Abstract
Background
Although traditional diagnostic techniques of infection are mature and price favorable at present, most of them are time-consuming and with a low positivity. Metagenomic next⁃generation sequencing (mNGS) was studied widely because of identification and typing of all pathogens not rely on culture and retrieving all DNA without bias. Based on this background, we aim to detect the difference between mNGS and traditional culture method, and to explore the relationship between mNGS results and the severity, prognosis of infectious patients.
Methods
109 adult patients were enrolled in our study in Shanghai Tenth People’s Hospital from October 2018 to December 2019. The diagnostic results, negative predictive values, positive predictive values, false positive rate, false negative rate, pathogen and sample types were analyzed by using both traditional culture and mNGS methods. Then, the samples and clinical information of 93 patients in the infected group (ID) were collected. According to whether mNGS detected pathogens, the patients in ID group were divided into the positive group of 67 cases and the negative group of 26 cases. Peripheral blood leukocytes, C-reactive protein (CRP), procalcitonin (PCT) and neutrophil counts were measured, and the concentrations of IL-2, IL-4, IL-6, TNF-α, IL-17A, IL-10 and INF-γ in the serum were determined by ELISA. The correlation between the positive detection of pathogens by mNGS and the severity of illness, hospitalization days, and mortality were analyzed.
Results
109 samples were assigned into infected group (ID, 92/109, 84.4%), non-infected group (NID, 16/109, 14.7%), and unknown group (1/109, 0.9%). Blood was the most abundant type of samples with 37 cases, followed by bronchoalveolar lavage fluid in 36 cases, tissue, sputum, pleural effusion, cerebrospinal fluid (CSF), pus, bone marrow and nasal swab. In the ID group, the majority of patients were diagnosed with lower respiratory system infections (73/109, 67%), followed by bloodstream infections, pleural effusion and central nervous system infections. The sensitivity of mNGS was significantly higher than that of culture method (67.4% vs 23.6%; P < 0.001), especially in sample types of bronchoalveolar lavage fluid (P = 0.002), blood (P < 0.001) and sputum (P = 0.037), while the specificity of mNGS was not significantly different from culture method (68.8% vs 81.3%; P = 0.41). The number of hospitals stays and 28-day-motality in the positive mNGS group were significantly higher than those in the negative group, and the difference was statistically significant (P < 0.05). Age was significant in multivariate logistic analyses of positive results of mNGS.
Conclusions
The study found that mNGS had a higher sensitivity than the traditional method, especially in blood, bronchoalveolar lavage fluid and sputum samples. And positive mNGS group had a higher hospital stay, 28-day-mortality, which means the positive of pathogen nucleic acid sequences detection may be a potential high-risk factor for poor prognosis of adult patients and has significant clinical value. MNGS should be used more in early pathogen diagnosis in the future.
Keywords: Next-generation sequencing, Sensitivity, Diagnostic, Infection, Survival
Background
Infectious diseases are a leading cause of morbidity and mortality worldwide and spread quickly. As the first-line of pathogen detection, microbiology laboratory plays an important role in infection control by means of microscopic examination, culture, identification, drug sensitivity and so on [1]. However, the limitation of molecular diagnosis and genotyping methods remain that pathogens are undetected in up to 60% of cases [2–4]. Failure to identify pathogens in time may delay the precise treatment of antibiotics, leading to unnecessary use of broad-spectrum antibiotics, inducing resistance, and increasing medical costs [5].
With the completion of the human genome project in the early twenty-first century and the rapid development of sequencing technology, high-throughput and low-cost second-generation sequencing technology emerged [6]. It had been used in whole genome sequencing, whole exome sequencing, macro gene sequencing and so on, among which metagenomic next⁃generation sequencing (mNGS) was studied most widely. The advantage of mNGS lies in the single run to obtain the sequence information of microbial nucleic acid fragments, through analysis and comparison of which to detect all microbial species and sequence [7]. Besides, mNGS can be used for the identification and typing of all pathogens because mNGS does not rely on culture and retrieve all DNA without bias [8]. Based on mNGS results, antimicrobial resistance, virulence, typing and other information can be used for epidemic investigation. It lays a theoretical foundation for the investigation of infectious diseases outbreak in hospital. Therefore, this technology may play a huge role in infection prevention and medical microbiology laboratory.
Thus, based on microbiome sequencing technology, we compared the sensitivity and specificity of mNGS method and traditional culture method to detect pathogens, and discussed the influence of mNGS detection results on the severity and prognosis of patients with infection in our study.
Methods
Study patients
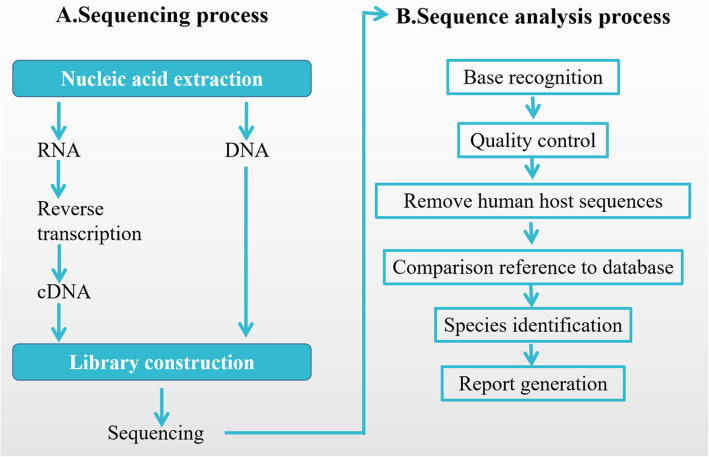
We retrospectively reviewed 161 cases suspected of acute or chronic infection from respiratory and critical care medicine department, geriatric department, emergency intensive care unit and emergency department at Shanghai 10th People’s Hospital in Shanghai, China, between October 2018 and December 2019. Excluding patients with pregnancy, mental illness and under the age of 18, 109 samples were included in our study and for analysis and then they were categorized into 3 groups, infectious disease (ID) group, noninfectious disease (NID) group, and unknown group according to final diagnosis. Specimens were subjected to mNGS testing (BGI, Intertek, Biotecan, China) and regular clinical microbiological assay in a pairwise manner and final diagnosis was determined by clinicians based on both of them and imaging, clinical feature of patients. Meanwhile, clinical data of all enrolled patients, including complete blood count, C-reactive protein (CRP), procalcitonin (PCT), neutrophil count, interleukin (IL)-2, IL-4, IL-6, Tumor Necrosis Factor-α (TNF-α), IL-17A, IL-10 and Interferon-γ (INF-γ) were collected. The flow diagram of cases inclusion and exclusion was shown in Fig. 1. This research had been approved by the ethics committee of the 10th People’s Hospital affiliated to Tongji University (No. SHSY-IEC-4.1/20–21/01).
Fig. 1.

Flow diagram of cases inclusion and exclusion
Metagenomic next-generation sequencing and analysis
Nucleic acid detection and sequencing were performed based on BGISEQ-50 platform (BGI-Tianjin, Tianjin, China) in this research. After the sample was taken, nucleic acid was extracted, the library was built and sequenced, and finally the data was analyzed by using the microbiome database (ftp://ftp.ncbi.nlm.nih.gov/genomes/). The experimental process was shown in Fig. 2.
