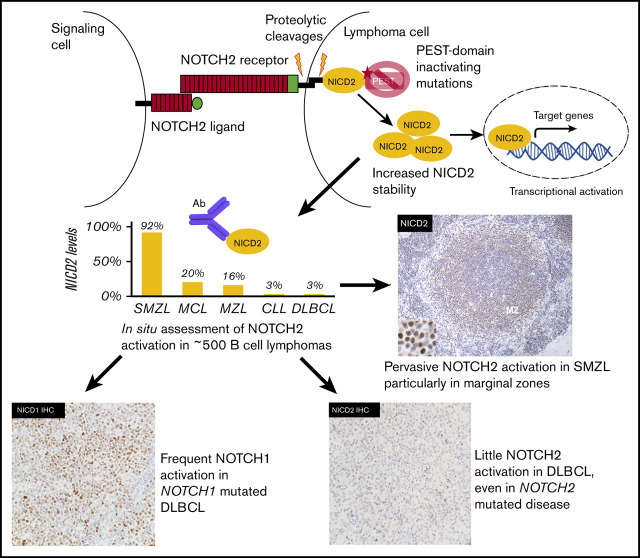
Key Points
NOTCH2 activation is common in SMZL and rare in other types of small B-cell lymphoma.
NOTCH2 activation is rare in DLBCL, including NOTCH2-mutated tumors, whereas a minor subset of DLBCLs has ongoing NOTCH1 activation.
Abstract
Notch receptors participate in a signaling pathway in which ligand-induced proteolysis frees the Notch intracellular domain (NICD), allowing it to translocate to the nucleus, form a transcription complex, and induce target gene expression. Chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL), splenic marginal zone B-cell lymphoma (SMZL), and distinct subsets of diffuse large B-cell lymphoma (DLBCL) are strongly associated with mutations in the 3′ end of NOTCH1 or NOTCH2 that disrupt a proline, glutamic acid, serine, and threonine (PEST) degron domain and stabilize NICD1 and NICD2. By contrast, mutations leading to constitutive Notch activation are rare in primary B-cell neoplasms, suggesting that Notch activation is confined to ligand-rich tumor microenvironments, or that cryptic strong gain-of-function mutations have been missed in prior analyses. To test these ideas, we used immunohistochemical stains to screen a broad range of B-cell tumors for Notch activation. Our analyses reveal that among small B-cell neoplasms, NICD2 is primarily detected in SMZL and is a common feature of both NOTCH2 wild-type and NOTCH2-mutated SMZLs, similar to prior findings with NOTCH1 in CLL/SLL. The greatest NOTCH2 activation was observed in NOTCH2-mutated SMZLs, particularly within splenic marginal zones. By contrast, little evidence of NOTCH2 activation was observed in DLBCL, even in NOTCH2-mutated tumors, suggesting that selective pressure for NOTCH2 activation is mainly confined to low-grade B-cell neoplasms, whereas DLBCLs with NOTCH1 mutations frequently showed evidence of ongoing NOTCH1 activation. These observations have important implications for the pathogenic role of Notch and its therapeutic targeting in B-cell lymphomas.
Visual Abstract
Introduction
Notch receptors are single-pass transmembrane proteins that are widely expressed in embryonic and adult tissues (for review, see Bray1). These receptors participate in a signaling pathway in which binding to a ligand of the Δ-like (DLL) or JAG family expressed on the surface of a neighboring cell induces successive proteolytic cleavages in Notch by ADAM10 and γ-secretase. The latter cleavage releases the Notch intracellular domain (NICD) from the plasma membrane, allowing it to move to the nucleus and form a Notch transcription complex that induces target gene expression. Notch target genes vary widely across lineages, and as a result the outcome of Notch activation is highly cell-context dependent.
This context-dependency is reflected in the pattern of Notch mutations in various cancers (for review, see Aster et al2). Loss-of-function mutations occur frequently in certain solid tumors (eg, in squamous cell carcinoma), reflecting tumor-suppressive functions. By contrast, gain-of-function mutations affecting the ectodomains of Notch receptors that lead to ligand-independent Notch receptor proteolysis are common in T-cell acute lymphoblastic leukemia and triple-negative breast cancer; these mutations consist of in-frame point substitutions or indels or Notch gene rearrangements that disrupt a juxtamembrane-negative regulatory region that holds Notch in an off state prior to engagement with ligand. A third pattern of gain-of-function mutations is observed in B-cell neoplasms, in which ectodomain mutations are rare and most mutations lie in the 3′ exons of NOTCH1 (mainly in chronic lymphocytic leukemia/small lymphocytic lymphoma [CLL/SLL]3-6 and a subset of diffuse large B-cell lymphoma [DLBCL]7-10) or NOTCH2 (mainly in splenic marginal zone lymphoma [MZL; SMZL]11,12 and another distinct subset of DLBCL7,10,13). These mutations result in the loss of C-terminal proline, glutamic acid, serine, and threonine (PEST) degron domains implicated in ubiquitinylation and turnover of NICD1 and NICD2 and should therefore elevate levels of active nuclear Notch. However, because the effects of PEST domain mutations are dependent on upstream factors that generate NICD (eg, access of tumor cells to ligand), they are predicted to only increase Notch signaling in ligand-rich microenvironments. This logic further predicts that B-cell neoplasms of the same type expressing wild-type Notch receptors should also show evidence of Notch activation in ligand-rich microenvironments, albeit at lower levels than tumors with PEST domain mutations.
It is also notable, however, that strong ligand-independent gain-of-function mutations consisting of Notch gene rearrangements that disrupt the expression of the negative regulatory region (NRR) may be missed by exome or even whole-genome sequencing.14 A clue to the existence of such rearrangements is identification of high levels of nuclear Notch in the absence of point substitutions or in-frame indels in the NRR-coding region. Previously, among B-cell tumors, intragenic Notch gene rearrangements that disrupt the NRR have been detected in 2 transformed mantle cell lymphoma (MCL) cell lines,15,16 both of which are highly sensitive to Notch pathway inhibitors, as well as in a case of CLL.17 Thus, a protein-based screen for evidence of cryptic strong Notch gain-of-function mutations (eg, Notch gene rearrangements) is merited in DLBCL, as detection of such events would have important pathogenic implications and would provide a rationale for trials of Notch inhibitors in this aggressive form of lymphoma. The latter idea is based on preclinical studies showing that one of the best predictors of tumor response to Notch pathway inhibitors is high levels of Notch activation,18 which can be directly assessed by immunohistochemical staining for nuclear Notch.15
Testing of these ideas requires reagents that specifically identify activated Notch receptors in situ. In past work using an immunohistochemical method that specifically detects NICD1, we observed that NOTCH1 activation was virtually ubiquitous in the nodal microenvironment in CLL/SLL, independent of NOTCH1 mutational status, consistent with selection for NOTCH1 signaling within the lymph node microenvironment in this tumor, whereas NICD1 staining was uncommon in other low-grade nodal B-cell neoplasms, such as MCL, as well as a small cohort of DLBCLs.15 To further evaluate Notch activation in B-cell neoplasms, here we describe studies using antibodies that recognize active, nuclear NOTCH2 in situ. We first assessed the distribution and frequency of NOTCH2 activation in SMZL. As predicted, we observed that, although NOTCH2-mutated SMZL has on average higher nuclear NOTCH2 levels, NOTCH2 activation is also readily detected in most NOTCH2 wild-type cases. NOTCH2 activation is highest in splenic marginal zones, consistent with mouse models showing that activation of Notch2 by the ligand Dll1 expressed on marginal zone stromal cells is critical for marginal zone B-cell specification and maintenance.19,20 We next investigated activation of NOTCH1 and NOTCH2 in a large cohort of DLBCLs, in which the presence of NOTCH1 and NOTCH2 pathway mutations defines distinct DLBCL subsets associated with different clinical outcomes.7,21 Nuclear NOTCH1 staining was largely confined to the N1 (NOTCH1-mutated) DLBCL subtype, whereas NOTCH2 activation was rare in DLBCL, even among tumors with NOTCH2 mutations leading to PEST domain deletion and others belonging to the BN2 subset associated with NOTCH2 pathway aberrations. Our findings indicate that among B-cell neoplasms, ongoing NOTCH signaling is largely restricted to CLL/SLL, SMZL, and a subset of NOTCH1-mutated DLBCLs.
Methods
Cell lines
Human 293T cells were obtained from American Tissue Type Collection; SU-DHL-4, WSU-DLCL2, OCI-Ly3, OCI-Ly10, and Ri-1 cells were obtained from the Cancer Cell Line Factory at the Broad Institute of MIT and Harvard. Cell lines were cultured at 37°C under 5% CO2 in Dulbecco’s modified Eagle medium or RPMI 1640 supplemented with 10% fetal calf serum, glutamine, and streptomycin/penicillin. Short-tandem-repeat testing was used to confirm the identity of cell lines.
