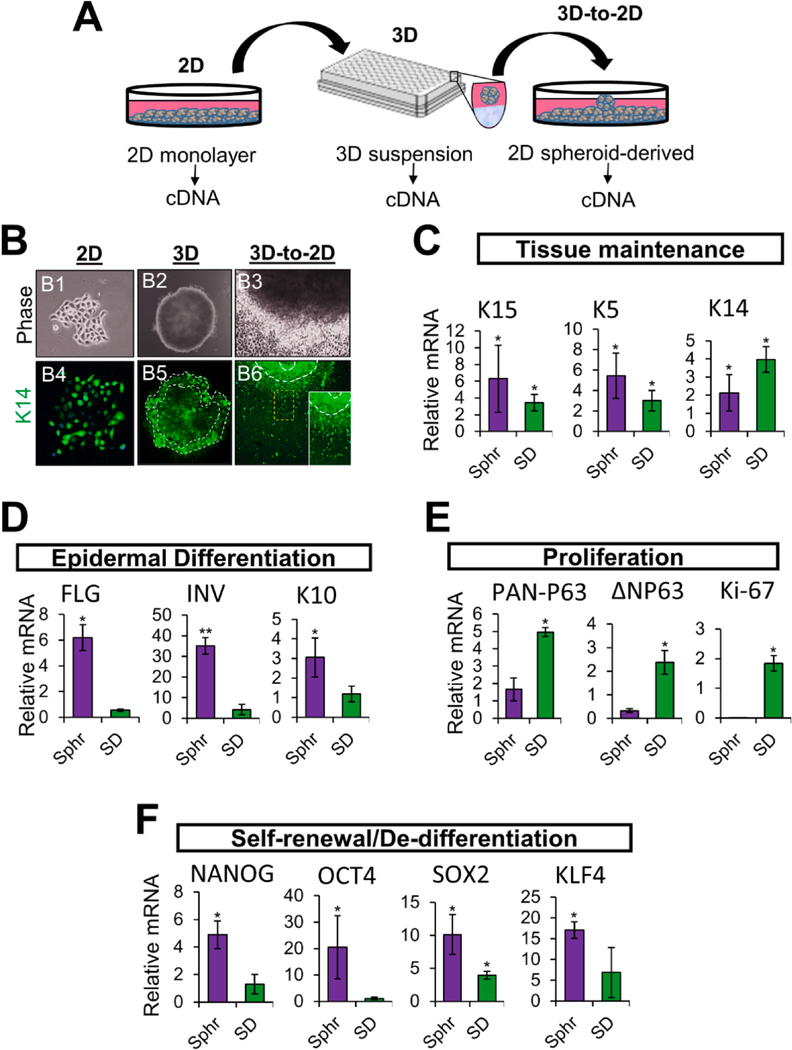
To gain further transcriptional insight into the lineage and differentiation fates of NHKc during suspension growth culture, we studied the expression of basal and suprabasal epidermal genes in 3-D spheroid suspensions and their corresponding SD-NHKc cultures, relative to primary 2-D monolayer cultures (Fig. 4A). First, we investigated the cytoplasmic intensities of basal K14 in NHKc during all three stages of the epidermal spheroid formation assay (Fig. 4B), i.e. 2-D monolayer mass culture, 3-D spheroid suspension culture, and 3-D-to-2-D generated SD-NHKc cultures. We found that NHKc colonies isolated from fresh skin explants expressed intense levels of K14 (Fig. 4B1, B4). K14 expression was also detected during 3-D spheroid culture, particularly in cells found along the edge of the suspension spheroid (Fig. 4B2, B5). Furthermore, when transferred into 2-D monolayer plastic culture, proliferating SD-NHKc located along the spheroid’s edge also maintained intense levels of cytokeratin 14 (Fig. 4B3, B6). These data revealed that despite the induction of suprabasal markers seen in spheroid suspensions, a fraction of basal K14+ keratinocytes are continuously maintained throughout the spheroid formation assay. Real time RT-PCR analysis revealed that spheroid cells exhibit a 2.12-fold increase in mRNAs encoding K14, while SD cultures expressed a 3.97-fold increase, in comparison with mass-cultured NHKc (Fig. 4C). Moreover, we observed a 6.29-fold upregulation in mRNAs encoding K15 in spheroid cells, while SD cultures expressed a 3.45-fold increase. Spheroid cells also expressed a 6.44-fold increase in mRNAs encoding K5, while SD cultures expressed a 3.02-fold increase. Notably, spheroid suspensions equally expressed significantly higher levels of the suprabasal epithelial markers filaggrin, involucrin, and K10, as compared to mass cultures and SD-NHKc (Fig. 4D), suggesting that keratinocytes cultivated as suspension spheroids undertake a variety of differentiation commitments, with some cells retaining basal features and others undergoing terminal differentiation. The marked reduction in suprabasal markers seen in SD cells suggest that cells able to proliferate from the spheroid after suspension culture are likely those that halt terminal differentiation processes. To determine how spheroid suspension culture affected epidermal proliferation programs, we measured the mRNA expression levels of pan-P63, delta NP63, and Ki-67 in cells collected at each stage of the spheroid re-plating assay. We found that SD-NHKc expressed a 4.95-fold increase in mRNA encoding pan-P63, a 2.37-fold increase in mRNA encoding delta NP63, and a 1.84-fold increase in mRNA encoding Ki-67 compared to corresponding mass cultures (Fig. 4E). Conversely, spheroid suspensions expressed a mere 1.67-fold increase in pan-P63, a 0.32-fold upregulation of delta NP63, and undetectable levels of Ki-67 (Fig. 4E). These data indicate that cell proliferation is substantially diminished in NHKc suspension cultures, which led us to question whether the reduced gene expression of proliferation markers seen in spheroid cultures was a mere indication of postmitotic cells or was also suggestive of the presence of slow-cycling self-renewing cells. To this aim, we measured relative mRNA