Fig. 2.
Flow diagram of Metagenomic Next-generation Sequencing and Analysis
Sample processing and library construction (Fig. 2a)
For infected patients or patients with fever of unknown cause, infected site samples or blood were collected according to standard procedures. Each blood, bronchoalveolar lavage Fluid (BALF) or urine sample was at least 5 ml (ml) and at least 3 ml of each sample of cerebrospinal fluid, sputum, or other sterile liquid. Blood must be collected in anticoagulant tube and stored at room temperature, the protective agent in anticoagulant tube is Ethylene Diamine Tetraacetic Acid (EDTA) anticoagulant and special deoxyribonucleic acid (DNA) protective agent. Other samples were collected in sterile tube and stored at − 80 °C. Blood was transported at room temperature, and other sterile samples were transported in drikold. Since most of the collected samples contain pathogenic pathogens, they were inactivated (56 °C, 30 min) before nucleic acid extraction. In addition to this, blood samples were centrifuged to separate plasma and leukocytes when intracellular bacterial infection was particularly suspicious by clinicians. Sputum samples were liquefied by using 0.1% dithiothreitol (DTT) for 30 min at room temperature after inactivation [5]. After that, DNA were extracted by TIANamp Micro DNA Kit (DP316, Tiangen Biotech) according to the manufacturer’s recommendation. DNA libraries were constructed in steps of DNA fragmentation by enzyme digestion, DNA supplementation terminal, dA tail and sequencing common connector connection. The constructed DNA library was used to obtain the sequence data of DNA fragments by gene sequencing instrument, and the results were analyzed by biological information software. Each trial included internal, negative and positive controls. Internal parameters is a specific molecular tag that is placed in the sample before nucleic acid extraction to track the entire process and to control the quality of DNA. The detection results of negative control products should be no pathogens detected. If there are relevant pathogens detected, it indicates that there may be DNA pollution sources in the environment. Positive contained specific microbic DNA.
Bioinformatic analysis (Fig. 2b)
Quality control
A. Sequencing subtracted of human host sequences need to be above 90%; B. Reads of microbial detection sequences need to be longer than 50 bp and the effective sequencing data volume should not be less than 20 M without removing the human genome component.
Data filtering
In order to obtain high quality sequence data, the qualified data was further filtered by bioinformatics analysis to remove low quality sequences. FASTQ format was used for analysis. The initial pretreatment steps include low quality read filtering, low- complexity read filtering and adapter trimming. Host subtraction was performed by mapping to host genome and/or transcriptome. The remaining unmapped reads are aligned directly with large reference databases, such as the National Center for Biotechnology Information (NCBI) GenBank database.
Sequences alignment
The filtered sequences were compared with the reference sequences in the pathogen database, which covers bacteria, fungi, viruses, protozoa and other pathogenic microorganisms. According to the final results of pathogen comparison, all parameters of detected pathogens were calculated, including sequence number, relative abundance, genome coverage and depth, etc.
Report generation
The species listed in the report were all the microorganisms detected in this test. They were classified by bacteria, viruses, fungi, parasites, mycoplasma, chlamydia and rickettsia. They were ranked from high to low according to their reads and the relative content of the former is higher. When the report goes to the clinic, whether the suspected pathogen detected is related to infection from the clinical dimension was judged, and the final diagnosis was determine by combining the detection parameters.
Determination of cytokines
Detection of TNF-a, IL-2, IL-4, IL-6, IL-8, IL-10, IL-17A and INF-r in serum was by solid phase, sandwich and chemiluminescence using the IMMULITE/IMMULIE 1000 analyzer. The analyzer and chemiluminescence kit were both from SIEMENS, Germany. The processed specimens were sent to the analyzer for testing according to the manufacturer’s instructions, and the corresponding cytokine concentrations were recorded.
Cell classification and count detection
Cells were classified using the automatic flow cytometer (Thermo Fisher SCIENTIFIC, American) and divided into total white blood cells, neutrophil count, CD4+ T cell count, CD8+ T cell count, B cells, and NK, T cell count.
Statistical analysis
Comparative analysis was conducted by Pearson χ2 test and t test. Data analysis was performed by using SPSS 22.0 software. P values < 0.05 were considered significant, and all tests were 2-tailed. Logistic regression analysis explored the risk factors associated with positive detection of mNGS.
Results
Sample and patient characteristics
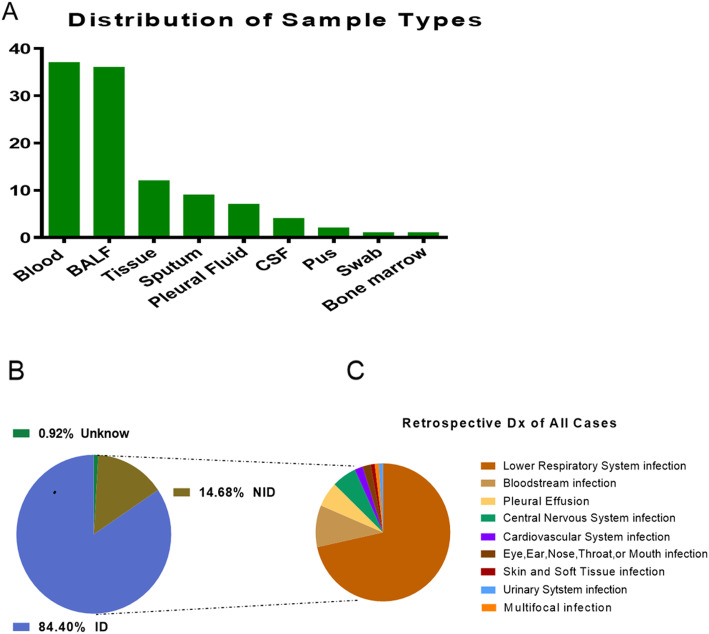
Demographic features of the patients were provided in Table 1. 87 males and 22 females participated in our study, whose average age was 61 years old, average length of stay was 17.5 days and the case fatality rate were 11.9%. Most (37/109, 33.9%) of our samples were from blood, 36 of 109 (33.0%) were from BALF, 12 of 109 (11.0%) were from tissue and 9 (8.3%) of 109 were from sputum, followed by pleural fluid (7, 6.4%), CSF (4, 3.7%), pus (2, 1.8%), bone marrow (1, 0.9%) and nasal swab (1, 0.9%) (Fig. 3a). In the study cohort, 92 (84.4%) patients diagnosed with confirmed pathogens by clinicians were assigned to ID group. The remaining specimens were subdivided into the NID (16/109, 14.7%) and unknown (1/109, 0.9%) groups (Fig. 3b). There were no statistical differences between ID group and NID group in age, gender, length of stay and case fatality rate (p > 0.05 in all). Most patients were diagnosed with respiratory system infections (73/109, 67.0%), followed by bloodstream infections (10/109, 9.17%), pleural effusion (6/109, 5.50%) and central nervous system infections (6/109, 5.50%) as shown in Fig. 3c.
Table 1.