Production of NICD2-specific antibody
An antibody that specifically recognizes γ-secretase cleaved NOTCH2 (NICD2) was developed and provided by Eli Lilly, Inc. Briefly, hybridomas produced from rabbits immunized with a peptide starting with the N terminus of NICD2 (amino acid 1697, Uniprot #Q04721) were screened by enzyme-linked immunosorbent assay and western blot for NICD2 immunoreactivity and counterscreened against NICD1, NICD3, and non–γ-secretase-cleaved forms of NOTCH2. Positive controls were created by transfecting 293T cells with a pcDNA3 plasmid encoding a truncated form of NOTCH2 that requires γ-secretase cleavage to generate NICD2.
Formalin-fixed paraffin-embedded tissues
Cases of low-grade B-cell neoplasms were retrieved from the pathology archives of Brigham and Women’s Hospital (BWH). DLBCL tissue microarrays (TMAs) were obtained from BC Cancer (BCC), Centre for Lymphoid Cancer. Diagnoses were made according to the 2016 World Health Organization (WHO) Classification of Lymphoid Neoplasms.22 Work was done under institutional review board protocols 2014P001256 (BWH) and H14-02304 (BCC). Patient-derived xenograft models of triple-negative breast cancers with or without NOTCH2 gene rearrangements were obtained from Champions Oncology, Inc.
Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) tissues were cut onto charged slides at 4-μm thickness and baked at 60°C for 1 hour. Staining was done on a Leica Bond III immunostainer following Epitope Retrieval 2 for 40 minutes. Staining with a rabbit monoclonal antibody specific for NICD2 was done by incubating at 1:50 for 60 minutes. Staining for NICD1 was done with rabbit monoclonal antibody clone D3B8 (Cell Signaling Technology) as described.15 Staining for total NOTCH2 was done with a rabbit monoclonal antibody (1:100; clone D76A6; Cell Signaling Technology) against an epitope centered on amino acid residue 2378 that is located ∼40 aa N-terminal of the NOTCH2 PEST domain. Staining was developed by incubation with secondary antibody linked to horseradish peroxidase followed by diaminobenzidine using the Polymer Refine Detection Kit (Leica). Slides were counterstained with hematoxylin. B-cell lymphoma 6 (BCL6) staining was performed separately on a Dako Omnis platform (Agilent Technologies, Inc, Santa Clara, CA) using mouse monoclonal antibody clone PG-B6p (GA625, ready to use; Dako). Scoring of IHC results was done independently by J.C.A., V.S., and J.W.C.; results represent a consensus score. Unless otherwise specified in the figure legends, all representative images were obtained using an Olympus BX41 microscope equipped with UPlanFLN semiapochromat objectives and an Olympus DP27 camera. All imaging was performed at room temperature and ambient air conditions at ×20 (numerical aperture, 0.50) or ×40 (numerical aperture, 0.75) original magnification. Olympus cellSens Entry software was used for white balancing and to capture images, and Adobe Photoshop was used to crop the images, adjust brightness and contrast, and highlight salient findings.
Image analysis
A subset of the SMZL cases (N = 20) subjected to next-generation sequencing (NGS) and stained for NICD2 were imaged with a ×40 objective using Aperio ScanScope. Five cases were not further analyzed because high levels of formalin pigment interfered with Aperio analysis (N = 4) or because extensive effacement of spleen precluded comparison of staining in red and white pulp regions (N = 1). In the remaining 15 cases (NOTCH2 mutated = 8; NOTCH2 wild type = 7) regions of interest were annotated using ImageScope (version 10.0.35.1800; Aperio Technology). The V9 nuclear algorithm was used to identify positively stained nuclei in 10 regions of interest in outer white pulp, inner white pulp, and red pulp. The inner white pulp was defined by a small ellipse with major and minor axis lengths half those of a larger ellipse, which defined the entire white pulp nodule. At least 4000 nuclei (mean, 32 642; range, 4030-110 595) were scored in each compartment and at least 20 000 nuclei (mean, 95 222; range, 23 143-220 435) were scored in each case.
Targeted NGS
NGS was performed on DNA isolated from 20 FFPE SMZLs and 1 MCL using Oncopanel,23,24 a hybrid capture assay covering exonic regions and selected intronic regions of 447 cancer genes (see supplemental Table 4 for full gene list). Following sequencing (Illumina HiSeq 2500), reads were aligned to human genome GRCh37 (hg19) with the Burrows-Wheeler Alignment tool,25 and sorted, duplicate marked, and indexed with Picard tools. Base-quality score calibration and alignments around indels were performed with the Genome Analysis Toolkit.26,27 Single-nucleotide variant calls were performed with MuTect.28 Copy-number alterations were identified with Robust CNV. Structural variants were detected using BreaKmer.24 Criteria for identifying likely pathogenic mutations are provided in supplemental Methods. RNA-sequencing (RNA-seq) data sets were described previously.29
DLBCL cohort
TMA construction, fluorescence in situ hybridization analyses, subtyping by IHC (Hans protocol), gene-expression profiling, and targeted DNA sequencing of presumed de novo DLBCLs have been described.29-32 From a cohort of 347 DLBCLs,30 314 tumors represented on TMAs that passed quality control measures were assigned molecular and genetic subtypes as described.33,34 Because of core dropout from TMAs, NICD2 (N = 313), NICD1 (N = 303), and total NOTCH2 (N = 302) staining was only evaluable in a subset of these cases. In 9 cases with NOTCH2 mutations identified by analysis of RNA-seq data and 7 cases with NOTCH1 mutations identified by DNA sequencing, staining of whole-tissue sections (at least 1 × 1 cm in size) was used to supplement or confirm consistency with TMA-staining results.
Gene-set enrichment analysis
RNA-seq data from 309 DLBCLs has been described.34 To assess expression of Notch target genes that are enriched in B-cell context, we used putative Notch target gene sets that were obtained from analysis of follicular lymphoma, MCL, and CLL. The “follicular lymphoma gene set” represents NOTCH2 target genes and genes encoding components of the Notch transcription complex that are inversely correlated with BCL6 expression in primary follicular lymphoma (from supplemental Table 4 in Valls et al35). The “MCL gene set” includes direct Notch target genes identified in MCL cell lines (from supplemental Table 4 in Ryan et al16). The “CLL gene set” includes genes induced by NICD1 in a CLL cell line (from supplemental Data set 9 in Fabbri et al36). After calculation of mean expression of Notch target genes in each tumor, expression levels were compared between Notch-mutated and unmutated tumors using the Kruskal-Wallis test and between NOTCH1-mutated, NOTCH2-mutated, and Notch wild-type tumors using pairwise Mann-Whitney U tests.
Results
NOTCH2 activation is pervasive in SMZL and uncommon in other indolent B-cell neoplasms
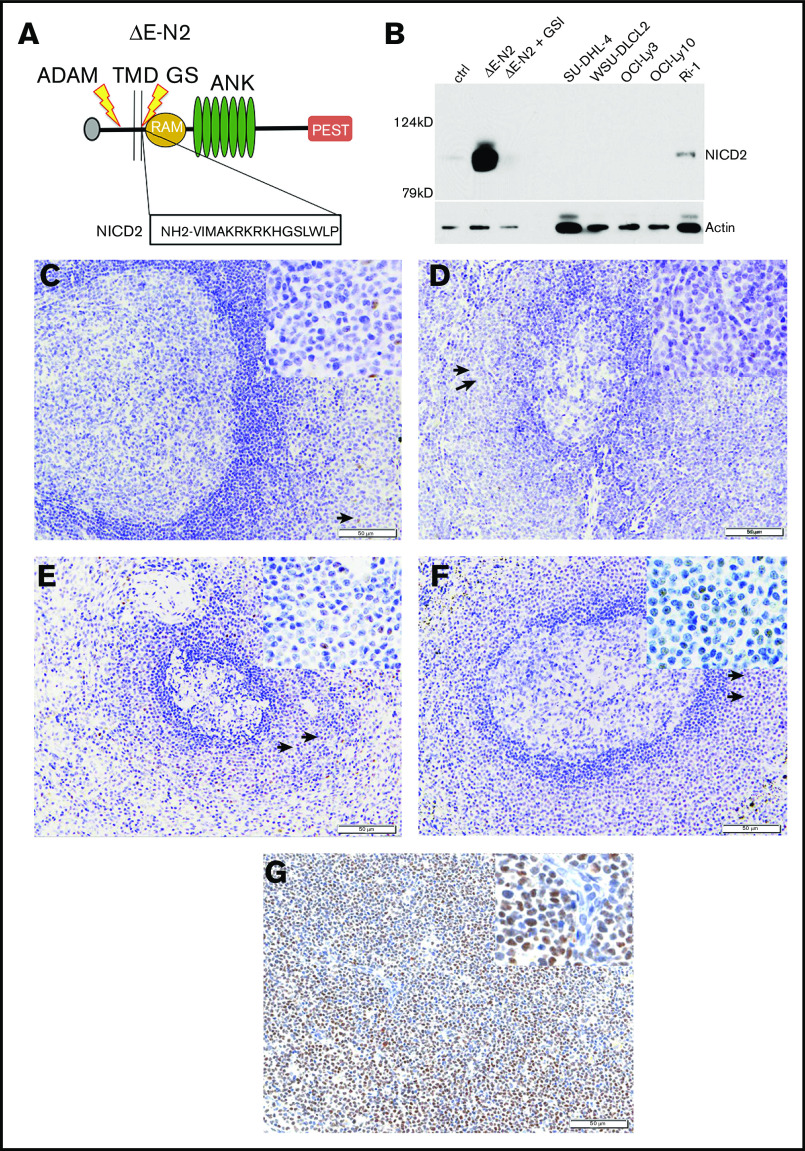
To detect NOTCH2 activation in archival samples, we used an antibody that specifically recognizes a neoepitope in NICD2 that is created by cleavage of NOTCH2 by γ-secretase (Figure 1A-B). Initial pilot studies with reactive tonsil (N = 8; Figure 1C), lymph node (N = 5; Figure 1D), and normal spleen with and without marginal zone B-cell expansion (N = 7; Figure 1E-F), showed weak to moderate nuclear NICD2 staining in a subset of cells found in normal splenic marginal zones. Notably, much more intense and pervasive nuclear NICD2 staining was seen in a case of NOTCH2-mutated SMZL (Figure 1G), consistent with the ability of our NICD2 stain to accurately gauge NOTCH2 activation in archival tissues.
