Yeast surface display was applied to a library of hematopoietic progenitor kinase 1 (HPK1) kinase domain variants in order to select those with an improved expression level and solubility. The HPK1 variant with the most improved properties contained two mutations and crystallized readily in complex with several small-molecule inhibitors.
Keywords: hematopoietic progenitor kinase 1, HPK1, homology modeling, yeast surface display, crystallizability
Abstract
Hematopoietic progenitor kinase 1 (HPK1) is an intracellular kinase that plays an important role in modulating tumor immune response and thus is an attractive target for drug discovery. Crystallization of the wild-type HPK1 kinase domain has been hampered by poor expression in recombinant systems and poor solubility. In this study, yeast surface display was applied to a library of HPK1 kinase-domain variants in order to select variants with an improved expression level and solubility. The HPK1 variant with the most improved properties contained two mutations, crystallized readily in complex with several small-molecule inhibitors and provided valuable insight to guide structure-based drug design. This work exemplifies the benefit of yeast surface display towards engineering crystallizable proteins and thus enabling structure-based drug discovery.
1. Introduction
Hematopoietic progenitor kinase 1 (HPK1; also called MAP4K1; UniProt Q92918), an STE20 kinase found in hematopoietic and progenitor cells (Hu et al., 1996 ▸), consists of a central SH3-binding proline-rich motif flanked by a kinase and a citron homology domain at the N- and C-terminus, respectively (Hernandez et al., 2018 ▸). Upon T-cell receptor (TCR) activation, HPK1 promotes the phosphorylation of adapter proteins (Sauer et al., 2001 ▸), leading to the breakup of TCR signaling clusters (Lasserre et al., 2011 ▸) and negative regulation of T-cell-mediated immune response, which is a hallmark of many cancers (Shui et al., 2007 ▸; Alzabin et al., 2010 ▸). Loss of HPK1 kinase activity reduces the inhibition of T-cell signaling, triggering an increase in antitumor immune-cell populations in the tumor environment and thus suppressing tumor growth (Liu et al., 2019 ▸). The antitumor effect is even greater when HPK1 inhibition is combined with antibody checkpoint inhibitors (Darvin et al., 2018 ▸; You et al., 2021 ▸). Thus, small-molecule inhibitors of HPK1 promise to augment current antibody-based immunotherapy in oncology (Hernandez et al., 2018 ▸).
Here, we report the use of yeast surface display (YSD), an in vitro protein-selection method, to enable the soluble expression and crystallization of the HPK1 kinase domain. In YSD, a gene encoding the protein of interest is cloned in a yeast expression vector as a fusion with a native yeast cell-wall protein. Upon expression, the cell-wall portion of the protein becomes embedded in the yeast cell wall, and the protein being engineered is displayed on the yeast surface (Boder & Wittrup, 1997 ▸; Cho et al., 1998 ▸; Rakestraw et al., 2011 ▸; Rhiel et al., 2014 ▸). Selection from a library of protein variants allows the enrichment and subsequent recovery of genes encoding variants with desirable properties. Early work by Wittrup and coworkers showed that the number of copies of a protein displayed on a yeast cell (the ‘display level’) correlates with its thermal stability and soluble secretion level (Shusta et al., 1999 ▸). Since then, YSD has been applied to engineering resistance to aggregation, an increased expression level and increased stability (Shusta et al., 2000 ▸; Traxlmayr et al., 2012 ▸, 2013 ▸; Pavoor et al., 2012 ▸), all of which are biochemical attributes that are known to favor protein crystallization.
We set out to re-engineer the HPK1 kinase domain with the goal of improving its biophysical properties while retaining its native-like fold and function. We expected the improved biophysical properties to lead to both a higher level of soluble expression and a greater ease of crystallization. Using a combination of YSD and site-directed mutagenesis, we identified a double mutant of HPK1 kinase with the desired properties. The crystal structure of this mutant confirmed that it had a wild type-like structure; this allowed us to use the structure as a starting point for the structure-based drug design of selective small-molecule inhibitors of HPK1 kinase.
2. Materials and methods
2.1. Library design and selection by yeast surface display
In order to improve the recombinant expression of an HPK1 fragment containing the kinase domain (residues 1–346), we set out to identify residues on the surface of the protein that might contribute to the potential aggregation or insolubility of this poorly expressing protein. In the absence of reported crystal structures of HPK1 at the time, we built homology models of the HPK1 kinase domain (Prime version 2014-4; Schrödinger, New York, USA) using the crystal structures of related kinases as templates: MST1 (PDB entry 3com), MST2 (PDB entry 4lg4), MST3 (PDB entry 3a7f), MST4 (PDB entry 3ggf) and HGK (PDB entry 4obo), as well as two additional related kinases (data not published). Visual inspection, spatial aggregation propensity (SAP) as implemented in BIOVIA Discovery Studio (version 2015; Dassault Systèmes, San Diego, California, USA) and surface-aggregation prediction using BioLuminate (version 2014-4; Schrödinger) were used to identify aggregation-prone sites on the surface of the protein. Each of the residues at the predicted aggregation-prone sites was virtually mutated to Asp, Asn, Thr, Ala, Glu, Gln or Ser and the protein stability and surface-aggregation potentials were calculated (BIOVIA; Chennamsetty et al., 2009 ▸). The eight residues for which mutations to less hydrophobic residues were predicted to lead to the greatest reduction in aggregation were Leu64, Leu84, Leu112, Leu170, Leu188, Leu221, Phe225 and Leu285. We designed a library in which each of these eight residues was mutated to Glu, Asp, Gln, Asn and Thr in all possible permutations, leading to a total of 1 679 616 possible mutants (Supplementary Fig. S1), which we were able to oversample by an order of magnitude after transforming the library into yeast using electroporation.