Demographic characteristic of samples
| Total | ID | NID | Unknown | P value between ID & NID | |
|---|---|---|---|---|---|
| Samples amount, n (%) | 109 (100%) | 92 (84.40) | 16 (14.68) | 1 (0.92) | / |
| Age, average years (range) | 61.02 (25–95) | 60.26 (25–95) | 66 (40–90) | 61(/) | 0.43 |
| Gender,male,n (%) | 87 (79.82) | 74 (80.43) | 12 (75.00) | 1 (100%) | 0.62 |
| Length of stay, average days (range) | 17.53 (1–70) | 16.88 (1–70) | 20.87 (6–61) | 22(/) | 0.31 |
| case fatality rate, % | 11.93 | 13.04 | 6.25 | 0 | 0.39 |
Abbreviations: ID infectious disease, NID noninfectious disease
Fig. 3.
Patients composition and samples types. a. In samples of this study, 33.9% were from blood which was the most, 33.0% from BALF, 11.0% from tissue and the others were from sputum (8.3%), pleural fluid (6.4%), CSF (3.7%), pus (1.8%), bone marrow (0.9%) and nasal swab (0.9%). b. Patients were subdivided into ID (92/109, 84.4%), NID (16/109, 14.7%) and unknown (1/109, 0.9%) groups according to their diagnosis by conventional technique. c. Infection sites of patients in ID group. Most were respiratory system infections (73/109, 67.0%) and followed by bloodstream infections (10/109, 9.17%), pleural effusion (6/109, 5.50%), central nervous system infections (6/109, 5.50%), cardiovascular system infection (2/109,1.83%), eye, ear, nose, throat, or mouth infection (2/109,1.83%), skin and soft tissue infection (1/109, 0.92%), multifocal infection (1/109, 0.92%), urinary system infection (1/109, 0.92%). Abbreviations: CSF, cerebrospinal fluid; BALF, bronchoalveolar lavage fluid
Diagnostic performance comparison of mNGS and culture
Comparison of diagnostic performance for differentiating ID from NID
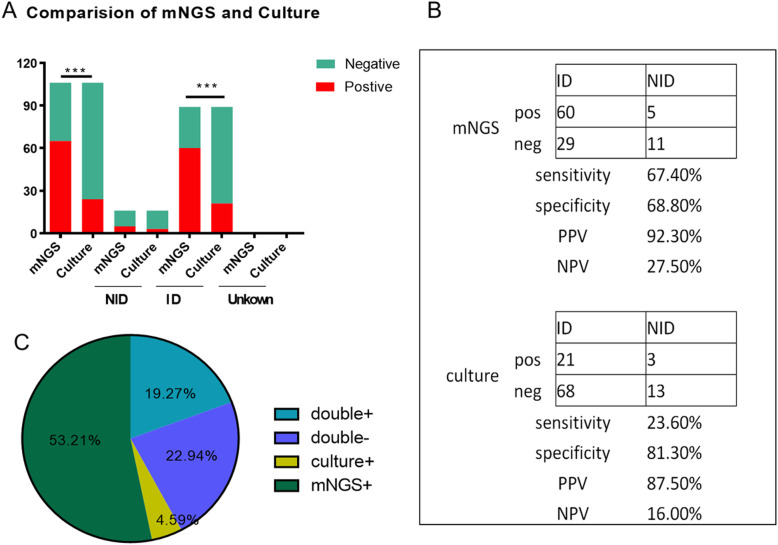
The cases of mNGS and culture tests in this study were illustrated in Fig. 4a. In the chi-square test of positive rate, there were statistical differences between mNGS and culture of all and of ID group, but no differences in NID and unknown group for the limited amounts. 105 samples were included for further study to compare the diagnostic efficiency for differentiating ID from NID. The positive predictive values and negative predictive values of diagnosing infectious disease by mNGS were 92.3 and 27.5%, respectively. The positive likelihood ratio and negative likelihood ratio being 2.16 and 0.47. The results showed that mNGS increased the sensitivity rate (positive number in ID/ID number) by approximately 44% compared with that of culture (67.4% vs 23.6%; P < 0.001) and decreased the specificity rate (negative number in NID /NID number) by 12.5% compared with that of culture (68.8% vs 81.3%; P = 0.41) (Fig. 4b).
Fig. 4.
Diagnostic Performance Comparison of mNGS and Culture. a. Positive and negative cases in all, ID, NID and unknown group of mNGS and the culture, respectively. There were statistical differences between mNGS and culture of all (P < 0.01) and of ID group (P < 0.01), but no differences in NID and unknown group for the limited amounts(P > 0.05). b. Contingency tables showed the sensitivity and specificity of mNGS were 67.4 and 68.8%, while those of culture were 23.6 and 81.3%. mNGS increased the sensitivity in comparison with that of culture (P < 0.001) while there were no differences in specificity between them (P = 0.41). c. Pie chart demonstrated the positivity distribution of mNGS and culture for all samples from 3 groups. 53.21% were positive by mNGS, 4.59% by culture, 19.27% by both and 22.94% were both negative. Abbreviations: NPV, negative predictive values; PPV, positive predictive values
Concordance between mNGS and culture for pathogen detection
In this study, mNGS and culture were both positive in 21 of 109 (19.3%) cases and were both negative in 25 of 109 (22.9%) cases. There were 58 cases (53.2%) were positive by mNGS only and 5 (4.6%) were positive only by culture. The 2 results in double-positive cases were completely matched (overlapped of all pathogens) in 3 of 21 and totally mismatched (overlapped of no pathogen) in 3 of 21 (Fig. 4c). The remaining 15 cases were found to at least one but not all overlapped of pathogens in polymicrobial results, which defined as “partly matched”.
“False positives” and “false negatives” of mNGS
In the ID group, three culturable pathogens were missed by mNGS. Among the three “mNGS false-negative” samples, there were 2 culture results paradoxical with clinical diagnosis, the other 1 was completely unidentified by mNGS. At the same time, the possible reasons for the 7 cases of “mNGS false-positive” in the NID group included potential concomitant infection with NIDs (3/7), overinterpretation (3/7) and unknown (1/7) (Table 2).
Table 2.