Figure 1.
Validation of a NICD2-specific antibody. (A) Schematic showing a “headless” form of NOTCH2 (ΔE-N2) retaining the leader peptide (L) and lacking the extracellular NRR that is sensitive to successive cleavages by ADAM10 (ADAM) and γ-secretase (GS), releasing NICD2 from the transmembrane domain (TMD) and generating an N-terminal neoepitope. (B) Western blot showing specificity of NICD2 antibody. Whole-cell extracts were prepared from (i) 293T cells transfected with empty vector or ΔE-N2 complementary DNA (cDNA) and treated with dimethyl sulfoxide (DMSO; vehicle control) or γ-secretase inhibitor (1 μM compound E; γsecretase inhibitor [GSI]) posttransfection, and (ii) B-cell lymphoma cell lines with (Ri-1) or without (SUDHL4, WSU-DLCL2, OCILy3, and OCILy10) gain-of-function mutations in NOTCH2. Note NICD2 staining is suppressed by GSI treatment in transfected 293T cells and is also observed in NOTCH2-mutated Ri-1 cells. (C-G) Representative images of NICD2 immunostaining in sections prepared from FFPE tonsil (C), lymph node (D), normal spleen (E), spleen with reactive marginal zone expansion (F), and spleen involved by NOTCH2-mutated SMZL (G). The sections in panels C-G were counterstained with hematoxylin (blue) after immunostaining using a method that produces a brown color. Tonsil and lymph node sections show only weak nuclear staining in subsets of lymphocytes in the perifollicular regions. Spleen sections show moderate nuclear staining of a subset of lymphocytes within marginal zones. Arrows in C-F highlight the position of cells that are shown at high power in the insets. The NOTCH2-mutated SMZL shows diffuse, strong nuclear staining in neoplastic cells. Original magnification, ×200; inset, ×400. ANK, ankyrin repeat domain; PEST, PEST degron domain; RAM, RBP-Jκ/CBF1-associated module.
We next determined the frequency and prevalence of NICD2 staining in 39 cases of SMZL, other low-grade B-cell neoplasms that are known to harbor Notch mutations (CLL/SLL, n = 43; MCL, n = 47), and non–splenic marginal zone B-cell lymphomas (n = 57), all diagnosed per WHO criteria.22 NICD2 staining was positive in 36 of 39 cases (92%) of SMZL, whereas such staining was uncommon in MCL (19%) and in nodal and extranodal MZL involving sites other than the spleen (16%) and was rare in CLL/SLL (2%) (Figure 2A). In each instance, staining of serial sections for the B-cell marker CD20 and the T-cell marker CD3 was used to ensure that scoring was performed in B-cell–rich areas (defined as >80% B cells). One outlier case of MCL with NICD2 staining in 40% of cells was further evaluated by performing targeted exon sequencing. This revealed the presence of a NOTCH2 p.2402Qfs*9 PEST domain mutation in 45% of reads (N = 38), consistent with prior studies showing that a small subset of MCL has gain-of-function mutations in NOTCH2.37,38
Figure 2.
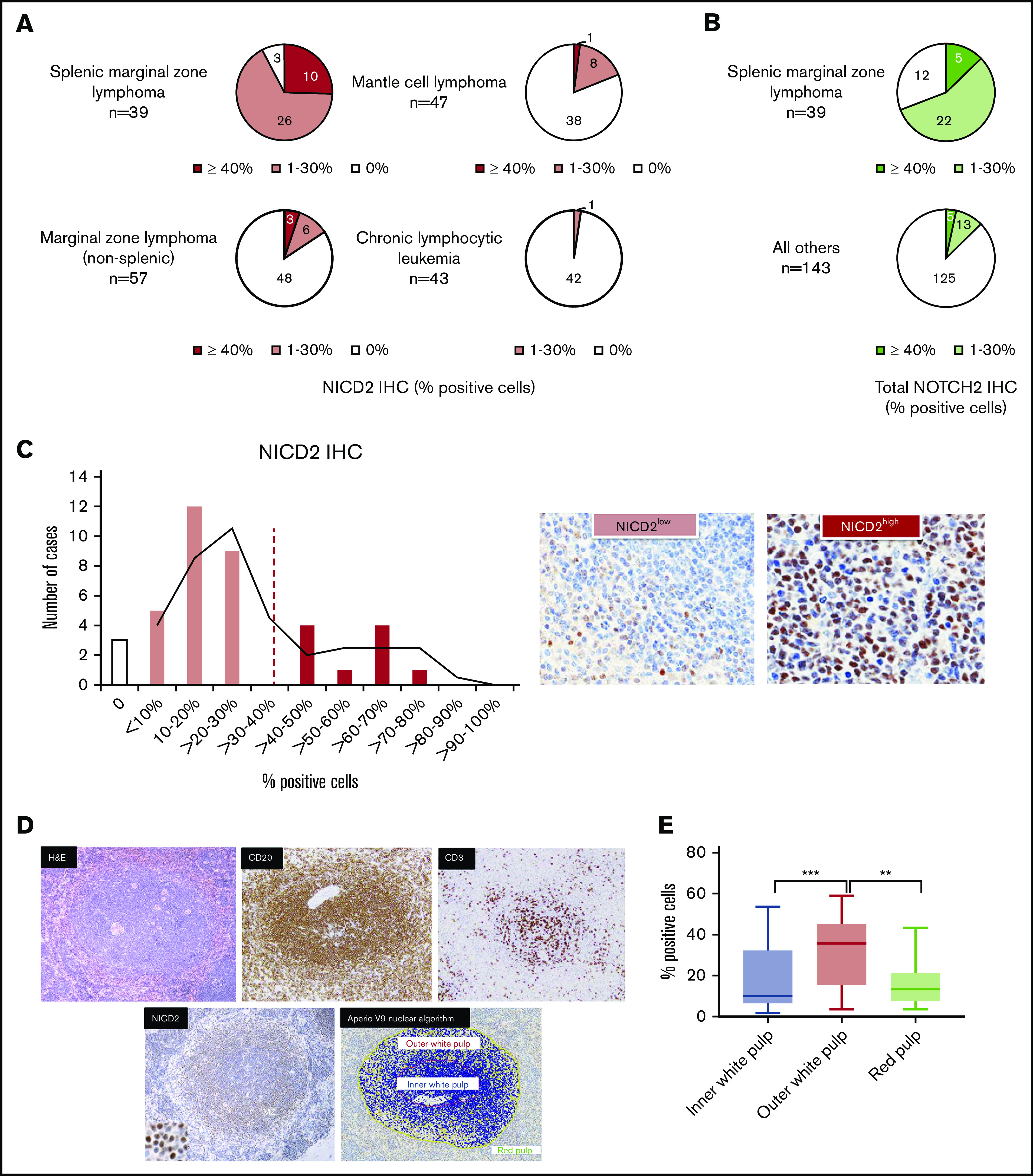
NOTCH2 activation is characteristic of SMZL and is preferentially seen in splenic marginal zones. (A) NICD2 staining in small B-cell lymphomas. (B) Total NOTCH2 staining in small B-cell lymphomas. (C) Histogram showing the percentage of NICD2+ cells across 39 cases of SMZL; representative images of tumors with high (≥40%) and low (<40%) levels of staining are shown. (D) Representative NICD2-stained case of SMZL (hematoxylin and eosin [H&E], CD3, CD20 stains also shown) and image-analyzed depiction of NICD2+ cells in the outer white pulp relative to the inner white pulp (original magnification: ×200; inset, ×400). Scoring was only performed in B-cell–rich areas (defined as >80% B cells). Yellow, positive cells; orange, strongly positive cells; blue, negative cells. (E) Box-and-whisker plots showing the frequency of NICD2+ cells in outer white pulp, red pulp, and inner white pulp in SMZL (***P < .001; **P < .01).