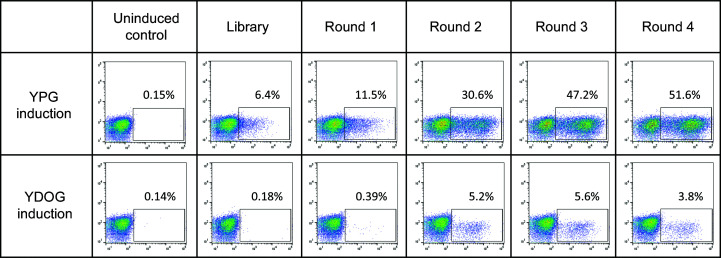
The yeast surface-display workflow was adapted from an earlier application to the selection of thermostable Adnectins (Lipovšek et al., 2018 ▸). The HPK1 library was assembled from pools of synthetic oligonucleotides that encoded the desired sequences using overlap extension polymerase chain reaction (Section S1, Supplementary Fig. S2). Next, library DNA was PCR-amplified with primers complementary to the cloning sites in yeast display vector pDV-154 (Section S2) and then co-transformed with linearized plasmid (Section S2.1) into yeast strain VWK18gal− by electroporation (Benatuil et al., 2010 ▸), resulting in an HPK1-c-Myc epitope-α-agglutinin 1 fusion. Transformed yeast cells were allowed to divide and were then induced using galactose (Section S3). To detect HPK1 display, the induced yeast was labeled with a combination of 9E10, a murine anti-c-Myc (BioLegend, San Diego, Calfornia, USA) primary antibody, and goat anti-mouse IgG Alexa Fluor 647 (Invitrogen, Carlsbad, California, USA), a fluorescently conjugated secondary antibody. During cell sorting, the yeast cells showing the highest level of fluorescence, which was presumably correlated with high levels of HPK1 display and high HPK1 solubility, were captured using a BD FACS ARIA II cell sorter (BD Bioscience, San Jose, California, USA). The captured yeast cells were grown, induced, labeled and then sorted three more times (Supplementary Table S1). The HPK1 display levels in yeast populations at different stages of selection were compared by analytical flow cytometry (Fig. 1 ▸). Plasmids containing the genes of enriched HPK1 variants were recovered from the post-round-3 and post-round-4 yeast populations using a Zymoprep Yeast Plasmid Miniprep Kit II (Zymo Research, Irvine, California, USA). The HPK1-variant genes from the enriched populations were amplified, subcloned into plasmid pET-9d and sequenced (Koide et al., 2012 ▸).
Figure 1.
Flow-cytometry analysis showing the increase in the display level of HPK1 on the surface of induced yeast after consecutive rounds of selection for high surface display. Yeast populations collected after each round of selection were regrown, induced and labeled with an epitope tag C-terminal to the HPK1 kinase domain. In the bivariate histograms, the vertical axis shows the mean fluorescence intensity (MFI) of yeast intrinsic fluorescence and the horizontal axis shows the MFI after labeling the epitope tag C-terminal to HPK1. Data panels in the two rows show the display of HPK1 induced using galactose in YPG (rich) medium versus YDOG (minimal) medium. The number in each panel corresponds to the percentage of yeast in each population that falls within the rectangular analytical gate; this number is a measure of the fraction of yeast cells that show detectable display of HPK1 variants. Regardless of the medium used in the selection, both the fraction of yeast displaying HPK1 and the MFI signal intensity of the labeled yeast increase with rounds of selection.
In this study, the wild-type human HPK1 kinase domain (residues 1–346) and all of the HPK1 variants chosen for characterization were codon-optimized for expression in baculovirus. The genes were synthesized as NdeI–XhoI fragments by GenScript (Piscataway, New Jersey, USA) and then cloned into a modified pFastBac1 vector (Invitrogen, Carlsbad) that included an N-terminal His tag and Tobacco vein mottling virus (TVMV) protease cleavage site.
2.2. Protein expression and characterization
For the constructs chosen for small-scale expression testing, the Bac-to-Bac system (Invitrogen, Carlsbad, California, USA) was used to generate recombinant HPK1 baculovirus. Sf9 insect cells were grown in ESF921 medium (Expression Systems, Davis, California, USA) to a density of 2.2 × 106 cells per millilitre. HPK1 recombinant baculovirus (1–2 × 107 virus particles per millilitre) was used to infect a 3 ml aliquot of Sf9 cells in a 24-deep-well block with shaking at 230 rev min−1 and 300 K for 65 h. Expressed proteins were purified from the cell pellets by one-step Ni–NTA batch purification and were analyzed by SDS–PAGE.