“False Positives” and “False Negatives” of mNGS
| Pathogens Detected Only by mNGS in the NID Group | ||||
|---|---|---|---|---|
| Sample No. | Specimen source | Diagnosis | mNGS result | Possible explanation |
| 2 | BALF | Hematencephalon | Acinetobacter baumannii, Klebsiella, Enterococcu | Unknown |
| 33 | Blood | Lymphoma | Pseudomonas, CMV | Potential cause of lymphoma |
| 62 | Blood | Aplastic anemia | Acinetobacter baumannii, Enterococcu | Overinterpretation |
| 67 | Blood | myelofibrosis | Phycomyces blakesleeanus | Overinterpretation |
| 74 | Pleural Fluid | Pleural effusion | Fusobacterium nucleatum, Streptococcus constellatus, Porphyromonas gingivalis | Potential cause of inflammation |
| 86 | Blood | Ulcerative Colitis | Porphyromonas gingivalis, HSV | Potential cause of inflammation |
| 88 | Blood | Lung cancer | Saccharomyces cerevisiae | Overinterpretation |
| Culturable Pathogens Missed by mNGS in the ID Group | ||||
| Microbe | Count | Possible explanation | ||
| MTB | 2 | Positive Not Detected | ||
| Pseudomonas | 1 | Microbes “Weak” | ||
Abbreviations: mNGS metagenomic next-generation sequencing, ID infectious disease, NID noninfectious disease, CMV metagenomic next-generation sequencing, HSV herpes simplex virus, MTB Mycobacterium tuberculosis
Comparison of mNGS and culture testing by pathogens and samples
Comparison analysis at the pathogen-type level
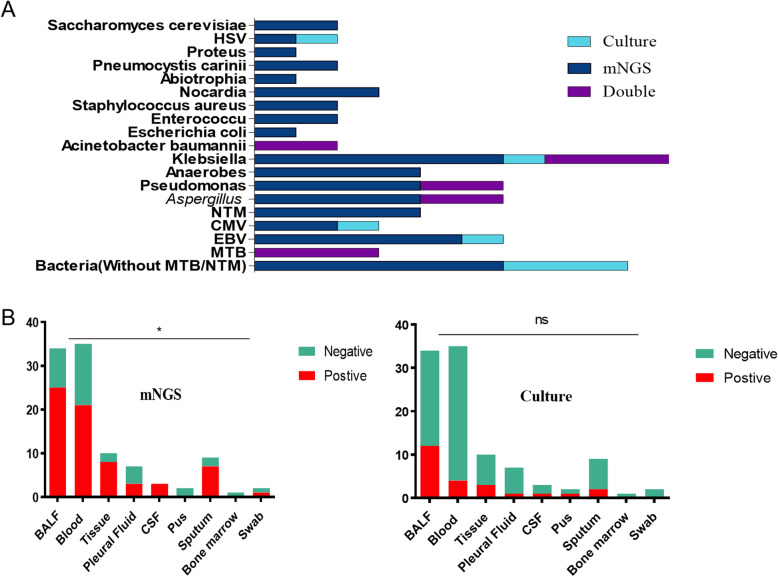
Klebsiella (10/69) was the most commonly detected pathogen among the 69 microbes isolated in mNGS and culture testing, followed by bacteria without MTB/NTM (9/69), Aspergillus (6/69), Pseudomonas (6/69) and EBV (6/69) (Fig. 5a). The percentage of mNGS-positive samples observed to have a higher yield rate than that of culture, but the differences were not significant (P > 0.05) in terms of Klebsiella, bacteria without MTB/NTM, EBV, CMV due to the small sample size. In Acinetobacter baumannii (n = 2) and MTB (n = 3), the number of mNGS-positive samples was equally with that of culture-positive samples. While only mNGS indicated positive results in NTM (n = 4), Anaerobes (n = 4), Saccharomyces cerevisiae (n = 2), Proteus (n = 1), Pneumocystis carinii (n = 2), Abiotrophia (n = 1), Nocardia (n = 3), Staphylococcus aureus (n = 2), Enterococcu (n = 2) and Escherichia coli (n = 1).
Fig. 5.
The overlap of positivity between mNGS and culture in pathogen and sample types. a. 19 pathogens detected in ID group with their corresponding frequencies were showed in histograms. Klebsiella, bacteria without MTB/NTM, EBV, CMV, NTM, Anaerobes, Saccharomyces cerevisiae, Proteus, Pneumocystis carinii, Abiotrophia, Nocardia, Staphylococcus aureus, Enterococcu and Escherichia coli demonstrated a trend of higher positivity rate in mNGS than that in culture with no statistical differences (P > 0.05). Acinetobacter baumannii and MTB were found equally in two groups. b. The overall sensitivity of mNGS in the different sample types were significantly different (P = 0.03) while sample types did not affect the sensitivity of pathogens in culture. Interestingly, especially in the types of BALF, blood and sputum samples, mNGS had significantly higher sensitivity than the culture (P = 0.002 for BALF, P < 0.001 for blood, P = 0.037 for sputum). Abbreviations: BALF, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; mNGS, metagenomic next-generation sequencing; HSV, herpes simplex virus; CMV, cytomegalovirus; EBV, Epstein-Barr virus; MTB, Mycobacterium tuberculosis; NTM, nontuberculous mycobacteria; ns, no significant difference
Comparison analysis at the sample-type level
In the types of BALF, tissue, blood and sputum samples, mNGS detection had significantly higher sensitivity than the culture method (P = 0.002 for BALF, P = 0.025 for tissue, P < 0.001 for blood, P = 0.018 for sputum), and the overall sensitivity of mNGS in the sample types was significantly different (P = 0.03). In the types of pleural fluid, CSF, pus, bone marrow and nasal swab, there were no significant differences in sensitivity between two methods (P > 0.05). In addition, in the culture method, the positive rate in BALF was higher than that in the whole blood (P = 0.019), and there was no difference in the overall sensitivity of the culture method in the sample type, as shown in Fig. 5b.
Comparison of infection indexes in positive and negative group by mNGS in ID
Classification and counting of leukocyte and lymphocyte in positive and negative group by mNGS
In this study, complete blood count, CRP and PCT tests were examined on the day of examination of pathogenic microorganisms to determine the differences in the total number of white blood cells, lymphocytes and neutrophils between the positive group and the negative group by mNGS. The results showed (Table 3) that there were no statistically differences in leukocyte and lymphocyte between positive and negative groups by mNGS (P > 0.05).
Table 3.
The counts of WBC, Cytokines and lymphocytes in positive and negative groups by mNGS
| Positive | Negative | P | |
|---|---|---|---|
| Cytokines pg/ml | |||
| IL-2 | 100.35 + 68.21 | 1.31 + 0.94 | 0.511 |
| IL-4 | 2.74 + 0.41 | 1.52 + 0.94 | 0.206 |
| IL-6 | 70.8 + 18.27 | 68.96 + 33.18 | 0.964 |
| TNF-α | 2.48 + 0.42 | 2.26 + 1.32 | 0.842 |
| IL-17a | 13.77 + 2.35 | 10.45 + 8.01 | 0.592 |
| IL-8 | 1154 + 0 | – | – |
| IL-10 | 26.14 + 7.75 | 8.29 + 3.33 | 0.044 |
| IFN-γ | 8.91 + 1.89 | 13.59 + 6.92 | 0.361 |
| Cellular Immunity % | |||
| CD4/CD8 | 1.42 + 0.23 | 2.06 + 0.44 | 0.185 |
| Th cell | 35 + 3.36 | 43.83 + 5.75 | 0.201 |
| Ts cell | 68.06 + 3.07 | 66.67 + 3.64 | 0.18 |
| NK cell | 15.71 + 2.17 | 15.5 + 1.89 | 0.958 |
| B cell | 13.53 + 2.94 | 12.83 + 3.73 | 0.899 |
| T cell | 68.06 + 3.07 | 66.67 + 3.64 | 0.958 |
| WBC ×109 | 8.32 + 0.52 | 7.36 + 0.48 | 0.283 |
| Neu× 109 | 6.99 + 0.58 | 5.38 + 0.48 | 0.109 |
| PCT ng/ml | 0.34 + 0.17 | 3.42 + 3.32 | 0.112 |
| CRP mg/l | 87.63 + 8.32 | 63.61 + 13.47 | 0.129 |
Abbreviations: mNGS metagenomic next-generation sequencing, WBC white blood cells, IL- interleukin-, IFN-γ Interferon-γ, TNF-α Tumor Necrosis Factor-α, CD4 Cluster of Differentiation 4 receptors, CD8 Cluster of Differentiation 8 receptors, Th helper T cell, Ts suppressor T cell, NK natural killer cell, Neu neutrophil, PCT procalcitonin, CRP C-reactive protein
Comparison of cytokine concentrations in positive and negative group by mNGS
In order to explore the correlation between the status of immune function in patients and the positive results of pathogen examination, this study detected and analyzed the peripheral blood (TNF-a, IL-2, IL-4, IL-6, IL-8, IL-10, IL-17A and INF-r) in infected patients. The results indicated that the peripheral blood concentrations of IL-10 in the positive group was higher than that in the negative group, and the differences were statistically significant (P = 0.044), while other cytokine showed no difference between groups as shown in Table 3.