To further confirm that SMZL is associated with NOTCH2 activation, we performed staining with an antibody against a NOTCH2 intracellular epitope located ∼40 aa N-terminal of the PEST degron domain. This antibody recognizes immature and mature full-length NOTCH2 receptors as well as NICD2, which in aggregate we refer to as “total NOTCH2.” A limitation of this antibody is that the epitope it recognizes (centered at residue 2378) is sometimes absent in polypeptides encoded by NOTCH2-mutated alleles; indeed, all 3 NOTCH2-mutated SMZLs with PEST mutations retaining the total NOTCH2 epitope showed nuclear staining, whereas nuclear staining was only seen in 1 of 6 NOTCH2-mutated SMZLs with PEST mutations predicted to remove this epitope (supplemental Figure 1). Despite this limitation, stains performed with this antibody again showed significantly more frequent nuclear NOTCH2 staining in SMZL (69%; 27 of 39) than in other low-grade B-cell lymphomas (13%; 18 of 143) (Fisher exact test; P < .05) (Figure 2B). Thus, NOTCH2 protein and NOTCH2 activation is readily detectable in a large majority of SMZL, a feature that, among small B-cell lymphomas, is relatively specific for SMZL.
NOTCH2 activation in SMZL is highest in splenic marginal zones
Further evaluation of NICD2 staining in SMZL demonstrated that the fraction of B cells that was NICD2+ showed a bimodal distribution, with 29 of 39 tumors (74%; NICD2 low) showing staining in 30% or fewer cells and the remainder (26%; NICD2 high) showing staining in 40% or more cells (Figure 2C). We reasoned that if NOTCH2 activation stemmed from engagement with DLL1 expressed on marginal zone stromal cells, the level of NOTCH2 activation would be highest in marginal zones. To test this idea, we used image analysis to assess NICD2 staining in white pulp and red pulp compartments in spleens involved by SMZL (Figure 2D). As described previously, staining of serial sections for the B-cell marker CD20 and the T-cell marker CD3 was used to ensure that scoring was performed in B-cell–rich areas. We noted that the frequency of NICD2+ cells was highest in the marginal zones of the white pulp and lowest in the red pulp of involved spleens (Figure 2E), consistent with the idea that NOTCH2 activation occurs preferentially within splenic marginal zones.
NOTCH2 mutations are associated with stronger and more pervasive NOTCH2 activation in SMZL
To explore the relationship between NOTCH2 activation and NOTCH2 mutational status, we performed targeted exome sequencing on a subset of our collection of SMZLs (10 NICD2-high and 10 NICD2-low cases) using a well-characterized clinical NGS platform. Nine of the 10 tumors with high NICD2 levels had NOTCH2 3′ aberrations consisting of gain-of-function frameshift or nonsense mutations that result in loss of the C-terminal NOTCH2 PEST domain (Figure 3A-C). Other aberrations seen in our case cohort included a variety of pathogenic mutations in other genes, many already described as recurrent abnormalities in SMZL.11,12,39-41 Included among these were mutations in TP53 (30%), KLF2 (25%), NFKBIE (15%), and TNFAIP3 (10%), as well as the following copy-number changes: del(7q) (35%), gain of 3q (25%), and del(17p) (20%) (summarized in Figure 3A and described in detail in supplemental Tables 1-4). Correlation of NOTCH2 mutation status with NICD2 staining revealed a significantly higher frequency of NICD2+ cells in mutated vs wild-type tumors (Figure 3D). NOTCH2 variant allele frequencies varied from 4% to 90% (Figure 3B), consistent with some tumors having subclonal NOTCH2 mutations and others having undergone selection for increases in mutant NOTCH2 copy number during tumor progression. Notably, cases such as case 16 and case 20, which lacked detectable NOTCH2 mutations yet showed NICD2 staining in a substantial fraction of cells (Figure 3B), had driver mutations in SUZ12 and TP53 with variant allele frequencies of 22% and 54%, respectively (supplemental Table 4). This makes it unlikely that the failure to detect NOTCH2 mutations in these cases stemmed from dilution of tumor DNA with nontumor DNA, and by inference supports the idea that NOTCH2 activation also occurs in SMZLs with wild-type NOTCH2 alleles. Interestingly, we also noted that NICD2 staining in NOTCH2-mutated cases was less restricted to the outer white pulp than in NOTCH2 wild-type cases (Figure 3E-F), possibly because the longer half-life of PEST-deleted NICD2 allows the protein to persist after cells egress from ligand-rich microenvironments like the outer white pulp zone.
Figure 3.
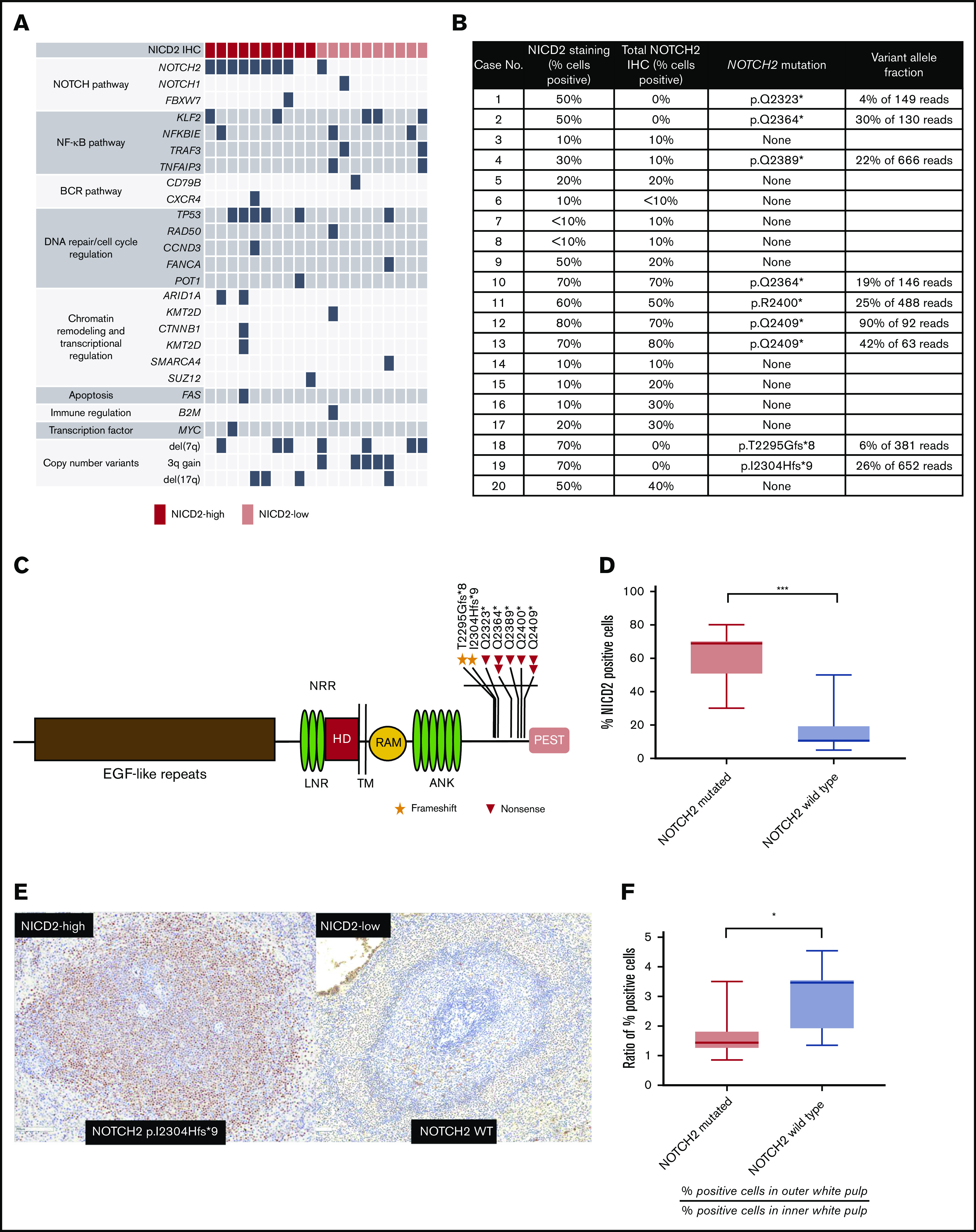
NOTCH2 mutation is associated with higher levels and wider distribution of NICD2 staining in SMZL. (A) Summary of pathogenic single-nucleotide, copy-number, and structural variants identified in 20 cases of SMZL. (B) Summary of NICD2 staining and NOTCH2 mutation status of genotyped SMZL cases. (C) Schematic of NOTCH2 mutations in SMZL. (D) Box-and-whisker plot showing frequency of NICD2+ cells in NOTCH2-mutated and wild-type SMZL (***P < .001). (E) Representative staining for NICD2 in NOTCH2-mutated and wild-type SMZL (original magnification, ×200). (F) Ratio of NICD2+ cells in outer and inner white pulp in NOTCH2-mutated vs wild-type cases (*P < .05). ANK, Ankyrin repeat; EGF, epidermal growth factor; HD, heterodimerization domain; LNR, Lin12/Notch repeat; RAM, RBP-Jκ/CBF1-associated module; TM, transmembrane domain.