Large-scale expression, 0.8–20 l, was carried out in either 2 l shake flasks with a vented cap or in the Wave Bioreactor System 20/50 EHT (GE Healthcare Life Sciences, Marlborough, Massachusetts, USA). Sf9 cells at a density of 2.2 × 106 cells per millilitre were infected with HPK1 recombinant baculovirus (1–1.5 × 108 virus particles per millilitre). Cultures in flasks were maintained at 130 rev min−1 and 300 K for 65 h, whereas Wave reactors were maintained at 300 K for 65 h with wave settings suited to the culture volumes. The cell pellets were harvested by centrifugation and stored at 193 K. Prior to purification, frozen Sf9 cells were resuspended in buffer A [50 mM Tris–HCl pH 8.5, 300 mM NaCl, 25 mM imidazole, 5 mM DTT, 5%(v/v) glycerol] plus cOmplete, EDTA-free protease-inhibitor tablets (Roche, Mannheim, Germany), lysed by nitrogen cavitation at 2.4 MPa for 40 min (Parr Instrument Co., Moline, Illinois, USA) and clarified by centrifugation. The supernatant was applied onto a 5 ml HisTrap FF column (GE Healthcare Life Sciences, Marlborough, Massachusetts, USA), washed and eluted with buffer A containing 300 mM imidazole. Peak UV absorbance fractions from the elution were pooled, concentrated (using Amicon Ultra-15 centrifugal filters, 10 kDa cutoff), applied onto a Superdex 200 column (26 × 600 mm; GE Healthcare Life Sciences) equilibrated with SEC buffer (25 mM Tris–HCl, 200 mM NaCl, 2 mM DTT, 0.5 mM EDTA 5% glycerol pH 8.25) and eluted. SEC peak fractions were combined, flash-frozen and stored at 193 K. The yield for most of the mutants was at least 3 mg purified protein per litre of culture. Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | Homo sapiens |
| DNA source | DNA fragment generated by synthesis |
| Expression vector | pFastBac1 |
| Expression host | Sf9 insect cells |
| Complete amino-acid sequence of the construct produced | MDVVDPDIFNRDPRDHYDLLQRLGGGTYGEVFKARDKVSGDLVALKMVKMEPDDDVSTLQKEILILKTCRHANIVAYHGSYLWLQKLWICMEFCGAGSLQDIYQVTGSLSELQISYVCREVLQGLAYLHSQKKIHRDIKGANILINDAGEVRLADFGISAQIGATLARRLSFIGTPYWMAPEVAAVALKGGYNELCDIWSLGITAIELAELQPPLFDVHPDRVLELMTKSGYQPPRLKEKGKWSAAFHNFIKVTLTKSPKKRPSATKMLSHQLVSQPGLNRGLILDLLDKLKNPGKGPSIGDIEDEEPELPPAIPRRIR |
The purified HPK1 variants were examined by SDS–PAGE under reducing conditions using a NuPAGE 4–12% acrylamide Bis-Tris Gel (Invitrogen, Carlsbad, California, USA) in MES running buffer and stained with SimplyBlue SafeStrain (Invitrogen). In general, a protein purity of greater than 95% was achieved. To determine the molecular mass of the purified proteins beyond SDS–PAGE, LC/MC analysis was performed using a 6230 TOF-MS system and the Mass Hunter BioConfirm deconvolution software (Agilent, Foster City, California, USA). Polydispersity, hydrodynamic radius, molecular weight and multimeric distribution were measured by dynamic light scattering (DLS) at 298 K using a DynaPro NanoStar 384-well plate reader. DLS data were analyzed using DYNAMICS version 7.8 (Wyatt Technologies, Santa Barbara, California, USA). Further analyses of the protein molecular mass and state of association were accomplished by SEC–MALS on an UFLC (Shimadzu, Columbia, Maryland, USA) coupled to multi-angle light scattering (MALS; Wyatt Technologies, Santa Barbara, California, USA) using ASTRA 6.
2.3. Crystallization
HPK1–inhibitor complexes were prepared by mixing the protein with ligand in a 1:10 molar ratio and incubating at 293 K for 2 h prior to crystallization-drop setup. Initial crystallization conditions for protein–ligand complexes were identified from the following sparse-matrix screens: MCSG1–4 (Anatrace, Maumee, Ohio, USA), Crystal Screen, Crystal Screen 2, Index, SaltRx (Hampton Research, Aliso Viejo, California, USA), Wizard (Rigaku, Bainbridge Island, Washington, USA), PACT and JCSG+ (Qiagen, Holland, Ohio, USA) using 96 × 2-well sitting-drop MRC UVP crystallization plates (Swissci, Neuheim, Switzerland). The screening drop (400 nl) was a 1:1 mixture of protein and precipitant solution and was equilibrated against an 80 µl reservoir. Initial crystal hits were observed in conditions from the MCSG3 and JCSG+ screens, both of which contained Bicine pH 9.0, 10% MPD. Final crystallization conditions after optimization are shown in Table 2 ▸. It took on average 3–5 days for the crystals to grow to their final size of about 100–200 µm. The crystals were cryoprotected by adding glycerol to the crystallization buffer to a final concentration of 25%(v/v).