Analysis of correlative factors for positive result of pathogen extraction by mNGS
In order to further explore the related risk factors of positive mNGS test in infected patients, this study used Logistic multivariate regression analysis to analyze the patients’ information and whether the pathogen was detected in the patients. After the confounding factors were removed, the variables that were significant for detection was age (P = 0.037, OR:1.076, 95% CI:1.005–1.152), which promoted the detection of pathogens (Table 4).
Table 4.
The analysis of the relevant factors of pathogens DNA positive in patients
| Values | B | SE (B) | Wald X2 | P | OR | 95% CI |
|---|---|---|---|---|---|---|
| Age | 0.073 | 0.035 | 4.367 | 0.037 | 1.076 | 1.005–1.152 |
| Sex | −0.545 | 1.157 | 0.222 | 0.637 | 0.58 | 0.06–5.601 |
| Read Number | −2.371 | 0.599 | 15.677 | 0.000 | 0.093 | 0.029–0.302 |
| HOD | −0.028 | 0.061 | 0.216 | 0.642 | 0.972 | 0.863–1.095 |
| Survival Time | −0.007 | 0.005 | 1.888 | 0.169 | 0.993 | 0.983–1.003 |
| Cytokines pg/ml | ||||||
| IL-2 | 0.171 | 1.115 | 0.023 | 0.878 | 1.186 | 0.133–10.553 |
| IL-4 | −1.299 | 0.893 | 2.116 | 0.146 | 0.273 | 0.047–1.57 |
| IL-6 | −0.005 | 0.019 | 0.077 | 0.781 | 0.995 | 0.957–1.033 |
| TNF-α | −0.374 | 0.373 | 1.003 | 0.316 | 0.688 | 0.331–1.430 |
| IL-17a | 0.202 | 0.137 | 2.165 | 0.141 | 1.223 | 0.935–1.6 |
| IL-10 | −2.64 | 0.206 | 1.639 | 0.201 | 0.768 | 0.513–1.151 |
| IFN-γ | 0.09 | 0.071 | 1.606 | 0.205 | 1.095 | 0.952–1.259 |
| Cellular Immunity % | ||||||
| CD4/CD8 | −0.488 | 0.965 | 0.256 | 0.613 | 0.614 | 0.093–4.067 |
| Th cell | 0.318 | 0.296 | 1.151 | 0.283 | 1.374 | 0.769–2.454 |
| Ts cell | 0.244 | 0.317 | 0.589 | 0.443 | 1.276 | 0.685–2.377 |
| NK cell | −0.223 | 0.211 | 1.121 | 0.29 | 0.800 | 0.529–1.209 |
| B cell | −0.26- | 0.245 | 1.172 | 0.279 | 0.767 | 0.227–1.475 |
| T cell | 0.5485 | 0.478 | 1.315 | 0.252 | 0.578 | 0.475–1.239 |
| WBC ×109 | −0.123 | 1.228 | 0.01 | 0.92 | 0.884 | 0.08–9.819 |
| Neu×109 | 0.141 | 1.39 | 0.01 | 0.919 | 1.151 | 0.076–17.535 |
| PCT ng/ml | −0.681 | 1.514 | 0.202 | 0.653 | 0.506 | 0.026–9.844 |
| CRP mg/l | −0.004 | 0.015 | 0.073 | 0.788 | 0.996 | 0.968–1.025 |
Abbreviations: HOD hospital day, WBC white blood cells, IL- interleukin-, IFN-γ Interferon-γ, TNF-α Tumor Necrosis Factor-α, CD4 Cluster of Differentiation 4 receptors, CD8 Cluster of Differentiation 8 receptors, Th helper T cell, Ts suppressor T cell, NK natural killer cell, Neu neutrophil, PCT procalcitonin, CRP C-reactive protein
Potential implications of clinical mNGS test
Potential inappropriate antibiotic usage for patients with virus isolates
There were 4 viruses identified by mNGS from 23 patients in this study, the majority of the identified viruses were herpes simplex virus (n = 15), followed by Epstein-Barr virus/ herpes simplex virus (n = 5), Epstein-Barr virus (n = 1), Hepatitis A virus (n = 1) and torque teno virus (n = 1). Nearly 50% of patients were diagnosed with a hospital-acquired infection (12/23) and 17 of 23 patients were given broad-spectrum antibiotics based on symptoms, imaging. 10 of 23 patients were suspected of inappropriate antibiotic usage, which means after broad-spectrum antibiotic treatment, patients’ symptoms did not improve or even worsened and after identifying the real pathogen through mNGS and adjusting the antibiotic use based on that, patients’ condition improved. 7 of 23 were considered immunocompromised hosts characterized by deficiency of the immune system or immune response caused by infectious factors, mycotoxins, drugs and nutritional deficiencies. (Table 5).
Table 5.
Clinical Characteristics of Patients with Virus Isolates (n = 23)
| Type of Virus | HAI | Immunosuppressed Patients |
Broad-spectrum Antibioticsa |
Suspected Inappropriate Antibiotic Usage | Treatment Responsive | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Yes | No | Yes | No | Yes | No | Yes | No | Yes | No | |
| HSV (n = 15) | 8 | 7 | 7 | 8 | 10 | 5 | 5 | 10 | 8 | 7 |
| HAV(n = 1) | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 |
| HSV/EBV (n = 5) | 3 | 2 | 0 | 5 | 4 | 1 | 2 | 3 | 3 | 2 |
| TTV(n = 1) | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 |
| EBV (n = 1) | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 |
| Total (N = 23) | 12 | 11 | 7 | 16 | 17 | 6 | 10 | 13 | 12 | 11 |
Abbreviations: EBV Epstein-Barr virus, HAI hospital-acquired infection, HSV herpes simplex virus, HAV herpes simplex virus, TTV torqueteno virus
The influence of positive by mNGS on the hospital days and survival of patients
As Table 6 showed, there were 67 samples in positive group with 57 males and 26 in negative group with 20 males. There was no significant difference in mean age between the two groups (59.70 yrs. vs 60.50 yrs., P = 0.84). Positive group had a longer hospital day (HOD, 176.63 days vs 150.96 days, P = 0.047) and a higher 28-day mortality (9.0% vs 0%, P = 0.049) than those of negative group, but there were no statistical differences in 14-day mortality (4.5% vs 0%, P = 0.278) and 90-day mortality (13.4% vs 3.9%, P = 0.180) between groups. The average survival time of two groups were 176.64 days and 150.96 days, respectively, but P value for t test between groups was 0.425, no statistical differences. The survival curves of the two groups were shown in Fig. 6. At the meantime, we analyzed the relationship between pathogens read number and HOD, 14-day-mortality, 28-day-mortality and 90-day-mortality, which showed that the higher pathogens read number, the higher 90-day-mortality and the longer HOD (Table 7).