Nuclear NOTCH2 staining is uncommon in BN2-subtype DLBCLs
Gain-of-function NOTCH2 mutations, generally 3′ mutations affecting the PEST domain that closely resemble those seen in SMZL, are found in 5% to 10% of DLBCL.7,8,21 These tumors most often belong to the BN2 genomic subgroup, are frequently associated with BCL6 gene rearrangements, and genetically resemble SMZL.34 An open question is what fraction of DLBCL have ongoing NOTCH2 activation and whether this is restricted to NOTCH2-mutated cases belonging to the BN2 subgroup.
To address this issue, we performed staining for NICD2 on a large cohort of previously characterized DLBCLs that had been subclassified according to the LymphGen algorithm.34 We observed NICD2 staining in DLBCL in only a minor subset of tumors (9 of 316; 2.8%), and then only in a minor subfraction (≤30%) of cells (Figure 4A-B). When NICD2 staining was identified, it was weaker than the staining observed in SMZLs (see representative NICD2 staining results in Figure 4H-I). Moreover, we failed to identify evidence of preferential NICD2 staining in BN2 tumors (3.3%) vs non-BN2 tumors (2.8%) (P = .6, 2-tailed Fisher exact test) (Figure 4A).
Figure 4.
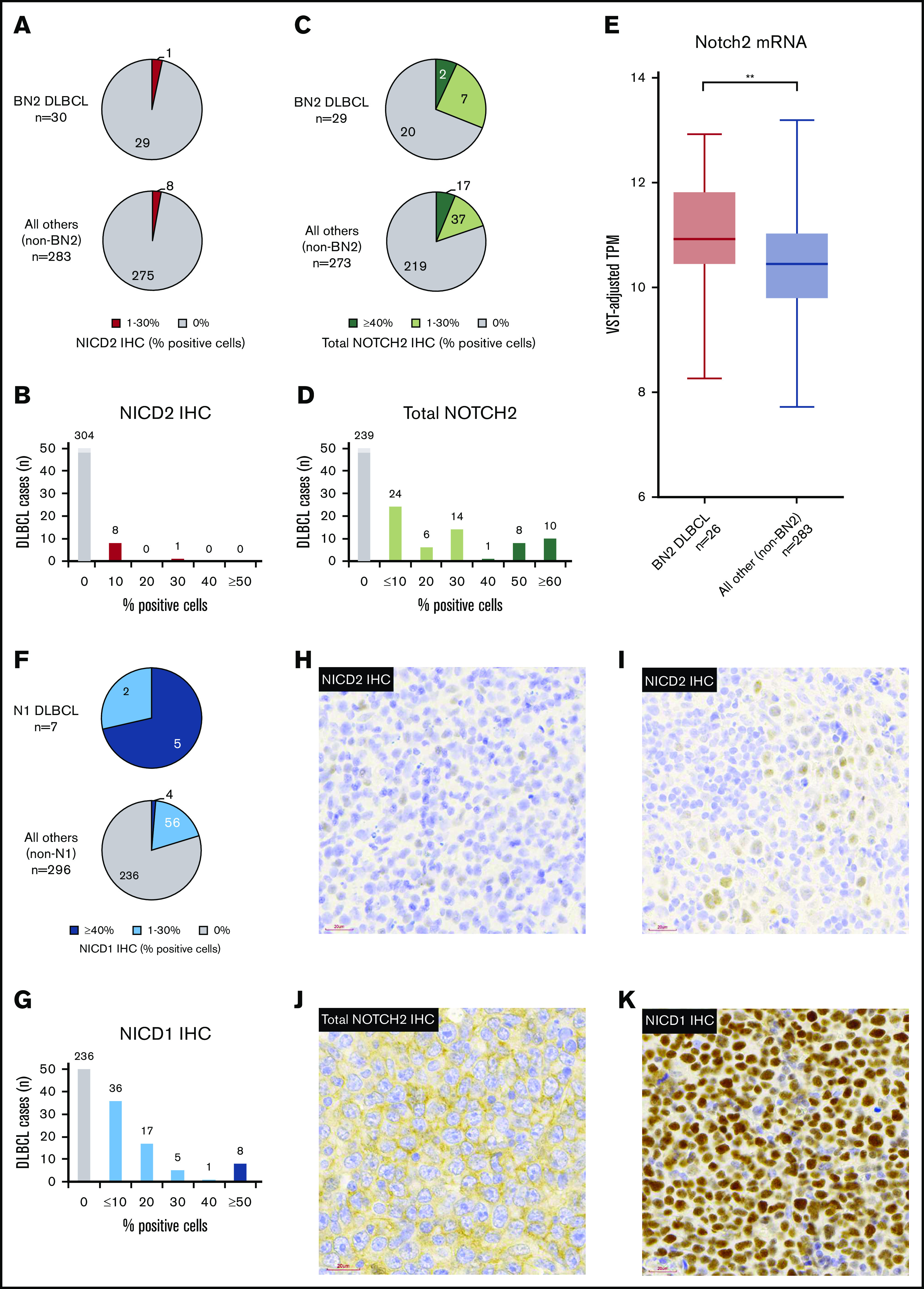
Nuclear staining for NOTCH2 is uncommon in BN2 DLBCLs, whereas nuclear staining for NOTCH1 is common in NOTCH1-mutated N1 DLBCLs. IHC was performed on a cohort of 314 DLBCLs. (A-B) Frequency and prevalence of NICD2 staining. (C-D) Frequency and prevalence of staining with an antibody that recognizes total NOTCH2. (E) NOTCH2 messenger RNA (mRNA) expression (variance stabilizing transformation [VST]-adjusted transcripts per kilobase million [TPM]) in BN2 DLBCL vs non-BN2 DLBCL cases (**P < .01). (F-G) Frequency and prevalence of NICD1 staining. (H-I) Representative images showing weak and limited NICD2 staining in DLBCL. (J) Example of DLBCL with diffuse membranous staining but absent nuclear staining for total NOTCH2. (K) Example of NOTCH1-mutated DLBCL with strong nuclear staining for NICD1. Images H-I were acquired using a MoticEasyScan Pro digital slide scanner (×40 magnification, standard mode; see https://moticeasyscan.com/product/moticeasyscan-pro/ for additional specifications) and Motic VM 3.0–Motic Digital Slide Assistant software (version 1.0.7.61b). All work was performed at room temperature in ambient air conditions. Fluorochromes were not required. Images were cropped using Adobe Photoshop Elements (version 14.0).
To further explore expression of NOTCH2 in DLBCL as well as the possibility that DLBCLs might harbor NOTCH2 rearrangements creating truncated genes that produce activated forms of NOTCH2 lacking the NICD2 neoepitope, we performed staining of DLBCLs for total NOTCH2. To confirm the ability of this antibody to detect nuclear NOTCH2 in tumors with NOTCH2 rearrangements, we first stained FFPE sections of triple-negative breast cancer patient-derived xenografts with or without NOTCH2 gene rearrangements. As expected, we noted intense nuclear and cytoplasmic staining in xenografts with NOTCH2 gene rearrangements, whereas no staining or only membrane staining was observed in xenografts with wild-type NOTCH2 alleles (see supplemental Figure 2 for representative results). When applied to our DLBCL cohort, total NOTCH2 staining was observed in a higher proportion of DLBCL than NICD2 staining (Figure 4C-D), with BN2 tumors trending toward having more frequent staining than non-BN2 tumors (31.0% vs 19.8%; P = .23, Fisher exact test). However, we failed to identify any DLBCLs with high levels of nuclear NOTCH2 staining with this second antibody.