Table 2. Crystallization.
| Method | Hanging drop |
| Plate type | EasyXtal 15-well (Qiagen) |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 10–12 |
| Buffer composition of protein solution | 25 mM Tris pH 8, 200 mM NaCl, 2 mM DTT, 0.5 mM EDTA, 5% glycerol |
| Composition of reservoir solution | 0.1 M Bicine pH 9, 7–10%(v/v) MPD, 3–5%(v/v) tert-butanol |
| Volume and ratio of drop | 2 µl, 1:1 |
| Volume of reservoir (µl) | 500 |
2.4. Data collection and processing
Data were processed using autoPROC (Vonrhein et al., 2011 ▸) with integration using XDS (Kabsch, 2010 ▸), isotropic scaling using XSCALE (Kabsch, 2010 ▸) and anisotropic scaling using STARANISO (Tickle et al., 2018 ▸). Data-collection and processing statistics are summarized in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | Beamline 17-ID, Advanced Photon Source | |
| Wavelength (Å) | 1.0 | |
| Temperature (K) | 100 | |
| Detector | PILATUS 6M | |
| Crystal-to-detector distance (mm) | 350 | |
| Rotation range per image (°) | 0.25 | |
| Total rotation range (°) | 360 | |
| Exposure time per image (s) | 0.168 | |
| Space group | P1 | |
| a, b, c (Å) | 55.8, 58.5, 60.5 | |
| α, β, γ (°) | 78.8, 79.8, 66.1 | |
| Mosaicity (°) | 0.32–0.43 | |
| Scaling | Isotropic | Anisotropic† |
| Resolution range (Å) | 50.0–2.50 (2.56–2.50) | 58.97–1.85 (2.19–1.85) |
| Total No. of reflections | 78465 (4111) | 93846 (4769) |
| No. of unique reflections | 22688 (1167) | 27255 (1363) |
| Completeness (%) | 95.8 (97.6) [spherical] | 85.2 (47.2) [ellipsoidal] |
| Multiplicity | 3.5 (3.5) | 3.5 (3.5) |
| 〈I/σ(I)〉 | 9.2 (2.3) | 8.0 (1.5) |
| R r.i.m. | 0.058 (0.617) | 0.045 (0.657) |
| CC1/2 | 0.996 (0.804) | 0.994 (0.274) |
| Overall B factor from Wilson plot (Å2) | 50.3‡ | |
Ellipsoid defined by three axes: 0.693a* + 0.630b* − 0.350c* extended to 2.43 Å; −0.057a* + 0.770b* + 0.636c* extended to 2.59 Å; 0.648a* − 0.056b* + 0.760c* extended to 1.84 Å; B 11 = −10.5 Å2, B 22 = 15.5 Å2, B 33 = −5.0 Å2, B 23 = 15.7 Å2, B 31 = −15.4 Å2, B 12 = 4.3 Å2.
Calculated by BUSTER (Bricogne et al., 2019 ▸).
2.5. Structure solution and refinement
Our original structure of HPK1 in complex with a compound not reported here was determined by molecular replacement using Phaser (McCoy et al., 2007 ▸). This crystal form contained two molecules in the asymmetric unit. The model was derived from PDB entry 3com (MST kinase) with the model divided into N-lobe and C-lobe components, but while both C-lobes were found, only one N-lobe was originally found. After sufficient rebuilding, the second N-lobe could be placed by using the previously built N-lobe as a model with Phaser. The structure reported here was placed using the rigid-body fitting function of the CCP4 (Winn et al., 2011 ▸) version of AMoRe (Navaza, 1994 ▸; Navaza & Vernoslova, 1995 ▸). The structure was refined with BUSTER (Bricogne et al., 2019 ▸; Smart et al., 2012 ▸) and the models were fitted into the electron density using Coot (Emsley & Cowtan, 2004 ▸; Emsley et al., 2010 ▸). The buried surface area was calculated using MS (Connolly, 1983 ▸) and the shape-complementarity statistic was calculated using SC (Lawrence & Colman, 1993 ▸; Winn et al., 2011 ▸). Refinement statistics are summarized in Table 4 ▸.
Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 58.97–1.84 (2.08–1.84) |
| Completeness† (%) | 46.1 (3.0) |
| σ Cutoff | 0 |
| No. of reflections, working set | 25932 (509) |
| No. of reflections, test set | 1333 (37) |
| Final R cryst | 0.217 (0.203) |
| Final R free | 0.232 (0.237) |
| No. of non-H atoms | |
| Protein | 4146 |
| Ligand | 68 |
| Water | 33 |
| Total | 4247 |
| R.m.s. deviations | |
| Bonds (Å) | 0.008 |
| Angles (°) | 0.9 |
| Average B factors (Å2) | |
| Protein | 59 |
| Ligand | 53 |
| Water | 45 |
| Ramachandran plot‡ | |
| Most favored (%) | 95.4 |
| Allowed (%) | 99.3 |
3. Results and discussion
3.1. Yeast-display selection
In YSD selection, highly displayed variants were enriched by fluorescence-activated cell sorting (FACS). Four rounds of selection for display led to an increase in the fraction of yeast cells that displayed HPK1, as well as to an increase in the magnitude of the display signal in the fraction of yeast that displayed HPK1 (Fig. 1 ▸). Sequencing of post-round-3 and post-round-4 populations showed enrichment of a different subset of designed mutations at each aggregation-prone site (Supplementary Table S2). As a result, 16 variants, M1–M16, were designed by fixing the positions Gln84, Glu112, Leu170 and Glu225 and by varying the positions 64, 188, 221 and 285, each with two enriched mutations, to generate 16 permutations (Supplementary Table S3). These variants were subsequently expressed in baculovirus-infected insect cells.
3.2. Revertant mutagenesis and characterization
11 of the 16 mutants (M1–M16) had a soluble expression level of between 3 and 10 mg l−1 (Supplementary Table S3). This compares favorably to more than 22 different constructs with the wild-type HPK1 kinase-domain sequence that had been tested previously and shown to express at up to 0.5 mg per litre in baculovirus-infected insect cells (data not shown). To determine which mutations in the selected variants were the most critical for improved expression, mutants M17–M23 were generated by reverting one mutation at a time to the wild type using M5 as a test case (Supplementary Table S3). SDS–PAGE analysis showed that when mutations at positions 221 or 225 were reverted to the wild type, soluble expression was significantly reduced. When both positions 221 and 225 were reverted to the wild type simultaneously, as designed in mutant M24 (Supplementary Table S3), expression of soluble HPK1 was undetectable in our expression tests. These observations led to the design of the double mutant HPK1(1–346) L221D F225E, designated M25. M25 grown in a 5 l Wave bioreactor yielded purified, soluble protein at 12 mg l−1 (an approximately threefold increase in expression relative to shake flasks) and formed a stable, noncovalent complex with a known small-molecule inhibitor of HPK1. A 3.45 Å resolution structure of this complex was subsequently determined (data not shown). In-depth SDS–PAGE and mass-spectrometric analysis of M25 expression and degradation fragments identified a species consistent with truncation after residue 319, suggesting a smaller, more stable kinase domain. This observation led to the design of HPK1(1–319) L221D F225E, which was used for high-resolution crystallization studies.
3.3. Structure of the HPK1–inhibitor complex
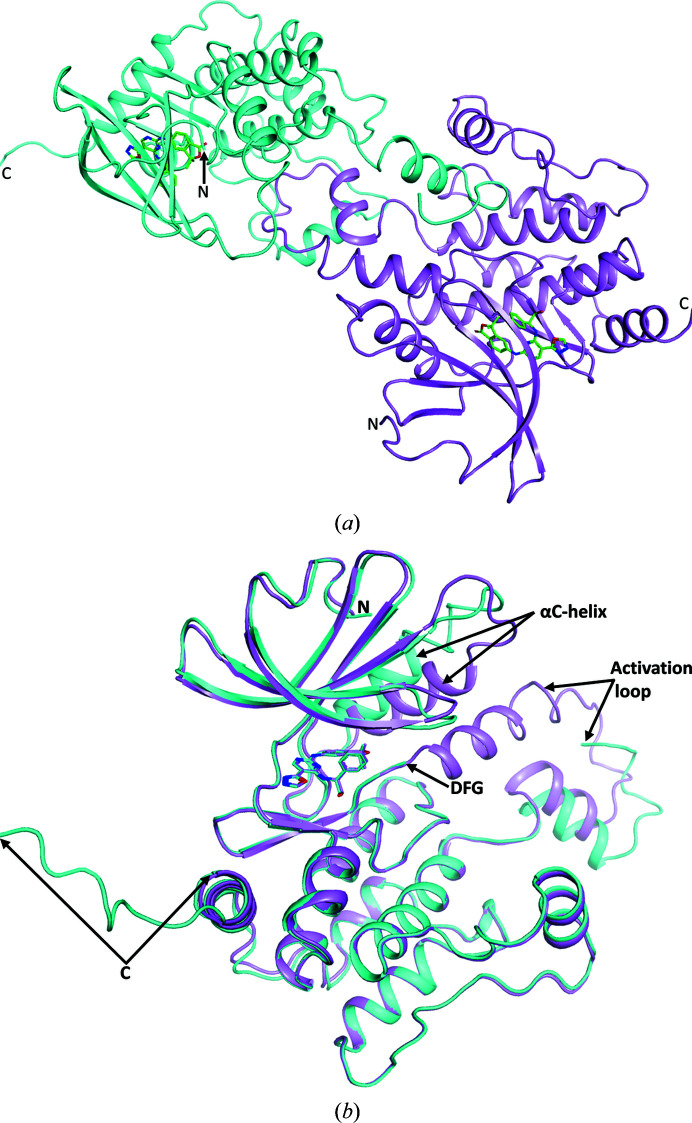
The HPK1 structure has two molecules in the asymmetric unit. As is typical of kinase domains, HPK1 has an N-lobe with a P-loop and a αC-helix, a hinge region and a mostly α-helical C-lobe, which includes an activation loop. The unusual feature of the activation loop is that it is domain-swapped (Fig. 2 ▸ a); that is, the activation loop of chain A binds to chain B and vice versa. In our construct, which runs from residues 1 to 319, we were able to interpret residues 7–156 and 172–305 in chain A and residues 7–295 in chain B. The segment of missing residues in chain A between the two interpreted segments includes the glycine of the DFG motif and extends through what is a four-turn α-helix in chain B at the beginning of the activation loop. The interpretation of additional residues at the C-terminus of chain A compared with chain B is probably owing to stabilization of these residues because they interact with the αC-helix of chain A of an adjacent unit cell. The interaction between the C-terminus of chain A and the αC-helix of chain A in an adjacent unit cell also leads to a repositioning of the αC-helix compared with chain B and other kinases (Fig. 2 ▸ b). The N-lobe of chain B is better defined by the electron density than is the N-lobe of chain A.