Table 6.
The basic demographic and clinical characteristics of initial and outcome patient variables
| Positive | Negative | P | |
|---|---|---|---|
| Sex | |||
| Female | 10 | 6 | 0.355 |
| Male | 57 | 20 | |
| Age | 59.70 + 2.16 | 60.50 + 3.06 | 0.84 |
| HOD | 176.63 + 17.70 | 150.96 + 103.14 | 0.047 |
| 14 days of death | 4.5% | 0 | 0.278 |
| 28 days of death | 9.0% | 0 | 0.049 |
| 90 days of death | 13.4% | 3.9% | 0.180 |
| Read Number | 5295.62 + 2507.26 | 16.67+ 4.79 | 0.039 |
| Survival time | 176.64 + 17.70 | 150.96+ 21.05 | 0.425 |
Abbreviation: HOD hospital day
Fig. 6.
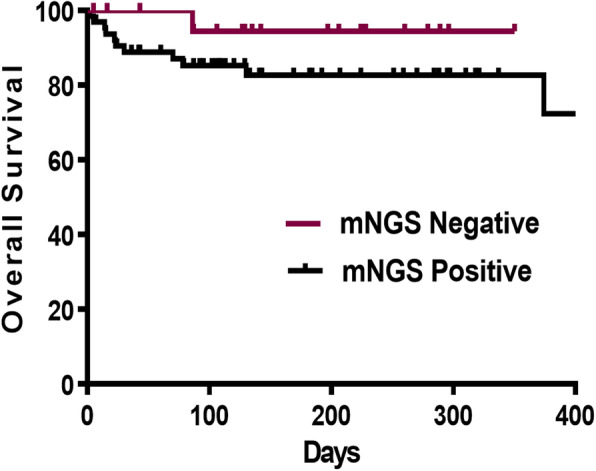
The survival curves of positive and negative group of mNGS in ID. The survival curves suggested that the overall survival rate declined faster in the positive group, however, there was no statistically differences between the two groups
Table 7.
The analysis between the pathogens read number and HOD,14, 28 and 90-day-mortality
| Read Number | |||||||
|---|---|---|---|---|---|---|---|
| 0 | 1–9 | 10–99 | 100–999 | 1000- | F | P | |
| No | 20 | 15 | 20 | 24 | 14 | ||
| HOD | 14.84 + 8.58 | 13.07 + 5.18 | 15.80 + 9.12 | 20.70 + 16.5 | 27.92 + 24.06 | 2.685 | 0.037 |
| 14-mortality | 0 | 0 | 0 | 0.04 | 0.14 | 1.898 | 0.118 |
| 28-mortality | 0 | 0.13 | 0.05 | 0.083 | 0.29 | 2.253 | 0.07 |
| 90-mortality | 0.05 | 0.2 | 0.05 | 0.083 | 0.36 | 2.598 | 0.042 |
| Survival Time | 169.74 + 102.68 | 138.40 + 100.27 | 158.70 + 125.83 | 185.45 + 124.82 | 194.71 + 216.79 | 0.424 | 0.791 |
Abbreviation: HOD hospital day
Discussion
The traditional clinical model for diagnosing infectious diseases is for doctors to make a differential diagnosis and then conduct a series of tests to try to identify the pathogen [9–12]. Traditional diagnostic techniques of microbiology laboratory ranges from smear microscopy, microorganisms’ culture, antigen antibody detection and PCR mainly. Whereas most traditional methods were often time-consuming and has a lower positive rate than mNGS [2–4]. Although molecular diagnostic assays are a quick way to diagnose the most common infections, almost all conventional microbial trials in use today only target a limited number of pathogens at a time or require successful culture of microorganisms from clinical samples [13]. While mNGS analyze the entire microbiome in patients’ samples [8] so it has been used to discover novel viral pathogens and diagnose viral infections in people widely [14–16]. Therefore, we explored the application and differences between traditional culture method and mNGS in clinical infectious diseases in adults. BALF, blood, sputum, tissue, CSF, pleural fluid, pus, bone marrow or nasal swab from 109 patients suspected of infection were collected and specimens were subjected to regular clinical microbiological assay and mNGS testing in a pairwise manner in our study. We then systematically compared the clinical features and test results of mNGS and traditional culture.
The results suggested that there were no significant differences in age, gender, length of stay and fatality rate between two groups and mNGS had an advantage in sensitivity rate compared with traditional culture method. A team of researchers also found that mNGS detected potential pathogenic bacteria, which had advantages in speed and sensitivity compared with culture and pathology [17], Miao’s team [5] showed that mNGS had a sensitivity of 50.7% for the diagnosis of infectious diseases, higher than traditional culture (50.7% vs 35.2%). In particular, the diagnosis of MTB, virus, anaerobic bacteria, nocardia and fungi has obvious advantages. The results were similar to our results, which showed that the sensitivity of mNGS was 67.4%, significantly higher than that of culture method (23.6%). High sensitivity of mNGS may because pathogen DNA has a long survival time in plasma, the use of antibiotics has a small impact on mNGS results, while traditional cultures are greatly affected by the use of antibiotics. Because of the small sample size, mNGS showed no statistical difference compared with culture method in pathogen types although there was a trend of superiority in Klebsiella, bacteria without MTB/NTM, EBV, CMV, NTM, Anaerobes, Saccharomyces cerevisiae, Proteus, Pneumocystis carinii, Abiotrophia, Nocardia, Staphylococcus aureus, Enterococcus and Escherichia coli. However, mNGS detection had a significantly higher sensitivity than the culture method in BALF (P = 0.002), tissue (P = 0.025), blood (P < 0.001) and sputum (P = 0.018) samples.
Based on the advantages shown by mNGS, we then investigated the influence of positive mNGS detection results on the severity and prognosis of patients with infection. By comparing the classification and counting of leukocyte, lymphocyte and cytokine concentrations in positive and negative groups, we found that IL-10 concentration in peripheral blood in the positive group was higher than that in the negative group and there were no statistically differences in other cytokine concentrations, leukocyte and lymphocyte. According to the results of correlative factors analysis for positive test of mNGS, patients’ age may promote the detection of pathogens. In the survival analysis, positive group had a higher 28-day mortality (9.0% vs 0%, P = 0.049) than that of negative group, but there were no statistical differences in average survival time. The pathogens read number by mNGS was positive related to the HOD and 90-day-mortality of patients with infectious diseases. All of that indicated older people were more likely to have positive results and positive results of mNGS detection may represent a worse outcome.