expression levels of genes encoding transcription factors involved in self-renewal and found that spheroid cultures expressed a 4.89-fold increase in Nanog, a 20.51-fold increase in OCT-4, a 10.12-fold increase in SOX2, and a 17.10-fold increase in KLF4, whereas these levels decreased dramatically in spheroid-derived cells cultivated as 2D monolayer cultures (Fig. 4F). Taken together, these findings reassert that spheroid formation is largely mediated by basal stem cell populations that readily suspend proliferation programs during suspension culture and reactivate them upon attachment to plastic culture. Furthermore, these findings demonstrate that in addition to inducing keratinocyte differentiation, self-assembling 3-D spheroid culture equally triggers cells to halt differentiation or to initiate de-differentiation programs associated with self-renewal.
Fig. 4. Transcriptional analysis of cells at each phase of the spheroid re-plating assay.
(A) Schematic illustrating the workflow employed to determine the gene expression profile of NHKc at different stages of the epidermal spheroid re-plating assay. (B) Phase contrast images of a (B1) 2-D monolayer mass cultured NHKc colony newly isolated from a skin explant, (B2) its corresponding 3-D suspension spheroid, and (B3) the subsequent spheroid-derived cultures obtained upon transfer of 3-D spheroids into 2-D monolayer culture. (B4-B6) corresponding cultures immunostained against Keratin 14. White contour lines indicate spheroid edge. (C) The expression of mRNAs encoding epidermal basal genes: K15, K5, K14; (D) Suprabasal differentiation genes: filaggrin, involucrin, K10; (E) epidermal proliferation genes: pan-P63, delta Np63, Ki-67; and (F) reprogramming genes: Nanog, Oct4, Sox2, Klf4 in suspension spheroids and SD cultures, relative to corresponding monolayer mass cultures as determined by RT-qPCR.
3.5. 2-D attached spheroids present gene signatures characteristic of epidermal skin formation
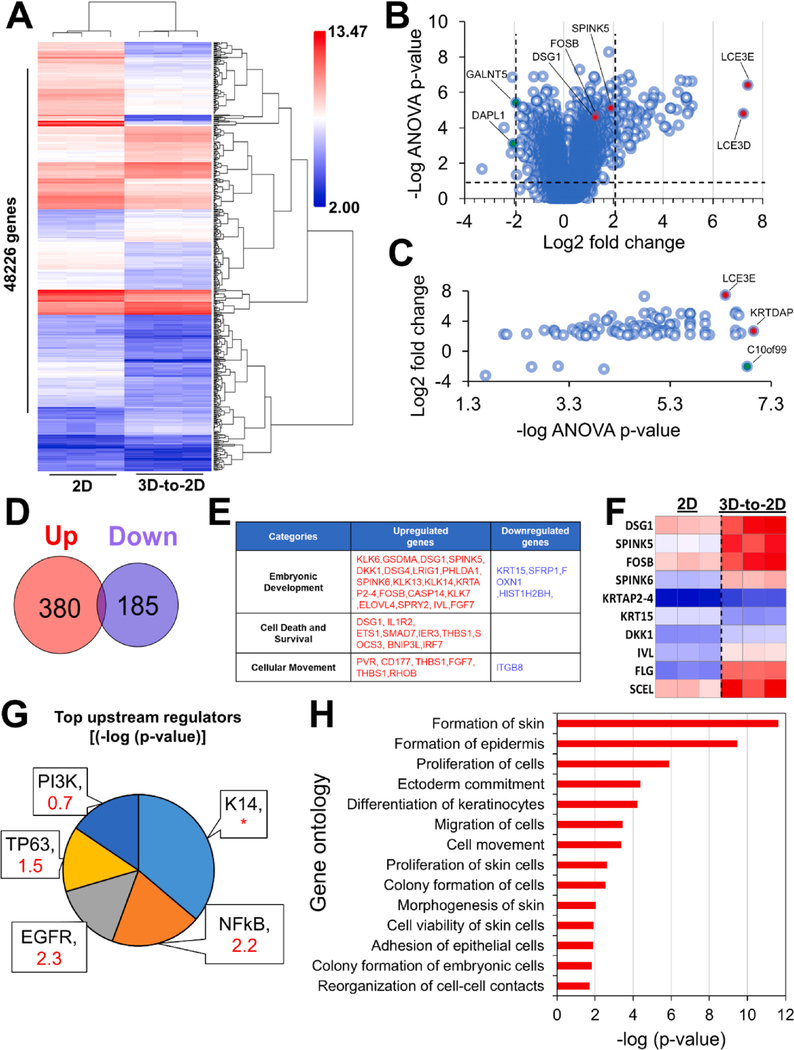
To better understand the transcriptional states and changes occurring in spheroids after their transfer in 2-D culture, we employed microarray analysis to examine gene expression profiles of 3-D-to-2-D transferred spheroids 24 h after attachment in monolayer culture compared to corresponding monolayer mass cultures maintained in 2-D culture throughout the study. Hierarchical clustering analysis detected 565 genes differentially expressed between 2-D adherent spheroids vs. monolayer mass cultures, with 380 genes upregulated and 185 genes downregulated (Fig. 5A–D). Several upregulated genes corresponded to signatures of hair and skin envelope formation, such as the marked induction of sciellin (SCEL), late cornified envelope (LCE3E), and SPINK5 (Fig. 5B, C, F). Furthermore, the profile of upregulated upstream regulators showed a significant induction of epidermal stratification markers, notably EGFR (z-core = 2.2) and P63 (Z-score = 1.5) (Fig. 5G). Signatures of mechanically induced cellular stress were also observed in 3-D-to-2-D transferred spheroids, such as the induction of DSG1 and the upstream activation of NFkB (Fig. 5F–G). Gene Ontology analysis identified several distinct epidermal developmental processes stimulated during spheroid transfer to 2-D monolayer culture, including ectoderm commitment, proliferation of skin cells, and keratinocyte differentiation (Fig. 5H). Taken together, these findings outline the varied transcriptional processes activated in multicellular spheroids upon transfer to 2-D monolayer culture and demonstrate that spheroid transfer to 2-D monolayer culture primes skin cells for epidermal reconstruction.