Figure 2.
(a) The contents of the asymmetric unit are shown with chain A in cyan and chain B in violet. Between the two monomers one can see the domain swapping of the activation loop. (b) Superposition of the two monomers (chain A in cyan and chain B in violet) showing that with the exception of the position of the αC-helix and surrounding residues, the activation loop and the extended C-terminus of chain A, the chains follow the same path.
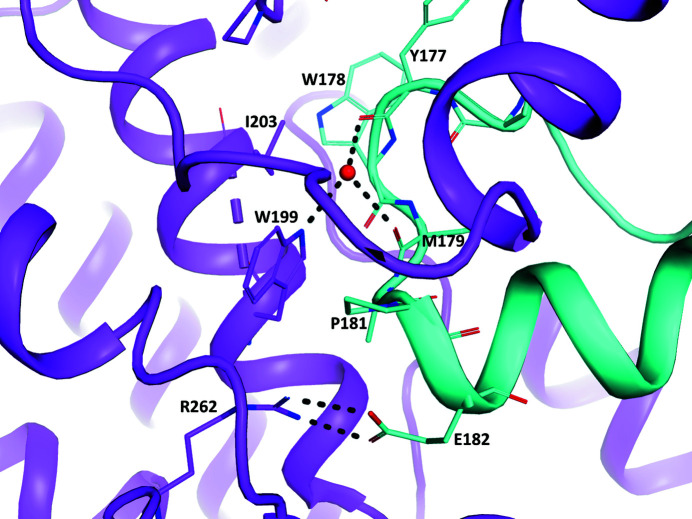
The interaction between the activation loop and the other monomer is quite intimate, with ∼1690 Å2 buried on each monomer and a surface-complementarity statistic of 0.75, which according to the analysis of Lawrence & Colman (1993 ▸) is within the realm of protease–protease inhibitor or oligomer interactions and above that for antibody–protein antigen interactions. The interaction encompasses the interaction loop of one chain (residues 157–194), a few residues between the hinge region and the DFG motif, and many residues in the rest of the C-lobe, particularly encompassing the E (residues 194–210) and F (residues 220–229) helices and the loop in between, and residues in the loop between the G (residues 245–254) and H (residues 265–270) helices. Particularly interesting interactions (Fig. 3 ▸) are a salt link between Glu182 of one chain and Arg262 of the other chain and the water-mediated hydrogen bonds from Trp199 NE1 in one chain to Tyr177 O and Met179 O in the other chain, although in this particular complex only the volume between TrpB199, TyrA177 and MetA179 has sufficient electron density to support the interpretation of a water molecule.
Figure 3.
Interactions in the activation loops of the two chains around Trp199, which includes a salt link between the ArgB262 and GluA182 side chains and water-mediated hydrogen bonds between the side chain of TrpB199, TyrA177 O and MetA197 O. The chain A cartoon and C atoms are shown in cyan and the chain B cartoon and C atoms are shown in violet; N atoms are blue, O atoms are red and S atoms are yellow.
The ligand (Supplementary Fig. S3) binds to the hinge region in each monomer and forms direct hydrogen bonds from one of the pyrimidinyl N atoms (N3) to Cys94 N, from the amino group (N9) to Glu92 O, from the hydroxyl group (O13) to Asp101 OD2 and from the carbonyl O atom (O30) to Asp155 N (in the DGF motif). The two N atoms (N33 and N34) of the oxadiazolyl are close enough to Gly95 O to form hydrogen bonds, but none of the atoms has an H atom attached.
The two residues mutated from wild-type HPK1 in M25, L221D and F225E, are on the surface. The electron density for the side chain of Glu225 is weak, especially in chain B, where the side chain is covered by −3 r.m.s.d. (−0.23 e−) difference density. The nearest residue in a different chain or molecule is the side chain of Leu188 in the activation loop, which is 3.6–4.0 Å distant from the Glu225 side chain and 7.5–8.0 Å from the Asp221 side chain. Leu188 in chain A is slightly closer to the residues in chain B. Comparison with PDB entry 6ng0, which has the native sequence at these two residues, shows that the side chains in both molecules occupy essentially the same volume.