Fortunately, mNGS has moved from scientific application to clinical practice and is changing the way disease diagnosed and treated [18–20]. In addition to what we mentioned above, mNGS also has merits in many other aspects. Firstly, mNGS does not need prior clinical information to detect infectious pathogens, and the results can be reported quickly and accurately, greatly shortening the diagnosis time of infectious pathogens. Early and rapid reporting of the results by mNGS provides clinical clues to the next step in diagnosis and treatment, especially avoiding overuse of antibiotics for viral infections [21, 22]. Rapid results reported by mNGS also can promote timely adjustment of treatment in clinical practice. As our data showed, almost one-half of patients with virus infection were suspected of inappropriate antibiotic usage. Secondly, mNGS was used in some rare infectious pathogens. It detected Naegleria fowleri [23], brucellosis [24], cysticercosis, taenia bocinea [25], gondii [26] in CSF, Hepatic tuberculosis in blood [27] in previous reports. Thirdly, studies have shown that mNGS can be used not only for pathogen identification, but also for microbiome characterization, parallel analyses of human host responses, drug resistance gene and virulence factor detection. All of these led to the rapid development of mNGS in immunodeficiency difficult-to-diagnose cases and immunocompromised patients [13]. Thirdly, antibiotic usage had little influence on mNGS results due to the long survival time of pathogen DNA in plasma, but traditional cultures were affected by antibiotic use [21, 22]. Higher sensitivity of mNGS than culture in this study may because that mNGS is less affected by prior antibiotic usage. However, mNGS still has some limitations at present, such as human background, background bacteria contamination, no uniform standards for detailed experimental procedures [2, 28–31], inability to distinguish infection and colonization, standardization of bioinformatics analysis process, and problem of report interpretation. The results must be interpreted in the context of the clinical situation. It’s worth noting that background microbial contamination is a common problem faced by mNGS technology, which can be partially eliminated through negative quality control, but it requires clinical familiarity with common background bacteria and better interpretation results combined with clinical practice [24].
In this study, we systematically compared mNGS and traditional culture method in sensitivity, specificity, pathogen type and sample type. On this basis, we also compared and analyzed the differences between the positive and negative groups of mNGS which was few at present. Patients of positive group found to have a trend of worse prognosis suggested need more attention clinically. Small sample size was the biggest deficiency of our study, so that there were many results indicated a certain trend without reaching statistical significance unfortunately. Therefore, more patients need to be included in the study in the future. Not randomized controled was also the limitation of study. As a retrospective study, this study has some limitations like limited data and data accumulation not controlled by the researcher. Besides, limit generalizability caused by single-center study, lack of a gold standard comparator for diagnostics, lack of antibiotic usage detail and classification bias were also the limitations of this study.
Conclusions
In summary, mNGS had a higher sensitivity than culture, especially in the types of BALF, blood and sputum samples, and there was a trend of higher sensitivity of Klebsiella, CMV and EBV detection. The worse trend of outcome in patients with positive mNGS results than negative group prompted more clinical attention to patients with positive mNGS results is required. Therefore, based on what we found above and other advantages of mNGS like quick results, less affected by prior antibiotic exposure and so on, we suggest that mNGS should be used more in early pathogen diagnosis in the future. Nonetheless, interpreting data of mNGS will be a challenge for doctors in guiding clinical treatment of infectious diseases.
Acknowledgments
We would like to thank all the patients who donate their biological samples.
Abbreviations
- mNGS
Metagenomic next-generation sequencing
- BALF
Bronchoalveolar lavage fluid
- CSF
Cerebrospinal fluid
- Dx
Diagnosis
- ID
Infectious disease
- NID
Noninfectious disease
- DNA
Deoxyribonucleic acid
- EDTA
Ethylene Diamine Tetraacetic Acid
- EBV
Epstein-Barr virus
- HAI
Hospital-acquired infection
- HboV
Human bocavirus
- HSV-1
Herpes simplex virus 1
- TTV
Torqueteno virus
- HAV
Hepatitis A virus
- HOD
Hospital day
- IL-2
Interleukin-2
- IL-4
Interleukin-4
- IL-6
Interleukin-6
- IL-10
Interleukin-10
- IL-8
Interleukin-10
- CD4
Cluster of Differentiation 4 receptors
- CD8
Cluster of Differentiation 8 receptors
- WBC
White blood cell
- Th
Helper T
- NK
Natural killer cell
- PCT
Procalcitonin
- CRP
C-reactive protein
- IFN-γ
Interferon-γ
- IL-17a
Interleukin-17a
- TNF-α
Tumor Necrosis Factor-α
- PPV
Positive predictive value
- NPV
Negative predictive value
- CMV
Cytomegalovirus
- MTB
Mycobacterium tuberculosis
- ns
No signicant difference
- NTM
Nontuberculous mycobacteria
- G +
Gram Positive
- G-
Gram Negative
Authors’ contributions
Conception and design: H.X.D, S.S.X, X. L, A.H.M. Acquisition, statistical analysis or interpretation of the data: P. L, Y. L, X.F.L, W.W.L. Drafting of the manuscript: C.H.W. All authors reviewed and approved the final version of the manuscript. All authors had read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81802262), the Fundamental Research Funds for the Central Universities (No. 22120180584), Shanghai Tenth Hospital’s Improvement Plan for NSFC (No. 04.03.17.032, 04.01.18.048, SYGZRPY2017014). Changhui Wang is the recipient of the foundations and was responsible for editing manuscript in our study.
Availability of data and materials
All data generated or analysed during this study are included in this published article. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
All experiments performed in this study are in accordance with the approved ethical standards by the ethics committee of the 10th People’s Hospital affiliated to Tongji University (No. SHSY-IEC-4.1/20–21/01). Formal consent is waived for this type of study.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hongxia Duan, Xuan Li and Aihong Mei contributed equally to this work.
Contributor Information
Changhui Wang, Email: wang-chang-hui@hotmail.com.
Shuanshuan Xie, Email: xieshuanshuan@aliyun.com.