Fig. 5. Gene expression and gene set enrichment analysis of 2-D attached spheroids and corresponding 2-D monolayer mass cultures.
(A) Unsupervised hierarchical clustering of log-transformed mean-centered mRNA expression levels for all transcripts differentially expressed greater ± 2-fold in 3-D-to-2-D attached spheroids compared to corresponding 2-D monolayer NHKc mass cultures. (B) Volcano plot of differentially regulated genes between 2-D attached spheroids and corresponding monolayer NHKc mass cultures. Vertical dashed lines denote greater ± 2-fold change and horizontal dashed lines denote ANOVA p-value < 0.05. Colored dots indicate top enriched (Red) or depleted (Green) genes with a GO annotation relating to hair formation (note marked depletion of cell death genes such as Death Associated Protein Like 1 (DAPL1). (C) Dot plot of differentially regulated genes with ≥ 2-fold change and = 0.05 ANOVA p-value in 2-D attached spheroids and corresponding monolayer cultures. Colored dots indicate top enriched (Red) or depleted (Green) genes with a GO annotation relating to skin barrier formation (D) Venn diagram depicting global numbers of deregulated and upregulated mRNAs expressed by 2D attached spheroids. (E) Table charting major gene changed in 2-D attached spheroids compared to corresponding monolayer cultures. (F) Heatmap depicting select genes changes implicated in skin formation (G) Pie chart of upstream regulators targeted for activation in 2-D attached spheroids. Pie chart segments are proportional to the -log of the overlap p-value. Respective activation z-score are indicated in red; * indicate undetermined activation pattern. (H) Downstream functional effects of gene transcripts upregulated and downregulated in 2-D attached spheroids (2-fold change FDR < 0.05). Gene Ontology analysis indicates activation of pathways related to cell proliferation and epidermis formation.
3.6. Spheroid-derived cells display enhanced regenerative properties in 3D organotypic skin culture
To investigate the functional output from transcriptional changes taking place in SD-NHKc, we performed skin regeneration assays with SD-NHKc and autologous 2D monolayer cultures using the 3D organotypic skin culture system. Despite the limited stratification of the epithelia in the skin equivalents, we found that SD-NHKc generated significantly thicker epidermal layers than autologous SF-NHKc monolayer cultures (Table 2; Supplemental Fig. 2C–D). Conversely, 3D suspensions from NF-NHKc were incapable of producing an epithelium in organotypic rafts (Table 2; Supplemental Fig. 2A). Interestingly, we did not observe a difference in epidermal regeneration abilities between monolayer cultures of NF and SF NHKc cells (Table 2; Supplemental Fig. 2B–D). Together, these functional studies suggest that SD-NHKc are a population of cells adapted for epidermal reconstruction with a greater capacity for tissue regeneration than cells continuously maintained as 2D monolayer cultures.