Nuclear NOTCH2 staining is uncommon in DLBCLs expressing NOTCH2 and in NOTCH2-mutated DLBCL
To further evaluate our DLBCL staining results, we compared total NOTCH2 staining with NOTCH2 expression based on RNA-seq data, which were available for 286 DLBCL tumors stained for total NOTCH2. We observed a significant correlation between normalized NOTCH2 transcript counts and total NOTCH2 staining (Spearman rank order correlation, ρ = 0.27, P = 2.44 × 10−6) and also noted that normalized NOTCH2 transcript counts were significantly higher in BN2 DLBCLs than in non-BN2 DLBCLs (2-tailed Student t test, P = .00705; Figure 4E). However, even in tumors with appreciable staining for total NOTCH2, nuclear staining (indicative of NOTCH2 activation) was almost always lacking (Figure 4J). Thus, although a relative increase in NOTCH2 expression is associated with the BN2 subgroup of DLBCL, NOTCH2 activation is rarely seen in these tumors.
The targeted exome-sequencing panel used to characterize our DLBCL cohort did not include NOTCH2, so, in order to correlate NICD2 and total NOTCH2 staining with NOTCH2 mutational status, we interrogated our RNA-seq data. Despite highly variable coverage (supplemental Figure 3), we identified 9 DLBCLs with NOTCH2 exon 34 frameshift or nonsense mutations that produce NOTCH2 PEST domain deletions (summarized in Table 1). Five of these 9 tumors harbor BCL6 rearrangements, and 2 of the remaining 4 cases are associated with an “unclassified” cell of origin, features that correlate with the presence of NOTCH2 mutations.21 However, none of these tumors stained positively for NICD2 on TMA sections, and only 3 of 9 showed convincing reactivity for total NOTCH2, always in a nonnuclear pattern (supplemental Figure 4). To exclude the possibility that regions sampled on the TMA were not representative, we also carried out NICD2 and total NOTCH2 staining on whole-tissue sections (at least 1 × 1 cm in size) on all NOTCH2-mutated DLBCLs and observed highly similar results to those obtained on the stained TMA (summarized in supplemental Table 5). Thus, NOTCH2 activation appears to be uncommon in DLBCL, even in tumors expressing NOTCH2 and bearing NOTCH2 gain-of-function mutations.
Table 1.
Molecular and IHC data for NOTCH2-mutated DLBCL cases
| DLBCL case ID | Notch2 sequence variant29 | Variant reads/total (%) | LymphGen subtype34 | Cell of origin32 | BCL6 translocation | BCL6 IHC* | NICD2 IHC, % POS cells* | Total Notch2 IHC, % POS cells* |
|---|---|---|---|---|---|---|---|---|
| DLC_0048 | p.H2293Efs*21 | 38/289 (13) | EZB/A53 | UNCL | NEG | POS (strong) | 0 | 0 |
| DLC_0060 | p.R2400* | 9/25 (36) | Other | GCB | POS | POS (strong) | 0 | 0 |
| DLC_0201 | p.Q2367* | 69/173 (40) | BN2 | ABC | POS | POS (weak, subset) | 0 | 0 |
| DLC_0231 | p.Q2361* | 6/21 (29) | BN2/A53 | ABC | POS | POS (strong) | 0 | 10 |
| DLC_0239 | p.I2304Hfs*9 | 292/400 (73) | Other | UNCL | NEG | NEG (weak, subset) | 0 | 30 |
| DLC_0242 | p.T2355Hfs*45 | 2/4 (50) | ST2 | GCB | NEG | POS (strong) | 0 | 0 |
| DLC_0269 | p.Y2383* | 93/194 (48) | BN2 | GCB | POS | POS (strong) | 0 | 30 |
| DLC_0319 | p.E2290Kfs*5 | 10/26 (38) | A53 | ABC | NEG | POS (weak, subset) | 0 | 0 |
| DLC_0330 | p.R2400* | 9/46 (20) | MCD | ABC | POS | POS (strong) | 0 | 0 |
ABC, activated B cell; GCB, germinal center B; ID, identifier; NEG, negative; POS, positive; UNCL, unclassifiable.
See supplemental Figure 4 for representative staining results.
NOTCH1 activation is common in NOTCH1-mutated, N1 DLBCLs
We next asked whether DLBCLs containing NOTCH1 3′ PEST domain mutations, corresponding to the N1 subtype, showed increased levels of NOTCH1 activation as compared with tumors without such mutations. We observed that NOTCH1-mutated DLBCLs (N = 7) were uniformly associated with NOTCH1 activation as assessed by NICD1 staining (Figure 4F), and that NOTCH1 activation was observed significantly more frequently than in NOTCH1–wild-type tumors (100% vs 20.2%, P < .05, Fisher exact test). Also, among NICD1+ tumors, widespread staining (defined as >30% of cells; Figure 4G) was more commonly seen in NOTCH1-mutated tumors than in NOTCH1–wild-type tumors (71.4% vs 1.4%, P < .05, Fisher exact test). A representative image of an NICD1 “high” N1 DLBCL is shown in Figure 4K. We conclude that unlike NOTCH2-mutated DLBCLs, NOTCH1-mutated DLBCLs frequently show evidence of ongoing Notch activation.
Notch target genes are upregulated in NOTCH1-mutated N1 DLBCL but not in NOTCH2-mutated DLBCL
A prediction of our immunohistochemical findings is that expression of Notch target genes should differ between NOTCH1- and NOTCH2-mutated DLBCL. To evaluate this, we performed gene-set enrichment analysis on our DLBCL RNA-seq data set using putative Notch target genes identified in 3 different B-cell contexts: follicular lymphoma,35 MCL,16 and CLL36 (Figure 5). We observed that although Notch target gene expression was significantly higher in Notch-mutated DLBCL than Notch wild-type DLBCL, this difference was confined to the NOTCH1-mutated N1 subtype, as Notch target gene expression in NOTCH2-mutated tumors and Notch wild-type tumors did not differ significantly (Figure 5A). Furthermore, Notch target gene expression was significantly lower in BN2 DLBCLs than in N1 DLBCLs and did not differ from DLBCLs belonging to other subgroups (Figure 5B). These analyses provide additional support for the idea that ongoing Notch signaling is a feature of NOTCH1-mutated DLBCLs and not NOTCH2-mutated or BN2 DLBCLs.
Figure 5.