References
- 1.Zhou K, Lokate M, Deurenberg RH, Tepper M, Arends JP, Raangs EG, Lo-Ten-Foe J, Grundmann H, Rossen JW, Friedrich AW. Use of whole-genome sequencing to trace, control and characterize the regional expansion of extended-spectrum beta-lactamase producing ST15 Klebsiella pneumoniae. Sci Rep. 2016;6:20840. doi: 10.1038/srep20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G, Professional Practice C, Committee on Laboratory Practices of the American Society for M, Microbiology Resource Committee of the College of American P. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med 2017; 141:776–786. [DOI] [PubMed]
- 3.Ewig S, Torres A, Angeles Marcos M, Angrill J, Rano A, de Roux A, Mensa J, Martinez JA, de la Bellacasa JP, Bauer T. Factors associated with unknown aetiology in patients with community-acquired pneumonia. Eur Respir J. 2002;20:1254–1262. doi: 10.1183/09031936.02.01942001. [DOI] [PubMed] [Google Scholar]
- 4.van Gageldonk-Lafeber AB, Heijnen ML, Bartelds AI, Peters MF, van der Plas SM, Wilbrink B. A case-control study of acute respiratory tract infection in general practice patients in the Netherlands. Clin Infect Dis. 2005;41:490–497. doi: 10.1086/431982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miao Q, Ma Y, Wang Q, Pan J, Zhang Y, Jin W, Yao Y, Su Y, Huang Y, Wang M, Li B, Li H, Zhou C, Li C, Ye M, Xu X, Li Y, Hu B. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical Practice. Clin Infect Dis. 2018;67:S231–S240. doi: 10.1093/cid/ciy693. [DOI] [PubMed] [Google Scholar]
- 6.Grumaz S, Stevens P, Grumaz C, Decker SO, Weigand MA, Hofer S, Brenner T, von Haeseler A, Sohn K. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8:73. doi: 10.1186/s13073-016-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecuit M, Eloit M. The diagnosis of infectious diseases by whole genome next generation sequencing: a new era is opening. Front Cell Infect Microbiol. 2014;4:25. doi: 10.3389/fcimb.2014.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefterova MI, Suarez CJ, Banaei N, Pinsky BA. Next-generation sequencing for infectious disease diagnosis and management: a report of the Association for Molecular Pathology. J Mol Diagn. 2015;17:623–634. doi: 10.1016/j.jmoldx.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Khare R, Espy MJ, Cebelinski E, Boxrud D, Sloan LM, Cunningham SA, Pritt BS, Patel R, Binnicker MJ. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol. 2014;52:3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leber AL, Everhart K, Balada-Llasat JM, Cullison J, Daly J, Holt S, Lephart P, Salimnia H, Schreckenberger PC, DesJarlais S, Reed SL, Chapin KC, LeBlanc L, Johnson JK, Soliven NL, Carroll KC, Miller JA, Dien Bard J, Mestas J, Bankowski M, Enomoto T, Hemmert AC, Bourzac KM. Multicenter evaluation of BioFire FilmArray meningitis/encephalitis panel for detection of Bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol. 2016;54:2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruggiero P, McMillen T, Tang YW, Babady NE. Evaluation of the BioFire FilmArray respiratory panel and the GenMark eSensor respiratory viral panel on lower respiratory tract specimens. J Clin Microbiol. 2014;52:288–290. doi: 10.1128/JCM.02787-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang YW, Gonsalves S, Sun JY, Stiles J, Gilhuley KA, Mikhlina A, Dunbar SA, Babady NE, Zhang H. Clinical evaluation of the Luminex NxTAG respiratory pathogen panel. J Clin Microbiol. 2016;54:1912–1914. doi: 10.1128/JCM.00482-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20:341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moustafa A, Xie C, Kirkness E, Biggs W, Wong E, Turpaz Y, Bloom K, Delwart E, Nelson KE, Venter JC, Telenti A. The blood DNA virome in 8,000 humans. PLoS Pathog. 2017;13:e1006292. doi: 10.1371/journal.ppat.1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rascovan N, Duraisamy R, Desnues C. Metagenomics and the human Virome in asymptomatic individuals. Annu Rev Microbiol. 2016;70:125–141. doi: 10.1146/annurev-micro-102215-095431. [DOI] [PubMed] [Google Scholar]
- 16.Somasekar S, Lee D, Rule J, Naccache SN, Stone M, Busch MP, Sanders C, Lee WM, Chiu CY. Viral surveillance in serum samples from patients with acute liver failure by metagenomic next-generation sequencing. Clin Infect Dis. 2017;65:1477–1485. doi: 10.1093/cid/cix596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Gao H, Meng H, Wang Q, Li S, Chen H, Li Y, Wang H. Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front Cell Infect Microbiol. 2018;8:205. doi: 10.3389/fcimb.2018.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houldcroft CJ, Beale MA, Breuer J. Clinical and biological insights from viral genome sequencing. Nat Rev Microbiol. 2017;15:183–192. doi: 10.1038/nrmicro.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlaberg R, Queen K, Simmon K, Tardif K, Stockmann C, Flygare S, Kennedy B, Voelkerding K, Bramley A, Zhang J, Eilbeck K, Yandell M, Jain S, Pavia AT, Tong S, Ampofo K. Viral pathogen detection by Metagenomics and Pan-viral group polymerase chain reaction in children with pneumonia lacking identifiable etiology. J Infect Dis. 2017;215:1407–1415. doi: 10.1093/infdis/jix148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, Salamat SM, Somasekar S, Federman S, Miller S, Sokolic R, Garabedian E, Candotti F, Buckley RH, Reed KD, Meyer TL, Seroogy CM, Galloway R, Henderson SL, Gern JE, DeRisi JL, Chiu CY. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes J, Hyder JA, Peruski LF, Fisher C, Jorakate P, Kaewpan A, Dejsirilert S, Thamthitiwat S, Olsen SJ, Dowell SF, Chantra S, Tanwisaid K, Maloney SA, Baggett HC. Antibiotic use in Thailand: quantifying impact on blood culture yield and estimates of pneumococcal bacteremia incidence. Am J Trop Med Hyg. 2010;83:301–306. doi: 10.4269/ajtmh.2010.09-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gosiewski T, Ludwig-Galezowska AH, Huminska K, Sroka-Oleksiak A, Radkowski P, Salamon D, Wojciechowicz J, Kus-Slowinska M, Bulanda M, Wolkow PP. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method - the observation of DNAemia. Eur J Clin Microbiol Infect Dis. 2017;36:329–336. doi: 10.1007/s10096-016-2805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo LY, Feng WY, Guo X, Liu B, Liu G, Dong J. The advantages of next-generation sequencing technology in the detection of different sources of abscess. J Inf Secur. 2019;78:75–86. doi: 10.1016/j.jinf.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Fan S, Ren H, Wei Y, Mao C, Ma Z, Zhang L, Wang L, Ge Y, Li T, Cui L, Wu H, Guan H. Next-generation sequencing of the cerebrospinal fluid in the diagnosis of neurobrucellosis. Int J Infect Dis. 2018;67:20–24. doi: 10.1016/j.ijid.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 25.Hu Z, Weng X, Xu C, Lin Y, Cheng C, Wei H, Chen W. Metagenomic next-generation sequencing as a diagnostic tool for toxoplasmic encephalitis. Ann Clin Microbiol Antimicrob. 2018;17:45. doi: 10.1186/s12941-018-0298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ai JW, Li Y, Cheng Q, Cui P, Wu HL, Xu B, Zhang WH. Diagnosis of local hepatic tuberculosis through next-generation sequencing: smarter, faster and better. Clin Res Hepatol Gastroenterol. 2018;42:178–181. doi: 10.1016/j.clinre.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Du B, Tao Y, Ma J, Weng X, Gong Y, Lin Y, Shen N, Mo X, Cao Q. Identification of sparganosis based on next-generation sequencing. Infect Genet Evol. 2018;66:256–261. doi: 10.1016/j.meegid.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Blauwkamp TA, Thair S, Rosen MJ, Blair L, Lindner MS, Vilfan ID, Kawli T, Christians FC, Venkatasubrahmanyam S, Wall GD, Cheung A, Rogers ZN, Meshulam-Simon G, Huijse L, Balakrishnan S, Quinn JV, Hollemon D, Hong DK, Vaughn ML, Kertesz M, Bercovici S, Wilber JC, Yang S. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4:663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 29.Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, Garcia-Cobos S, Kooistra-Smid AM, Raangs EC, Rosema S, Veloo AC, Zhou K, Friedrich AW, Rossen JW. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol. 2017;243:16–24. doi: 10.1016/j.jbiotec.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 30.Gargis AS, Kalman L, Lubin IM. Assuring the quality of next-generation sequencing in clinical microbiology and public health laboratories. J Clin Microbiol. 2016;54:2857–2865. doi: 10.1128/JCM.00949-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller S, Naccache SN, Samayoa E, Messacar K, Arevalo S, Federman S, Stryke D, Pham E, Fung B, Bolosky WJ, Ingebrigtsen D, Lorizio W, Paff SM, Leake JA, Pesano R, DeBiasi R, Dominguez S, Chiu CY. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29:831–842. doi: 10.1101/gr.238170.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article. The data that support the findings of this study are available from the corresponding author upon reasonable request.