Table 2.
Epidermal thickness of skin equivalents established from spheroid-derived NHKc.
| NF-NHKc | NF-NHKc | SF-NHKc | SD-NHKc | |
|---|---|---|---|---|
| Culture condition | 3D | 2D | 2D | 3D-to-2D |
| Thickness (μM) | 6 (± 2.65) | 14.33 (± 9.29) | 23.37 (± 1.48) | 45.03 (± 13.17) |
(±) denotes standard deviation.
3.7. Autonomous 3-D spheroid cultures exhibit transcriptional profiles reminiscent of skin diseases
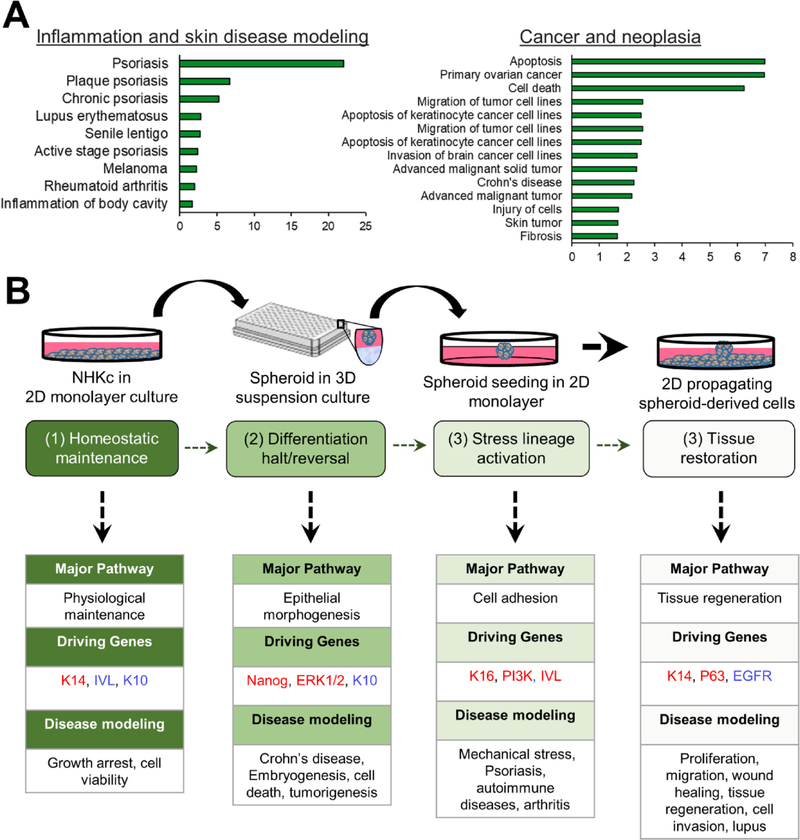
To explore the gene expression profiles of the spheroid-derived cells, we performed Ingenuity Pathway Analysis (IPA) on all genes differentially expressed in 2-D attached spheroids relative to monolayer mass cultures. Our results indicate that these cells have gene expression profiles reminiscent of skin diseases, inflammation, and cancer (Fig. 6A) and share gene changes with various disease/processes involving epithelial cells, in high throughput ex-vivo systems (Fig. 6B).
Fig. 6. Hypothetical modeling of epithelial disease states using the epidermal spheroid re-plating assay.
(A) Tissue abnormalities simulated by the functional effects of gene transcripts upregulated and downregulated in 2D attached spheroids (2-fold change FDR < 0.05) as indicated by Ingenuity Pathway Analysis (IPA). Note skin disease and neoplasia as highly simulated disease types in the spheroid re-plating assay. (B) Schematic of the in vitro epidermal spheroid re-plating assay outlining sequential modeling of the various phases of epidermal tissue regeneration and associated diseases.
4. Discussion
In this study we describe a methodology to cultivate human skin epidermal spheroids in 3-D suspension culture and demonstrate that certain strains of neonatal human keratinocytes are intrinsically resistant to suspension-induced cell death (anoikis). Although we did not find major differences in colony forming efficiencies between freshly-isolated primary cultures from spheroid forming or spheroid non-forming NHKc strains, we found that spheroid-forming ability was generally greater in primary cultures that expressed lower cell surface levels of EGFR. These observations strengthen earlier reports by our group describing interindividual variability in epidermal NHKc EGFR levels (Akerman et al., 2001; Woappi et al., 2018; Bheda et al., 2008), and corroborate several findings depicting EGFRlow expressing NHKc as spheroid-forming keratinocyte stem cells (KSC) (Vollmers et al., 2012; Bheda et al., 2008; Fortunel et al., 2003; Le Roy et al., 2010; Takeuchi et al., 2001; Zyzak et al., 1994; La Fleur et al., 2012; Sun et al., 2005).