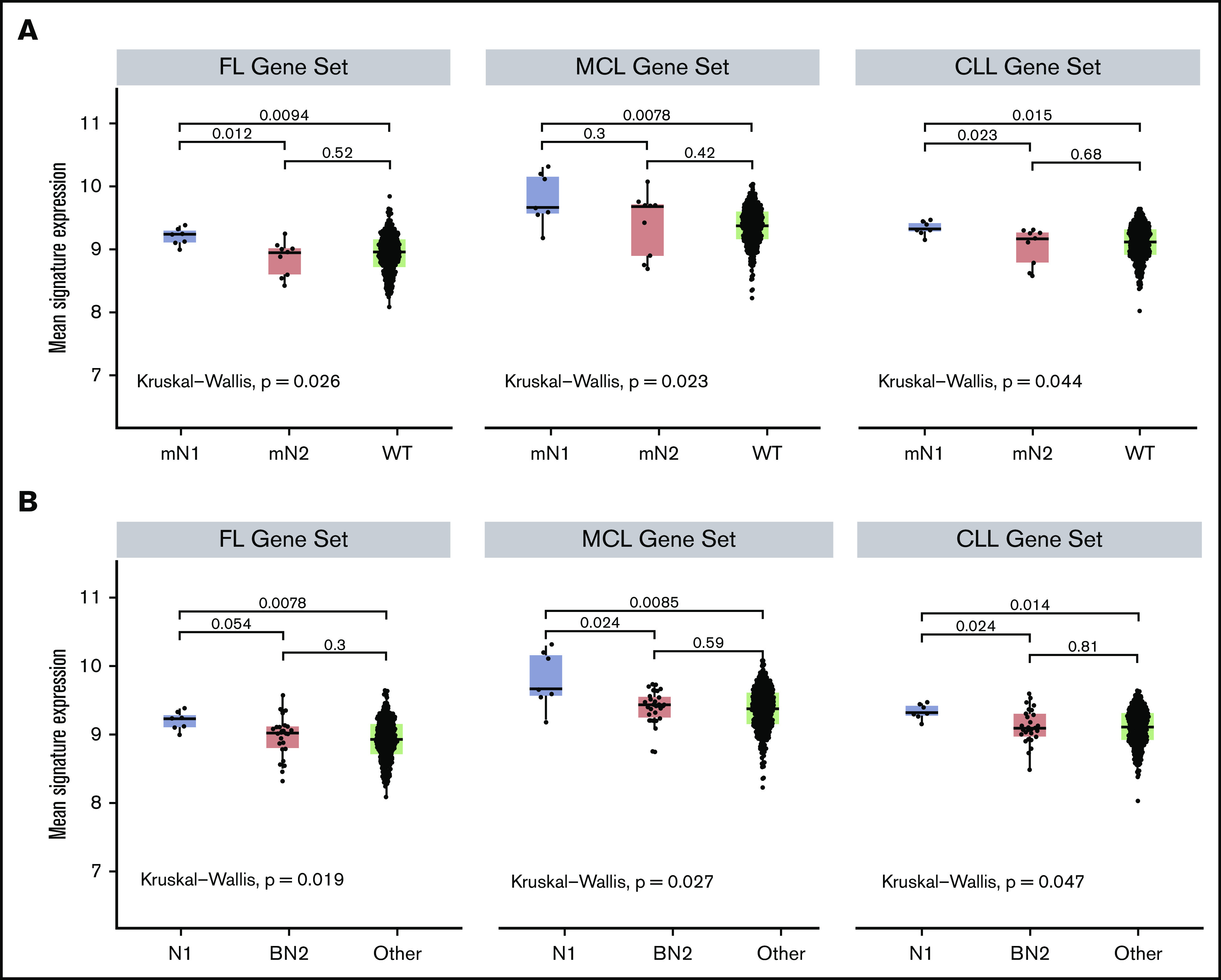
Notch target gene expression is elevated in NOTCH1-mutated DLBCLs and not in NOTCH2-mutated or BN2 DLBCLs. Three hundred nine DLBCLs with RNA-seq data were sorted into (A) NOTCH1-mutated (mN1, N = 7), NOTCH2-mutated (mN2, N = 9), and Notch wild-type (WT, N = 293) subgroups, or (B) N1, BN2, and “other” subgroups, and then compared for expression of Notch target genes using Notch-sensitive genes identified in follicular lymphoma (FL), MCL, and CLL. The Kruskal-Wallis test was used to evaluate gene expression in Notch-mutated and Notch wild-type tumors, whereas the Mann-Whitney U test was used to do pairwise evaluations of gene expression across all 3 DLBCL subgroups.
Discussion
Mutational data implicate Notch signaling in the pathogenesis of mature B-cell neoplasms,5-7,9,11-14,40,42 and the presence of gain-of-function mutations in NOTCH1/2 has been associated with worse outcomes in CLL/SLL,43-45 MCL,14 and SMZL11,46 as well as transformation of CLL4,44 and follicular lymphoma.47 These findings indicate that Notch is an important oncogenic driver in B-cell neoplasms and, taken at face value, suggest that it is also a rational therapeutic target. This may still prove true, at least in a subset of B-cell neoplasms, but our work here studying Notch activation in diverse B-cell tumors and its relationship to the microenvironment and Notch mutational status suggests a complex, nuanced role for Notch in these diseases.
In situ staining of cohorts of small B-cell neoplasms indicates that NOTCH2 activation is nearly ubiquitous in SMZL and less common in other low-grade B-cell neoplasms, in line with past work showing that NOTCH2 mutations are also significantly more frequent in SMZL.46 We observed the highest levels of activated NOTCH2 in involved splenic marginal zones, consistent with the idea that even in NOTCH2-mutated tumors, receptor activation depends on engagement with a ligand, presumably DLL1 expressed on splenic marginal zone stromal cells. This is analogous to prior work in which we observed that in CLL/SLL, levels of activated NOTCH1 were highest in lymph node as compared with extranodal locations.15 Our observations are in line with the idea that 3′ mutations affecting the Notch PEST domain in B-cell neoplasms serve to increase and sustain signaling in a specific microenvironment in which ligand-mediated Notch activation has favorable effects on some pro-oncogenic facet of tumor cell behavior. Because strong gain-of-function mutations in the NRR-coding regions of NOTCH1 and NOTCH2 that lead to ligand-independent Notch activation have not been identified in primary B-cell neoplasms, selective pressure for Notch signaling in these neoplasms may only exist in certain ligand-rich microanatomic locations. A corollary that follows is that Notch mutations in small B-cell neoplasms such as SMZL and CLL/SLL are unlikely to be initiating events, but instead provide a selective advantage in tumors that have already established themselves in ligand-rich microenvironments. Further evaluation of these ideas will benefit from the development of reliable reagents and methods for studying the expression of Notch ligands such as DLL1 and DLL4 in human FFPE tissues and deeper characterization of the stromal compartments in which these tumors preferentially grow.
The idea that NOTCH1 and NOTCH2 mutations in small B-cell neoplasms are likely to be secondary events associated with disease progression, based on the logic outlined herein, also has implications for the observation that Notch mutations are consistently associated with worse clinical outcomes in these neoplasms.11,43-46 Rather than more aggressive disease being a consequence of increased Notch signaling per se, as progression events, mutations in NOTCH1 and NOTCH2 may instead identify tumors that are, on average, more genetically heterogeneous and therefore more likely by chance to harbor subclones that are more aggressive and/or convey resistance to therapy.
Our studies also suggest that the connection between Notch signaling and the ontogeny of DLBCL is complex. We observed no association between NOTCH2 activation and the BN2 subtype of DLBCL, which is defined in part by the presence of NOTCH2 mutations, nor did we identify significant NOTCH2 activation or evidence of upregulation of Notch target genes in DLBCLs with NOTCH2 PEST mutations. In fact, none of the DLBCLs screened had levels of NOTCH2 activation approximating those seen commonly in SMZL. We speculate that the explanation for this seeming clash between tumor genetics and NOTCH2 activation status relates to the evolutionary history of NOTCH2-mutated DLBCLs. One plausible explanation is for a large fraction of NOTCH2-mutated DLBCLs to arise from transformation of unrecognized overt SMZL or subclinical SMZL-like precursor cells. Evidence supporting this idea includes the observation that DLBCLs arising in the setting of hepatitis C infection, a risk factor for SMZL, are also strongly associated with NOTCH2 PEST domain mutations,42 and the existence of certain commonalities among genes/pathways that are frequently mutated along with NOTCH2 in DLBCL and SMZL, particularly components of the NF-κB–signaling pathway.48 A striking genetic distinction between NOTCH2-mutated SMZL and NOTCH2-mutated DLBCL is that the latter is strongly associated with BCL6 translocations,7,21 which could serve as a transforming event that lessens or eliminates dependence on NOTCH2 expression and signaling. Consistent with this possibility, BCL6 has been reported to antagonize NOTCH2 function in B cells by repressing its expression and function.35Further studies, including use of NOTCH2-mutated mouse models, are needed to evaluate these ideas.
By contrast to NOTCH2-mutated DLBCLs, NOTCH1-mutated DLBCLs do show evidence of ongoing Notch signaling. Notably, the observed differences between NOTCH1- and NOTCH2-mutated DLBCL are unlikely to be explained by inherent functional differences between activated NOTCH1 and activated NOTCH2, as mice in which the intracellular domain of NOTCH1 is swapped with that of NOTCH2, or vice versa, are normal.49 Other factors that may contribute to this distinction include differences in tumor cell context and in the tumor microenvironment. The outcome of Notch signaling is highly context dependent and, given that NOTCH1- and NOTCH2-mutated DLBCL show substantial differences in gene expression and associated genomic alterations, it is likely that phenotypes produced by Notch activation diverge in these DLBCL subsets. A second possible factor is related to the identity of Notch ligands that are expressed in the nodal microenvironment. Biochemical studies have shown that Notch ligand DLL4 has higher affinity for NOTCH1 than NOTCH2,50 and preferential expression of DLL4 in lymph nodes, where DLL4 has important roles in specifying T-follicular helper cell fate51 and generating alloimmune responses,52 could also contribute to the apparent difference in NOTCH1 vs NOTCH2 activation in nodal DLBCLs. Characterization of lymph node stromal fractions and expression of Notch ligands by these cells will be necessary to evaluate this latter possibility.
Finally, our work has implications for the prospects of Notch-directed therapeutics in B-cell neoplasms. Prior preclinical and clinical studies have shown that response of cancers (including B-cell neoplasms) to single-agent Notch pathway inhibitor therapy is restricted to tumors with high levels of nuclear Notch16,18; thus, our work strongly suggests that NOTCH2-mutated DLBCLs are unlikely to be responsive to Notch-directed therapeutics. By contrast, Notch inhibitors may have a role in SMZL and NOTCH1-mutated DLBCL, particularly in combination with other agents. Further work is needed to test these ideas.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the Cancer Cell Line Factory at the Broad Institute of MIT and Harvard for providing the DLBCL cell lines.