4. Discussion
Since we originally determined the structure of HPK1, two groups have reported structures of HPK1 (Wu et al., 2019 ▸; Johnson et al., 2019 ▸). Broadly, our observation of a dimer with domain-swapped activation loops recapitulates the previously reported results. Specifically, our crystal form is essentially identical to PDB entry 6ng0 (Johnson et al., 2019 ▸), despite our construct and that reported in PDB entry 6ng0 being different lengths (our construct, residues 1–319; PDB entry 6ng0, residues 1–309) and having different mutations (our construct, L221D and F225E; PDB entry 6ng0, T165E and S171E). As found by Johnson et al. (2019 ▸), we were able to co-crystallize a wide variety of inhibitors in this crystal form, and structures of these complexes provided useful feedback to structure-based design efforts.
We showed that yeast surface display can be used when native protein sequences fail to yield soluble or otherwise biophysically well behaved proteins. In the case of HPK1, two mutations in the kinase domain were sufficient to improve its properties from a protein that was very poorly expressed in the soluble fraction to one that was soluble, abundant and amenable to crystallization.
Our readout of the HPK1 yeast surface-display selection used traditional Sanger sequencing. In more recent projects, we have switched to next-generation sequencing (NGS), which allows the detection of at least 1000 times more variants of the protein being engineered, as well as detailed quantification of how much each variant is enriched in each round. The downside of NGS is the current limitation on the length of each sequence read, which would not have allowed us to define the full-length HPK1 sequence.
We believe that yeast surface display adds another method to the crystallographer’s armamentarium to produce well behaved proteins for structural studies.
5. Related literature
The following references are cited in the supporting information for this article: Gnügge & Rudolf (2017 ▸) and Zakian et al. (1979 ▸).
Supplementary Material
PDB reference: HPK1, complex with inhibitor, 7kac
Supporting information including Supplementary Tables and Figures. DOI: 10.1107/S2053230X20016015/rl5189sup1.pdf
Acknowledgments
We thank Drs Stanley Krystek and John Tokarski for providing molecular-modeling advice. We thank Godwin Kumi and Dr Andrew P. Degnan for the synthesis of, and for providing us with, Compound K.
References
- Alzabin, S., Pyarajan, S., Yee, H., Kiefer, F., Suzuki, A., Burakoff, S. & Sawasdikosol, S. (2010). Cancer Immunol. Immunother. 59, 419–429. [DOI] [PMC free article] [PubMed]
- Benatuil, L., Perez, J. M., Belk, J. & Hsieh, C. M. (2010). Protein Eng. Des. Sel. 23, 155–159. [DOI] [PubMed]
- Boder, E. & Wittrup, K. D. (1997). Nat. Biotechnol. 15, 553–557. [DOI] [PubMed]
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Smart, O., Vonrhein, C. & Womack, T. (2019). BUSTER-TNT 2.11.7. Global Phasing Ltd, Cambridge, United Kingdom.
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Chennamsetty, N., Voynov, V., Kayser, V., Helk, B. & Trout, B. L. (2009). Proc. Natl Acad. Sci. USA, 106, 11937–11942. [DOI] [PMC free article] [PubMed]
- Cho, B. K., Kieke, M. C., Boder, E. T., Wittrup, K. D. & Kranz, D. M. (1998). J. Immunol. Methods, 220, 179–188. [DOI] [PubMed]
- Connolly, M. L. (1983). J. Appl. Cryst. 16, 548–558.
- Darvin, P., Toor, S. M., Sasidharan Nair, V. & Elkord, E. (2018). Exp. Mol. Med. 50, 1–11. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gnügge, R. & Rudolf, F. (2017). Yeast, 34, 205–221. [DOI] [PubMed]
- Hernandez, S., Qing, J., Thibodeau, R. H., Du, X., Park, S., Lee, H., Xu, M., Oh, S., Navarro, A., Roose-Girma, M., Newman, R. J., Warming, S., Nannini, M., Sampath, D., Kim, J. M., Grogan, J. L. & Mellman, I. (2018). Cell Rep. 25, 80–94. [DOI] [PubMed]
- Hu, M. C.-T., Qiu, W. R., Wang, X., Meyer, C. F. & Tan, T.-H. (1996). Genes Dev. 10, 2251–2264. [DOI] [PubMed]
- Johnson, E., McTigue, M., Gallego, R. A., Johnson, T. W., Timofeevski, S., Maestre, M., Fisher, T. S., Kania, R., Sawasdikosol, S., Burakoff, S. & Cronin, C. N. (2019). J. Biol. Chem. 294, 9029–9036. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Koide, S., Koide, A. & Lipovšek, D. (2012). Methods Enzymol. 503, 135–156. [DOI] [PubMed]
- Lasserre, R., Blecher-Gonen, R., Libman, E., Biquand, E., Danckaert, A., Yablonski, D., Alcover, A. & Di Bartolo, V. (2011). J. Cell Biol. 195, 839–853. [DOI] [PMC free article] [PubMed]
- Lawrence, M. C. & Colman, P. M. (1993). J. Mol. Biol. 234, 946–950. [DOI] [PubMed]
- Lipovšek, D., Carvajal, I., Allentoff, A. J., Barros, A. Jr, Brailsford, J., Cong, Q., Cotter, P., Gangwar, S., Hollander, C., Lafont, V., Lau, W. L., Li, W., Moreta, M., O’Neil, S., Pinckney, J., Smith, M. J., Su, J., Terragni, C., Wallace, M. A., Wang, L., Wright, M., Marsh, H. N. & Bryson, J. W. (2018). Protein Eng. Des. Sel. 31, 159–171. [DOI] [PMC free article] [PubMed]
- Liu, J., Curtin, J., You, D., Hillerman, S., Li-Wang, B., Eraslan, R., Xie, J., Swanson, J., Ho, C. P., Oppenheimer, S., Warrack, B. M., McNaney, C. A., Nelson, D. M., Blum, J., Kim, T., Fereshteh, M., Reily, M., Shipkova, P., Murtaza, A., Sanjuan, M., Hunt, J. T. & Salter-Cid, L. (2019). PLoS One, 14, e0212670. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Navaza, J. & Vernoslova, E. (1995). Acta Cryst. A51, 445–449.