Somewhat expectedly, we found that skin from neonatal tissue have a greater ability to generate multicellular spheroids compared to a cell line derived from adult skin, corroborating observations made in equine and mouse epidermis (Borena et al., 2014; Vollmers et al., 2012). Moreover, the epidermal spheroid re-plating assay models the phenomenon of epithelial spread (epiboly, in the embryo) (Lei et al., 2017; Luo et al., 2011) mimicking a more primitive differentiation state. What makes human newborn skin specimens unique for epidermal stem cell studies is their inherently greater reserves of EpSCs and the virtual absence of genomic insults that frequently accumulate in adult tissue (Gago et al., 2009; Pirisi et al., 1988; Van Der Schueren et al., 1980). These attributes allow for more genetically precise investigations of factors driving epidermal cell activation in normal human skin. Interestingly, 2-D attached spheroids equally recapitulated the in vivo spatial orientation of skin development, with basal K14+ cells localized underneath the adherent spheroid and the differentiated cells limited to the superficial surface of the attached spheroid. It is likely that the attachment of spheroids onto a plain 2-D polystyrene cell culture dish is interpreted as a ‘loss of local confluence’ (Guo and Jahoda, 2009; Roshan et al., 2016) by SD-NHKc, thus triggering mechanotransduction resulting in colony initiation and epidermal restoration. Functional studies of SD-NHKc revealed that cells most proximal to the explant were smaller in size, divided more frequently, and stained positively for basal epidermal markers, corroborating similar reports by (Mazzalupo et al., 2002) (Mazzalupo et al., 2002). However, we did not observe that cells distal to the spheroid were much larger or flattened. Contrarily, distal cells also remained small in size and stained positive for keratin 14 and P63, mimicking the wound-edge regeneration dynamics of human skin in vivo. Several of the phenotypic features of SD-NHKc showed great similarities with wound edge keratinocytes (Fitsialos et al., 2007; Patel et al., 2006).
We found that spheroid suspensions maintained elevated expressions of mRNAs encoding pluripotent reprogramming factors, corroborating observations in esophageal and oral keratinocytes 3-D spheroid and organoid cultures (DeWard et al., 2014; Klicks et al., 2019). Gene Ontology analysis also revealed ectodermal commitment as a pathway significantly activated in surface-attached spheroids, indicative of the presence of a primitive cell population extant within neonatal spheroid cultures, while the broad array of gene signatures present within NHKc spheroids reflects a mosaic of cell fate commitments (Mo et al., 2013; Pastrana et al., 2011; Rybak et al., 1813). These findings indicate that spontaneous spheroid formation favors embryonic-like cell de-differentiation in neonatal NHKc (Wong et al., 2019). This process bestows functional regenerative advantages, as displayed by SD-NHKc cells’ greater ability to generate organotypic skin equivalents. The shear tension created during the self-assembling of cells in suspension likely triggers a topologically dependent feedback similar to the one described in skin organoids (Yu et al., 2019; Grebenyuk and Ranga, 2019). Our findings closely resembled the classic proliferative zone maps of 3D non-scaffold spheroids and the epithelial planarization patterns outlined by several investigators (Luo et al., 2011; Costa et al., 2016). This is reminiscent of stretch-induced keratinocyte expansion patterns (Aragona et al., 2020) and reveals that spheroid cultures retain spaciotemporal inclinations that direct cellular fate in culture. It is likely that epidermal stem cells are sensitive to the post-embryonic stage of the tissue of origin. These findings reinforce earlier reports demonstrating that young skin can more quickly repair wounding in a manner consistent with a post-embryonic clock, and not due to healing by wound contraction (Lei et al., 2017; Keyes et al., 2017).
We also observed a cystic conformation in spheroids after several days in suspension. In this conformation, cells undergoing apoptosis were found around the center, while K14+ outer cells encircled the aggregate, similarly to previous reports (Lei et al., 2017; Halpern et al., 1966). It is possible that uncommitted basal stem/progenitor populations present within primary NHKc cultures are signaled to remain in, or revert to, a less differentiated state during suspension culture, while committed progenitors (CP) are directed into terminal differentiation. A precise ratio between EpSCs, early TAC, and CP may therefore be critical in calibrating spheroids’ regenerative potency and lineage choice in culture. Our data add to the growing body of evidence demonstrating that progenitor cells can switch between quiescence, de-differentiation, and activation depending on microenvironmental signals and tissue regenerative needs (Roshan et al., 2016; Clayton et al., 2007; Doupé et al., 2012; Horsley et al., 2008; Tumbar et al., 2004), and corroborate reports that 3-D cultivation of mammalian cells acts as a reprogramming tool capable of reverting differentiated cells to a more primitive state (Borena et al., 2014; Rybak et al., 1813; Liu et al., 2009; Shamir and Ewald, 2014).