This work was supported by a grant from the Leukemia & Lymphoma Society (J.C.A.).
Footnotes
RNA-sequencing data sets are available at https://www.ebi.ac.uk/ega/studies/EGAS00001002657.
Detailed protocols for immunohistochemical staining methods are available upon request.
Authorship
Contribution: V.S., J.W.C., D.W.S., L.K.H., M.H.N., C.K.R., and S.L. analyzed results; K.F. generated expression constructs and western blot data; V.S. and J.W.C. made the figures; B.F. developed and validated immunohistochemical assays; and V.S., J.W.C., and J.C.A. designed the research and wrote the paper.
Conflict-of-interest disclosure: J.C.A. is a consultant for Cellestia, Inc, and for Ayala Pharmaceuticals (the work for these companies is not in conflict with the work described herein). D.W.S. is a consultant for AbbVie, AstraZeneca, Celgene, and Janssen; has research funding from Janssen and NanoString Technologies; and is a named inventor on a patent licensed to NanoString Technologies (his work for these companies is not in conflict with the work described herein). The remaining authors declare no competing financial interests.
Correspondence: Jon C. Aster, Department of Pathology, Brigham and Women’s Hospital, Boston, MA 02115; e-mail: jaster@rics.bwh.harvard.edu.
References
- 1.Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17(11):722-735. [DOI] [PubMed] [Google Scholar]
- 2.Aster JC, Pear WS, Blacklow SC. The varied roles of Notch in cancer. Annu Rev Pathol. 2017;12:245-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Ianni M, Baldoni S, Rosati E, et al. . A new genetic lesion in B-CLL: a NOTCH1 PEST domain mutation. Br J Haematol. 2009;146(6):689-691. [DOI] [PubMed] [Google Scholar]
- 4.Fabbri G, Rasi S, Rossi D, et al. . Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208(7):1389-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puente XS, Beà S, Valdés-Mas R, et al. . Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519-524. [DOI] [PubMed] [Google Scholar]
- 6.Puente XS, Pinyol M, Quesada V, et al. . Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapuy B, Stewart C, Dunford AJ, et al. . Molecular subtypes of diffuse large B-cell lymphoma are associated with distinct pathogenic mechanisms and outcomes [published correction appears in Nat Med. 2018;24(8):1290-1291]. Nat Med. 2018;24(5):679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karube K, Enjuanes A, Dlouhy I, et al. . Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia. 2018;32(3):675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohr JG, Stojanov P, Lawrence MS, et al. . Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109(10):3879-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright GW, Wilson WH, Staudt LM. Genetics of diffuse large B-cell lymphoma [letter]. N Engl J Med. 2018;379(5):493-494. [DOI] [PubMed] [Google Scholar]
- 11.Kiel MJ, Velusamy T, Betz BL, et al. . Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med. 2012;209(9):1553-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi D, Trifonov V, Fangazio M, et al. . The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209(9):1537-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SY, Kumano K, Nakazaki K, et al. . Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci. 2009;100(5):920-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kridel R, Meissner B, Rogic S, et al. . Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119(9):1963-1971. [DOI] [PubMed] [Google Scholar]
- 15.Kluk MJ, Ashworth T, Wang H, et al. . Gauging NOTCH1 activation in cancer using Immunohistochemistry. PLoS One. 2013;8(6):e67306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan RJH, Petrovic J, Rausch DM, et al. . A B cell regulome links Notch to downstream oncogenic pathways in small B cell lymphomas. Cell Rep. 2017;21(3):784-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crotty R, Dias-Santagata D, Aster JC, Nardi V. A novel SEC22B-NOTCH2 fusion in chronic lymphocytic leukemia. Human Pathol Case Rep. 2020;21:200408. [Google Scholar]
- 18.Stoeck A, Lejnine S, Truong A, et al. . Discovery of biomarkers predictive of GSI response in triple-negative breast cancer and adenoid cystic carcinoma. Cancer Discov. 2014;4(10):1154-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hozumi K, Negishi N, Suzuki D, et al. . Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol. 2004;5(6):638-644. [DOI] [PubMed] [Google Scholar]
- 20.Witt CM, Won WJ, Hurez V, Klug CA. Notch2 haploinsufficiency results in diminished B1 B cells and a severe reduction in marginal zone B cells. J Immunol. 2003;171(6):2783-2788. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz R, Wright GW, Huang DW, et al. . Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swerdlow SH, Campo E, Pileri SA, et al. . The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagle N, Berger MF, Davis MJ, et al. . High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abo RP, Ducar M, Garcia EP, et al. . BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res. 2015;43(3):e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DePristo MA, Banks E, Poplin R, et al. . A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A, Hanna M, Banks E, et al. . The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cibulskis K, Lawrence MS, Carter SL, et al. . Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arthur SE, Jiang A, Grande BM, et al. . Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat Commun. 2018;9(1):4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ennishi D, Mottok A, Ben-Neriah S, et al. . Genetic profiling of MYC and BCL2 in diffuse large B-cell lymphoma determines cell-of-origin-specific clinical impact. Blood. 2017;129(20):2760-2770. [DOI] [PubMed] [Google Scholar]
- 31.Scott DW, King RL, Staiger AM, et al. . High-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with diffuse large B-cell lymphoma morphology. Blood. 2018;131(18):2060-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott DW, Mottok A, Ennishi D, et al. . Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J Clin Oncol. 2015;33(26):2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott DW, Wright GW, Williams PM, et al. . Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123(8):1214-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright GW, Huang DW, Phelan JD, et al. . A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37(4):551-568.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valls E, Lobry C, Geng H, et al. . BCL6 antagonizes NOTCH2 to maintain survival of human follicular lymphoma cells. Cancer Discov. 2017;7(5):506-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabbri G, Holmes AB, Viganotti M, et al. . Common nonmutational NOTCH1 activation in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2017;114(14):E2911-E2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beà S, Valdés-Mas R, Navarro A, et al. . Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci USA. 2013;110(45):18250-18255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang P, Zhang W, Wang J, Liu Y, An R, Jing H. Genomic landscape and prognostic analysis of mantle cell lymphoma. Cancer Gene Ther. 2018;25(5-6):129-140. [DOI] [PubMed] [Google Scholar]
- 39.Clipson A, Wang M, de Leval L, et al. . KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia. 2015;29(5):1177-1185. [DOI] [PubMed] [Google Scholar]
- 40.Parry M, Rose-Zerilli MJ, Gibson J, et al. . Whole exome sequencing identifies novel recurrently mutated genes in patients with splenic marginal zone lymphoma. PLoS One. 2013;8(12):e83244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spina V, Khiabanian H, Messina M, et al. . The genetics of nodal marginal zone lymphoma. Blood. 2016;128(10):1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arcaini L, Rossi D, Lucioni M, et al. . The NOTCH pathway is recurrently mutated in diffuse large B-cell lymphoma associated with hepatitis C virus infection. Haematologica. 2015;100(2):246-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Giudice I, Rossi D, Chiaretti S, et al. . NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 2012;97(3):437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossi D, Rasi S, Fabbri G, et al. . Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood. 2012;119(2):521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willander K, Dutta RK, Ungerbäck J, et al. . NOTCH1 mutations influence survival in chronic lymphocytic leukemia patients. BMC Cancer. 2013;13:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campos-Martín Y, Martínez N, Martínez-López A, et al. . Clinical and diagnostic relevance of NOTCH2-and KLF2-mutations in splenic marginal zone lymphoma. Haematologica. 2017;102(8):e310-e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karube K, Martínez D, Royo C, et al. . Recurrent mutations of NOTCH genes in follicular lymphoma identify a distinctive subset of tumours. J Pathol. 2014;234(3):423-430. [DOI] [PubMed] [Google Scholar]
- 48.Rossi D, Deaglio S, Dominguez-Sola D, et al. . Alteration of BIRC3 and multiple other NF-κB pathway genes in splenic marginal zone lymphoma. Blood. 2011;118(18):4930-4934. [DOI] [PubMed] [Google Scholar]
- 49.Liu Z, Brunskill E, Varnum-Finney B, et al. . The intracellular domains of Notch1 and Notch2 are functionally equivalent during development and carcinogenesis. Development. 2015;142(14):2452-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrawes MB, Xu X, Liu H, et al. . Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J Biol Chem. 2013;288(35):25477-25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fasnacht N, Huang HY, Koch U, et al. . Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J Exp Med. 2014;211(11):2265-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung J, Ebens CL, Perkey E, et al. . Fibroblastic niches prime T cell alloimmunity through Delta-like Notch ligands. J Clin Invest. 2017;127(4):1574-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.