- Pavoor, T. V., Wheasler, J. A., Kamat, V. & Shusta, E. V. (2012). Protein Eng. Des. Sel. 25, 625–630. [DOI] [PMC free article] [PubMed]
- Rakestraw, J. A., Aird, D., Aha, P. M., Baynes, B. M. & Lipovsek, D. (2011). Protein Eng. Des. Sel. 24, 525–530. [DOI] [PubMed]
- Rhiel, L., Krah, S., Günther, R., Becker, S., Kolmar, H. & Hock, B. (2014). PLoS One, 9, e114887. [DOI] [PMC free article] [PubMed]
- Sauer, K., Liou, J., Singh, S. B., Yablonski, D., Weiss, A. & Perlmutter, R. M. (2001). J. Biol. Chem. 276, 45207–45216. [DOI] [PubMed]
- Shui, J.-W., Boomer, J. S., Han, J., Xu, J., Dement, G. A., Zhou, G. & Tan, T.-H. (2007). Nat. Immunol. 8, 84–91. [DOI] [PubMed]
- Shusta, E. V., Holler, P. D., Kieke, M. C., Kranz, D. M. & Wittrup, K. D. (2000). Nat. Biotechnol. 18, 754–759. [DOI] [PubMed]
- Shusta, E. V., Kieke, M. C., Parke, E., Kranz, D. M. & Wittrup, K. D. (1999). J. Mol. Biol. 292, 949–956. [DOI] [PubMed]
- Smart, O. S., Womack, T. O., Flensburg, C., Keller, P., Paciorek, W., Sharff, A., Vonrhein, C. & Bricogne, G. (2012). Acta Cryst. D68, 368–380. [DOI] [PMC free article] [PubMed]
- Tickle, I. J., Flensburg, C., Keller, P., Paciorek, W., Sharff, A., Vonrhein, C. & Bricogne, G. (2018). STARANISO. Global Phasing Ltd, Cambridge, United Kingdom.
- Traxlmayr, M. W., Faissner, M., Stadlmayr, G., Hasenhindl, C., Antes, B., Rüker, F. & Obinger, C. (2012). Biochim. Biophys. Acta, 1824, 542–549. [DOI] [PMC free article] [PubMed]
- Traxlmayr, M. W., Lobner, E., Antes, B., Kainer, M., Wiederkum, S., Hasenhindl, C., Stadlmayr, G., Rüker, F., Woisetschläger, M., Moulder, K. & Obinger, C. (2013). Protein Eng. Des. Sel. 26, 255–265. [DOI] [PubMed]
- Vonrhein, C., Flensburg, C., Keller, P., Sharff, A., Smart, O., Paciorek, W., Womack, T. & Bricogne, G. (2011). Acta Cryst. D67, 293–302. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Wu, P., Sneeringer, C. J., Pitts, K. E., Day, E. S., Chan, B. K., Wei, B., Lehoux, I., Mortara, K., Li, H., Wu, J., Franke, Y., Moffat, J. G., Grogan, J. L., Heffron, T. P. & Wang, W. (2019). Structure, 27, 125–133. [DOI] [PubMed]
- You, D., Hillerman, S., Locke, G., Chaudhry, C., Stromko, C., Murtaza, A., Fan, Y., Koenitzer, J., Chen, Y., Briceno, S., Bhadra, R., Duperret, E., Gullo-Brown, J., Degnan, A. P., Kumi, G., Wittman, M., Johnson, B. M., Parrish, K., Gokulrangan, G., Morrison, J. T., Hunt, J. T., Salter-Cid, L., Lees, E., Sanjuan, M. & Liu, J. (2021). J. Immunother. Cancer, 9, e001402. [DOI] [PMC free article] [PubMed]
- Zakian, V. A., Brewer, B. J. & Fangman, W. L. (1979). Cell, 17, 923–934. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: HPK1, complex with inhibitor, 7kac
Supporting information including Supplementary Tables and Figures. DOI: 10.1107/S2053230X20016015/rl5189sup1.pdf