5. Conclusion
Taken together, our findings have many implications. First, the complex behaviors exhibited by primary NHKc during suspension culture provide new insight into elements that influence basal cell homeostasis and outlines the broad array of cell plasticity stimulated during keratinocyte spheroid culture. Moreover, most EpSC studies have been conducted in mice with a single genetic background. Whereas studies performed on a variety of human newborn skin are better representative of the genetic diversity found across humans. This may have a more significant impact on mammalian epidermal studies than previously appreciated. For instance, the stark differences seen in spheroid-forming abilities between NHKc strains isolated from different individuals may be evidence of intrinsic, potentially congenital traits influencing person-to-person basal stem cell densities or propensity for basal stem/progenitor cell activation. 3-D epidermal spheroid re-plating assay using neonatal keratinocytes also presents an opportunity for numerous cell-based study of several dermatological ailments including psoriasis, alopecia, wound healing, and squamous cell carcinoma (Ratushny et al., 2012). For example, multifaceted wound repair modeling may be achieved by co-culturing of epidermospheres with various populations of cells found in the tissue microenvironment, such as tissue-resident immune cells, then performing epidermal spheroid re-plating assays with them. This approach might enable multiplexed modeling of the regulatory impact of specific immune cell populations on epidermal keratinocytes at various stages of wound healing or psoriasis, which is largely driven by immune dysregulation of epidermal stem cells (Li et al., 2020). Moreover, the epidermal spheroid re-plating assay enables rapid assessment of neoplastic events such as colorectal cancers (Costa et al., 2016) and HPV-mediated transformation (Woappi et al., 2018). As such, the 3D-to-2D re-plating phase of the assay, which models stress cell lineage activation, has served as an experimental setpoint at which to introduce oncogenic HPV16 DNA, advancing our understanding of the link between keratinocyte stem cells and individual HKc strain responsiveness to HPV-mediated transformation (Woappi et al., 2018). Nevertheless, additional studies are required to assess the usefulness of this system in multiple disease types, and to understand the genetic and signaling networks regulating pathologies driven by aberrant basal stem/progenitor cell activation.
Supplementary Material
Acknowledgements
Microarray experiments and data analysis were performed at the UofSC College of Pharmacy Microarray Core Facility (MCF). The UofSC School of Medicine Instrumentation Resource Facility (IRF) provided access to imaging and cell sorting equipment and technical assistance. This work was supported in part by grant 1R21CA201853. The MCF and the IRF receive partial support from NIH grant P20GM103499, SC INBRE. The MCF also receives support from NIH grant P20GM109091. Yvon Woappi was supported in part by NIH grants 2R25GM06652606A1 (PREP) and R25GM076277 (IMSD), and by a Fellowship by the Grace Jordan McFadden Professors Program (GJMP, formerly the African American Professors Program, AAPP) at UofSC. Also, special thanks to Justin Vercellino, Geraldine Ezeka, and Sean Bloos for their technical assistance.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2020.102048.
References
- Green H, 1977. Terminal differentiation of cultured human epidermal cells. Cell 11, 405–416. [DOI] [PubMed] [Google Scholar]
- Wakita H, Takigawa M, 1999. Activation of epidermal growth factor receptor promotes late terminal differentiation of cell-matrix interaction-disrupted keratinocytes. J. Biol. Chem. 274, 37285–37291. [DOI] [PubMed] [Google Scholar]
- Guo A, Jahoda CA, 2009. An improved method of human keratinocyte culture from skin explants: cell expansion is linked to markers of activated progenitor cells. Exp. Dermatol. 18, 720–726. [DOI] [PubMed] [Google Scholar]
- Zong Z, Li N, Ran X, Su Y, Shen Y, Shi CM, Cheng TM, 2012. Isolation and characterization of two kinds of stem cells from the same human skin back sample with therapeutic potential in spinal cord injury. PLoS ONE 7, e50222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma JG, McKenzie IA, Bagli D, Miller FD, 2005. Isolation and characterization of multipotent skin-derived precursors from human skin. Stem Cells 23, 727–737. [DOI] [PubMed] [Google Scholar]
- Borena BM, Meyer E, Chiers K, Martens A, Demeyere K, Broeckx SY, Duchateau L, Spaas JH, 2014. Sphere-forming capacity as an enrichment strategy for epithelial-like stem cells from equine skin. Cell. Physiol. Biochem. 34, 1291–1303. [DOI] [PubMed] [Google Scholar]
- Higgins CA, Richardson GD, Ferdinando D, Westgate GE, Jahoda CA, 2010. Modelling the hair follicle dermal papilla using spheroid cell cultures. Exp. Dermatol. 19, 546–548. [DOI] [PubMed] [Google Scholar]
- Kang BM, Kwack MH, Kim MK, Kim JC, Sung YK, 2012. Sphere formation increases the ability of cultured human dermal papilla cells to induce hair follicles from mouse epidermal cells in a reconstitution assay. J, Invest. Dermatol. 132, 237–239. [DOI] [PubMed] [Google Scholar]
- Vollmers A, Wallace L, Fullard N, Höher T, Alexander MD, Reichelt J, 2012. Twoand three-dimensional culture of keratinocyte stem and precursor cells derived from primary murine epidermal cultures. Stem Cell Rev Rep 8, 402–413. [DOI] [PubMed] [Google Scholar]
- Akerman GS, Tolleson WH, Brown KL, Zyzak LL, Mourateva E, Engin TS, Basaraba A, Coker AL, Creek KE, Pirisi L, 2001. Human papillomavirus type 16 E6 and E7 cooperate to increase epidermal growth factor receptor (EGFR) mRNA levels, overcoming mechanisms by which excessive EGFR signaling shortens the life span of normal human keratinocytes. Cancer Res. 61, 3837–3843. [PubMed] [Google Scholar]
- Woappi Y, Hosseinipour M, Creek KE, Pirisi L, 2018. Stem cell properties of normal human keratinocytes determine transformation responses to human papillomavirus 16 DNA. J. Virol. 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A, Kashleva H, 2006. Development of a highly reproducible three-dimensional organotypic model of the oral mucosa. Nat. Protoc. 1, 2012–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Chua AW, Yang E, Teo P, Ting Y, Song C, Lane EB, Lee ST, 2015. Breast cancer resistance protein identifies clonogenic keratinocytes in human interfollicular epidermis. Stem Cell Res. Ther. 6, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheda A, Creek KE, Pirisi L, 2008. Loss of p53 induces epidermal growth factor receptor promoter activity in normal human keratinocytes. Oncogene 27, 4315–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunel NO, Hatzfeld JA, Rosemary PA, Ferraris C, Monier MN, Haydont V, Longuet J, Brethon B, Lim B, Castiel I, Schmidt R, Hatzfeld A, 2003. Long-term expansion of human functional epidermal precursor cells: promotion of extensive amplification by low TGF-beta1 concentrations. J. Cell Sci. 116, 4043–4052. [DOI] [PubMed] [Google Scholar]
- Le Roy H, Zuliani T, Wolowczuk I, Faivre N, Jouy N, Masselot B, Kerkaert JP, Formstecher P, Polakowska R, 2010. Asymmetric distribution of epidermal growth factor receptor directs the fate of normal and cancer keratinocytes in vitro. Stem Cells Dev. 19, 209–220. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Kato M, Suzuki H, Akhand AA, Wu J, Hossain K, Miyata T, Matsumoto Y, Nimura Y, Nakashima I, 2001. Acrolein induces activation of the epidermal growth factor receptor of human keratinocytes for cell death. J. Cell. Biochem. 81, 679–688. [DOI] [PubMed] [Google Scholar]
- Zyzak LL, MacDonald LM, Batova A, Forand R, Creek KE, Pirisi L, 1994. Increased levels and constitutive tyrosine phosphorylation of the epidermal growth factor receptor contribute to autonomous growth of human papillomavirus type 16 immortalized human keratinocytes. Cell Growth Differ. 5, 537–547. [PubMed] [Google Scholar]
- La Fleur L, Johansson AC, Roberg K, 2012. A CD44high/EGFRlow subpopulation within head and neck cancer cell lines shows an epithelial-mesenchymal transition phenotype and resistance to treatment. PLoS ONE 7, e44071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Goderie SK, Temple S, 2005. Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron 45, 873–886. [DOI] [PubMed] [Google Scholar]
- Lei M, Schumacher LJ, Lai Y-C, Juan W-T, Yeh C-Y, Wu P, Jiang T-X, Baker RE, Widelitz RB, Yang L.i., Chuong CM., 2017. Self-organization process in newborn skin organoid formation inspires strategy to restore hair regeneration of adult cells. PNAS 114 (34), E7101–E7110. 10.1073/pnas.1700475114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Yufit T, Carson P, Fiore D, Falanga J, Lin X, Mamakos L, Falanga V, 2011. Differential keratin expression during epiboly in a wound model of bioengineered skin and in human chronic wounds. Int J Low Extrem Wounds 10, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago N, Pérez-López V, Sanz-Jaka JP, Cormenzana P, Eizaguirre I, Bernad A, Izeta A, 2009. Age-dependent depletion of human skin-derived progenitor cells. Stem Cells 27, 1164–1172. [DOI] [PubMed] [Google Scholar]
- Pirisi L, Creek KE, Doniger J, DiPaolo JA, 1988. Continuous cell lines with altered growth and differentiation properties originate after transfection of human keratinocytes with human papillomavirus type 16 DNA. Carcinogenesis 9, 1573–1579. [DOI] [PubMed] [Google Scholar]
- Van Der Schueren B, Cassiman JJ, Van Den Berghe H, 1980. Morphological characteristics of epithelial and fibroblastic cells growing out from biopsies of human skin. J, Invest. Dermatol. 74, 29–35. [DOI] [PubMed] [Google Scholar]
- Roshan A, Murai K, Fowler J, Simons BD, Nikolaidou-Neokosmidou V, Jones PH, 2016. Human keratinocytes have two interconvertible modes of proliferation. Nat. Cell Biol. 18, 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzalupo S, Wawersik MJ, Coulombe PA, 2002. An Ex Vivo Assay to Assess the Potential of Skin Keratinocytes for Wound Epithelialization. J, Invest. Dermatol. 118, 866–870. [DOI] [PubMed] [Google Scholar]
- Fitsialos G, Chassot AA, Turchi L, Dayem MA, LeBrigand K, Moreilhon C, Meneguzzi G, Buscà R, Mari B, Barbry P, Ponzio G, 2007. Transcriptional signature of epidermal keratinocytes subjected to in vitro scratch wounding reveals selective roles for ERK1/2, p38, and phosphatidylinositol 3-kinase signaling pathways. J. Biol. Chem. 282, 15090–15102. [DOI] [PubMed] [Google Scholar]
- Patel GK, Wilson CH, Harding KG, Finlay AY, Bowden PE, 2006. Numerous keratinocyte subtypes involved in wound re-epithelialization. J, Invest. Dermatol. 126, 497–502. [DOI] [PubMed] [Google Scholar]
- DeWard AD, Cramer J, Lagasse E, 2014. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep 9, 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klicks J, Maßlo C, Kluth A, Rudolf R, Hafner M, 2019. A novel spheroid-based coculture model mimics loss of keratinocyte differentiation, melanoma cell invasion, and drug-induced selection of ABCB5-expressing cells. BMC Cancer 19, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J, Sun B, Zhao X, Gu Q, Dong X, Liu Z, Ma Y, Zhao N, Liu Y, Chi J, Sun R, 2013. The in-vitro spheroid culture induces a more highly differentiated but tumorigenic population from melanoma cell lines. Melanoma Res. 23, 254–263. [DOI] [PubMed] [Google Scholar]
- Pastrana E, Silva-Vargas V, Doetsch F, 2011. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 8, 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak AP, He L, Kapoor A, Cutz JC, Tang D, 1813. Characterization of sphere-propagating cells with stem-like properties from DU145 prostate cancer cells. Biochim. Biophys. Acta, Mol. Cell. Biol. Lipids 2011, 683–694. [DOI] [PubMed] [Google Scholar]
- Wong CW, LeGrand CF, Kinnear BF, Sobota RM, Ramalingam R, Dye DE, Raghunath M, Lane EB, Coombe DR, 2019. In Vitro Expansion of Keratinocytes on Human Dermal Fibroblast-Derived Matrix Retains Their Stem-Like Characteristics. Sci. Rep. 9, 18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Hunziker W, Choudhury D, 2019. Engineering Microfluidic Organoid-on-a-Chip Platforms. Micromachines (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenyuk S, Ranga A, 2019. Engineering Organoid Vascularization. Front. Bioeng. Biotechnol. 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa EC, Moreira AF, de Melo-Diogo D, Gaspar VM, Carvalho MP, Correia IJ, 2016. 3D tumor spheroids: an overview on the tools and techniques used for their analysis. Biotechnol. Adv. 34, 1427–1441. [DOI] [PubMed] [Google Scholar]
- Aragona M, Sifrim A, Malfait M, Song Y, Van Herck J, Dekoninck S, Gargouri S, Lapouge G, Swedlund B, Dubois C, Baatsen P, Vints K, Han S, Tissir F, Voet T, Simons BD, Blanpain C, 2020. Mechanisms of stretch-mediated skin expansion at single-cell resolution. Nature 584, 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes BE, Liu S, Asare A, Naik S, Levorse J, Polak L, Lu CP, Nikolova M, Pasoli HA, Fuchs E, 2017. Repair in Aged Skin 167, 1323–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern B, Pejsachowicz B, Febvre HL, Barski G, 1966. Differences in patterns of aggregation of malignant and non-malignant mammalian cells. Nature 209, 157–159. [DOI] [PubMed] [Google Scholar]
- Clayton E, Doupé DP, Klein AM, Winton DJ, Simons BD, Jones PH, 2007. A single type of progenitor cell maintains normal epidermis. Nature 446, 185–189. [DOI] [PubMed] [Google Scholar]
- Doupé DP, Alcolea MP, Roshan A, Zhang G, Klein AM, Simons BD, Jones PH, 2012. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science 337, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E, 2008. NFATc1 balances quiescence and proliferation of skin stem cells. Cell 132, 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E, 2004. Defining the epithelial stem cell niche in skin. Science 303, 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Clem B, Zuba-Surma EK, El-Naggar S, Telang S, Jenson AB, Wang Y, Shao H, Ratajczak MZ, Chesney J, Dean DC, 2009. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell Stem Cell 4, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir ER, Ewald AJ, 2014. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 15, 647–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny V, Gober MD, Hick R, Ridky TW, Seykora JT, 2012. From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest 122, 464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Yuan CW, Xu S, Zu T, Woappi Y, Lee CAA, Abarzua P, Wells M, Ramsey MR, Frank NY, Wu X, Mandinova A, Frank MH, Lian CG, Murphy GF, 2020. Loss of the Epigenetic Mark 5-hmC in Psoriasis: Implications for Epidermal Stem Cell Dysregulation. J, Invest. Dermatol. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.